
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:KCNAB1-LRTM1 (FusionGDB2 ID:41346) |
Fusion Gene Summary for KCNAB1-LRTM1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: KCNAB1-LRTM1 | Fusion gene ID: 41346 | Hgene | Tgene | Gene symbol | KCNAB1 | LRTM1 | Gene ID | 7881 | 57408 |
| Gene name | potassium voltage-gated channel subfamily A member regulatory beta subunit 1 | leucine rich repeats and transmembrane domains 1 | |
| Synonyms | AKR6A3|KCNA1B|KV-BETA-1|Kvb1.3|hKvBeta3|hKvb3 | HT017 | |
| Cytomap | 3q25.31 | 3p14.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | voltage-gated potassium channel subunit beta-1K(+) channel subunit beta-1K+ channel Beta1a chainpotassium channel beta 3 chainpotassium channel beta3 subunitpotassium channel shaker chain beta 1apotassium channel, voltage gated subfamily A regulator | leucine-rich repeat and transmembrane domain-containing protein 1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q14722 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000497291, ENST00000302490, ENST00000389634, ENST00000389636, ENST00000471742, ENST00000490337, | ENST00000273286, ENST00000493075, | |
| Fusion gene scores | * DoF score | 14 X 14 X 3=588 | 3 X 2 X 3=18 |
| # samples | 14 | 4 | |
| ** MAII score | log2(14/588*10)=-2.0703893278914 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(4/18*10)=1.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: KCNAB1 [Title/Abstract] AND LRTM1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | KCNAB1(156233009)-LRTM1(54952919), # samples:1 KCNAB1(156233009)-LRTM1(54959242), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | KCNAB1 | GO:1901379 | regulation of potassium ion transmembrane transport | 7890032 |
 Fusion gene breakpoints across KCNAB1 (5'-gene) Fusion gene breakpoints across KCNAB1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
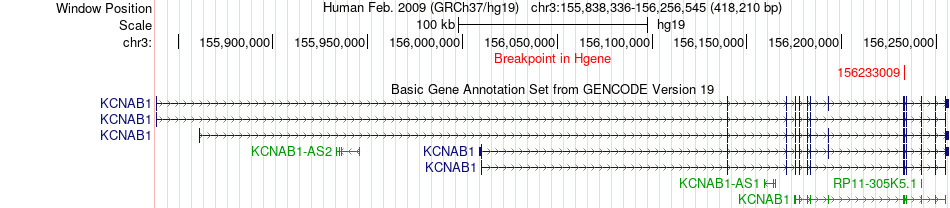 |
 Fusion gene breakpoints across LRTM1 (3'-gene) Fusion gene breakpoints across LRTM1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
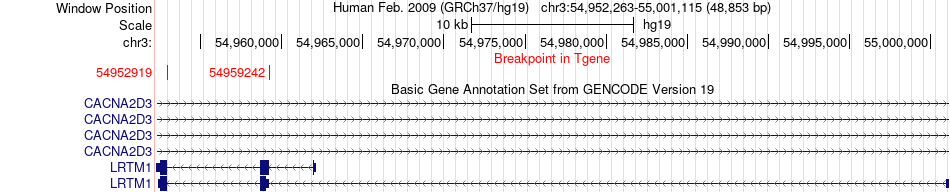 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PCPG | TCGA-QT-A5XO-01A | KCNAB1 | chr3 | 156233009 | + | LRTM1 | chr3 | 54952919 | - |
| ChimerDB4 | PCPG | TCGA-QT-A5XO-01A | KCNAB1 | chr3 | 156233009 | - | LRTM1 | chr3 | 54959242 | - |
Top |
Fusion Gene ORF analysis for KCNAB1-LRTM1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000497291 | ENST00000273286 | KCNAB1 | chr3 | 156233009 | + | LRTM1 | chr3 | 54952919 | - |
| 3UTR-3CDS | ENST00000497291 | ENST00000273286 | KCNAB1 | chr3 | 156233009 | - | LRTM1 | chr3 | 54959242 | - |
| 3UTR-3CDS | ENST00000497291 | ENST00000493075 | KCNAB1 | chr3 | 156233009 | + | LRTM1 | chr3 | 54952919 | - |
| 3UTR-5UTR | ENST00000497291 | ENST00000493075 | KCNAB1 | chr3 | 156233009 | - | LRTM1 | chr3 | 54959242 | - |
| 5CDS-5UTR | ENST00000302490 | ENST00000493075 | KCNAB1 | chr3 | 156233009 | - | LRTM1 | chr3 | 54959242 | - |
| 5CDS-5UTR | ENST00000389634 | ENST00000493075 | KCNAB1 | chr3 | 156233009 | - | LRTM1 | chr3 | 54959242 | - |
| 5CDS-5UTR | ENST00000389636 | ENST00000493075 | KCNAB1 | chr3 | 156233009 | - | LRTM1 | chr3 | 54959242 | - |
| 5CDS-5UTR | ENST00000471742 | ENST00000493075 | KCNAB1 | chr3 | 156233009 | - | LRTM1 | chr3 | 54959242 | - |
| 5CDS-5UTR | ENST00000490337 | ENST00000493075 | KCNAB1 | chr3 | 156233009 | - | LRTM1 | chr3 | 54959242 | - |
| Frame-shift | ENST00000302490 | ENST00000273286 | KCNAB1 | chr3 | 156233009 | + | LRTM1 | chr3 | 54952919 | - |
| Frame-shift | ENST00000302490 | ENST00000493075 | KCNAB1 | chr3 | 156233009 | + | LRTM1 | chr3 | 54952919 | - |
| Frame-shift | ENST00000389634 | ENST00000273286 | KCNAB1 | chr3 | 156233009 | + | LRTM1 | chr3 | 54952919 | - |
| Frame-shift | ENST00000389634 | ENST00000493075 | KCNAB1 | chr3 | 156233009 | + | LRTM1 | chr3 | 54952919 | - |
| Frame-shift | ENST00000389636 | ENST00000273286 | KCNAB1 | chr3 | 156233009 | + | LRTM1 | chr3 | 54952919 | - |
| Frame-shift | ENST00000389636 | ENST00000493075 | KCNAB1 | chr3 | 156233009 | + | LRTM1 | chr3 | 54952919 | - |
| Frame-shift | ENST00000471742 | ENST00000273286 | KCNAB1 | chr3 | 156233009 | + | LRTM1 | chr3 | 54952919 | - |
| Frame-shift | ENST00000471742 | ENST00000493075 | KCNAB1 | chr3 | 156233009 | + | LRTM1 | chr3 | 54952919 | - |
| Frame-shift | ENST00000490337 | ENST00000273286 | KCNAB1 | chr3 | 156233009 | + | LRTM1 | chr3 | 54952919 | - |
| Frame-shift | ENST00000490337 | ENST00000493075 | KCNAB1 | chr3 | 156233009 | + | LRTM1 | chr3 | 54952919 | - |
| In-frame | ENST00000302490 | ENST00000273286 | KCNAB1 | chr3 | 156233009 | - | LRTM1 | chr3 | 54959242 | - |
| In-frame | ENST00000389634 | ENST00000273286 | KCNAB1 | chr3 | 156233009 | - | LRTM1 | chr3 | 54959242 | - |
| In-frame | ENST00000389636 | ENST00000273286 | KCNAB1 | chr3 | 156233009 | - | LRTM1 | chr3 | 54959242 | - |
| In-frame | ENST00000471742 | ENST00000273286 | KCNAB1 | chr3 | 156233009 | - | LRTM1 | chr3 | 54959242 | - |
| In-frame | ENST00000490337 | ENST00000273286 | KCNAB1 | chr3 | 156233009 | - | LRTM1 | chr3 | 54959242 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000490337 | KCNAB1 | chr3 | 156233009 | - | ENST00000273286 | LRTM1 | chr3 | 54959242 | - | 2182 | 929 | 64 | 1959 | 631 |
| ENST00000389636 | KCNAB1 | chr3 | 156233009 | - | ENST00000273286 | LRTM1 | chr3 | 54959242 | - | 2065 | 812 | 34 | 1842 | 602 |
| ENST00000471742 | KCNAB1 | chr3 | 156233009 | - | ENST00000273286 | LRTM1 | chr3 | 54959242 | - | 2302 | 1049 | 217 | 2079 | 620 |
| ENST00000302490 | KCNAB1 | chr3 | 156233009 | - | ENST00000273286 | LRTM1 | chr3 | 54959242 | - | 2935 | 1682 | 820 | 2712 | 630 |
| ENST00000389634 | KCNAB1 | chr3 | 156233009 | - | ENST00000273286 | LRTM1 | chr3 | 54959242 | - | 1977 | 724 | 0 | 1754 | 584 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000490337 | ENST00000273286 | KCNAB1 | chr3 | 156233009 | - | LRTM1 | chr3 | 54959242 | - | 0.00912302 | 0.990877 |
| ENST00000389636 | ENST00000273286 | KCNAB1 | chr3 | 156233009 | - | LRTM1 | chr3 | 54959242 | - | 0.012769613 | 0.98723036 |
| ENST00000471742 | ENST00000273286 | KCNAB1 | chr3 | 156233009 | - | LRTM1 | chr3 | 54959242 | - | 0.008130875 | 0.9918691 |
| ENST00000302490 | ENST00000273286 | KCNAB1 | chr3 | 156233009 | - | LRTM1 | chr3 | 54959242 | - | 0.015036717 | 0.98496324 |
| ENST00000389634 | ENST00000273286 | KCNAB1 | chr3 | 156233009 | - | LRTM1 | chr3 | 54959242 | - | 0.013663016 | 0.986337 |
Top |
Fusion Genomic Features for KCNAB1-LRTM1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
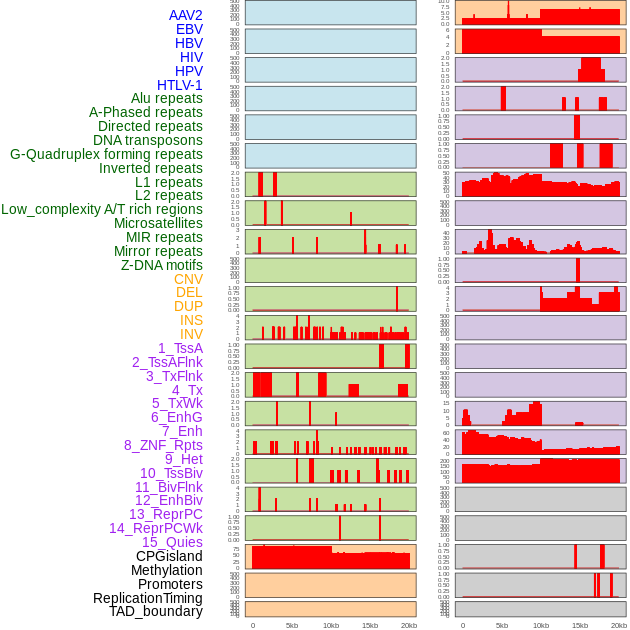 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for KCNAB1-LRTM1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:156233009/chr3:54952919) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| KCNAB1 | . |
| FUNCTION: Cytoplasmic potassium channel subunit that modulates the characteristics of the channel-forming alpha-subunits (PubMed:7499366, PubMed:7603988, PubMed:17156368,PubMed:17540341, PubMed:19713757). Modulates action potentials via its effect on the pore-forming alpha subunits (By similarity). Promotes expression of the pore-forming alpha subunits at the cell membrane, and thereby increases channel activity (By similarity). Mediates closure of delayed rectifier potassium channels by physically obstructing the pore via its N-terminal domain and increases the speed of channel closure for other family members (PubMed:9763623). Promotes the closure of KCNA1, KCNA2 and KCNA5 channels (PubMed:7499366, PubMed:7890032, PubMed:7603988, PubMed:7649300, PubMed:8938711, PubMed:12077175, PubMed:12130714, PubMed:15361858, PubMed:17540341, PubMed:19713757). Accelerates KCNA4 channel closure (PubMed:7890032, PubMed:7649300, PubMed:7890764, PubMed:9763623). Accelerates the closure of heteromeric channels formed by KCNA1 and KCNA4 (PubMed:17156368). Accelerates the closure of heteromeric channels formed by KCNA2, KCNA5 and KCNA6 (By similarity). Isoform KvB1.2 has no effect on KCNA1, KCNA2 or KCNB1 (PubMed:7890032, PubMed:7890764). Enhances KCNB1 and KCNB2 channel activity (By similarity). Binds NADPH; this is required for efficient down-regulation of potassium channel activity (PubMed:17540341). Has NADPH-dependent aldoketoreductase activity (By similarity). Oxidation of the bound NADPH strongly decreases N-type inactivation of potassium channel activity (By similarity). {ECO:0000250|UniProtKB:P63143, ECO:0000250|UniProtKB:P63144, ECO:0000269|PubMed:12077175, ECO:0000269|PubMed:12130714, ECO:0000269|PubMed:15361858, ECO:0000269|PubMed:17156368, ECO:0000269|PubMed:17540341, ECO:0000269|PubMed:19713757, ECO:0000269|PubMed:7499366, ECO:0000269|PubMed:7603988, ECO:0000269|PubMed:7649300, ECO:0000269|PubMed:7890032, ECO:0000269|PubMed:7890764, ECO:0000269|PubMed:8938711, ECO:0000269|PubMed:9763623, ECO:0000305}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KCNAB1 | chr3:156233009 | chr3:54959242 | ENST00000302490 | - | 10 | 14 | 108_109 | 270 | 402.0 | Nucleotide binding | NADP |
| Hgene | KCNAB1 | chr3:156233009 | chr3:54959242 | ENST00000302490 | - | 10 | 14 | 240_241 | 270 | 402.0 | Nucleotide binding | NADP |
| Hgene | KCNAB1 | chr3:156233009 | chr3:54959242 | ENST00000471742 | - | 10 | 14 | 108_109 | 277 | 409.0 | Nucleotide binding | NADP |
| Hgene | KCNAB1 | chr3:156233009 | chr3:54959242 | ENST00000471742 | - | 10 | 14 | 240_241 | 277 | 409.0 | Nucleotide binding | NADP |
| Hgene | KCNAB1 | chr3:156233009 | chr3:54959242 | ENST00000490337 | - | 10 | 14 | 108_109 | 288 | 420.0 | Nucleotide binding | NADP |
| Hgene | KCNAB1 | chr3:156233009 | chr3:54959242 | ENST00000490337 | - | 10 | 14 | 240_241 | 288 | 420.0 | Nucleotide binding | NADP |
| Tgene | LRTM1 | chr3:156233009 | chr3:54959242 | ENST00000273286 | 0 | 3 | 180_234 | 2 | 346.0 | Domain | Note=LRRCT | |
| Tgene | LRTM1 | chr3:156233009 | chr3:54959242 | ENST00000273286 | 0 | 3 | 28_50 | 2 | 346.0 | Domain | Note=LRRNT | |
| Tgene | LRTM1 | chr3:156233009 | chr3:54959242 | ENST00000493075 | 0 | 3 | 180_234 | 0 | 270.0 | Domain | Note=LRRCT | |
| Tgene | LRTM1 | chr3:156233009 | chr3:54959242 | ENST00000493075 | 0 | 3 | 28_50 | 0 | 270.0 | Domain | Note=LRRNT | |
| Tgene | LRTM1 | chr3:156233009 | chr3:54959242 | ENST00000273286 | 0 | 3 | 123_144 | 2 | 346.0 | Repeat | Note=LRR 4 | |
| Tgene | LRTM1 | chr3:156233009 | chr3:54959242 | ENST00000273286 | 0 | 3 | 147_168 | 2 | 346.0 | Repeat | Note=LRR 5 | |
| Tgene | LRTM1 | chr3:156233009 | chr3:54959242 | ENST00000273286 | 0 | 3 | 51_72 | 2 | 346.0 | Repeat | Note=LRR 1 | |
| Tgene | LRTM1 | chr3:156233009 | chr3:54959242 | ENST00000273286 | 0 | 3 | 75_96 | 2 | 346.0 | Repeat | Note=LRR 2 | |
| Tgene | LRTM1 | chr3:156233009 | chr3:54959242 | ENST00000273286 | 0 | 3 | 99_120 | 2 | 346.0 | Repeat | Note=LRR 3 | |
| Tgene | LRTM1 | chr3:156233009 | chr3:54959242 | ENST00000493075 | 0 | 3 | 123_144 | 0 | 270.0 | Repeat | Note=LRR 4 | |
| Tgene | LRTM1 | chr3:156233009 | chr3:54959242 | ENST00000493075 | 0 | 3 | 147_168 | 0 | 270.0 | Repeat | Note=LRR 5 | |
| Tgene | LRTM1 | chr3:156233009 | chr3:54959242 | ENST00000493075 | 0 | 3 | 51_72 | 0 | 270.0 | Repeat | Note=LRR 1 | |
| Tgene | LRTM1 | chr3:156233009 | chr3:54959242 | ENST00000493075 | 0 | 3 | 75_96 | 0 | 270.0 | Repeat | Note=LRR 2 | |
| Tgene | LRTM1 | chr3:156233009 | chr3:54959242 | ENST00000493075 | 0 | 3 | 99_120 | 0 | 270.0 | Repeat | Note=LRR 3 | |
| Tgene | LRTM1 | chr3:156233009 | chr3:54959242 | ENST00000273286 | 0 | 3 | 28_288 | 2 | 346.0 | Topological domain | Extracellular | |
| Tgene | LRTM1 | chr3:156233009 | chr3:54959242 | ENST00000273286 | 0 | 3 | 310_345 | 2 | 346.0 | Topological domain | Cytoplasmic | |
| Tgene | LRTM1 | chr3:156233009 | chr3:54959242 | ENST00000493075 | 0 | 3 | 28_288 | 0 | 270.0 | Topological domain | Extracellular | |
| Tgene | LRTM1 | chr3:156233009 | chr3:54959242 | ENST00000493075 | 0 | 3 | 310_345 | 0 | 270.0 | Topological domain | Cytoplasmic | |
| Tgene | LRTM1 | chr3:156233009 | chr3:54959242 | ENST00000273286 | 0 | 3 | 289_309 | 2 | 346.0 | Transmembrane | Helical | |
| Tgene | LRTM1 | chr3:156233009 | chr3:54959242 | ENST00000493075 | 0 | 3 | 289_309 | 0 | 270.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KCNAB1 | chr3:156233009 | chr3:54959242 | ENST00000302490 | - | 10 | 14 | 295_300 | 270 | 402.0 | Nucleotide binding | NADP |
| Hgene | KCNAB1 | chr3:156233009 | chr3:54959242 | ENST00000302490 | - | 10 | 14 | 375_381 | 270 | 402.0 | Nucleotide binding | NADP |
| Hgene | KCNAB1 | chr3:156233009 | chr3:54959242 | ENST00000471742 | - | 10 | 14 | 295_300 | 277 | 409.0 | Nucleotide binding | NADP |
| Hgene | KCNAB1 | chr3:156233009 | chr3:54959242 | ENST00000471742 | - | 10 | 14 | 375_381 | 277 | 409.0 | Nucleotide binding | NADP |
| Hgene | KCNAB1 | chr3:156233009 | chr3:54959242 | ENST00000490337 | - | 10 | 14 | 295_300 | 288 | 420.0 | Nucleotide binding | NADP |
| Hgene | KCNAB1 | chr3:156233009 | chr3:54959242 | ENST00000490337 | - | 10 | 14 | 375_381 | 288 | 420.0 | Nucleotide binding | NADP |
Top |
Fusion Gene Sequence for KCNAB1-LRTM1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >41346_41346_1_KCNAB1-LRTM1_KCNAB1_chr3_156233009_ENST00000302490_LRTM1_chr3_54959242_ENST00000273286_length(transcript)=2935nt_BP=1682nt AGTTCCAACTGTTGTGCTGCAGCAGATTTGGTCTGAGTCTGGGCAGAGCCCCTTTTTATGGAGCAACCCTCCCGATTTGTTACAGCAAGA GCCTGGTGCAAGTGATCTTTGCCTTCCCATCGCACACAAAGGCAAAGAGCTCCTTAAAGGGACCTGCCTGCTCCTCCGTGCAGCTGCAAA CCCGTGTCGGTGCAGGCTCAGCAACCCAGGCTTTCGCTCGAGAATGAGTGGGGAGGGGTCTTGAGGAGCCCCCACGTCCTCCCGGAGCGG AGGCGGGAGGGAGCTGACAGCCGGAGTGGATGGAGACCACCAGTCCGAGGGGACCCGCTTGCGATTAGCTGCTTCTTGGGATGCTCCGCG CGCCAGTTGAGCATCCTGCCTGTGGTCCAGGGGATGCTGGCGGCGCATCTTGCGGTAAGCCTGCGCTGATTTGAGCCATCGCCCAGAAGC GAGCCCGCATTCAGACACCGCGGACCGGCTCCGCGAAGGGCTGCGTGCAGTCACCATGAATGCTCAGCTCTAGGCTTCTGTAAAAACCTC CCCCACTGCATGGATTTGGCTGTTTAATCAGCCGAGATTACAGCCCGGGAAGATTTGCAAACCTGATTCTCCCTCCAGCTGCTGTGACAA CTTCGGGGCCGTCTGTTTACTTCTCTGGGTCCCCGGGGACTCTTTCCTGCATCGCAGCCCCTTGTGCCACTTTGTGGGCTTGGCTCGAAA GTCAGCCGCCAGGACTCCTCTGACGGCATCCCCAGGAAGGGGGAGCAAGGAGGGCTTAAAAGAAAAGCAGAAATCCCCGCAACTGCTCAA CCTCTCGTGACTGCCTGTGTTCTGGGGTTCTGAGAGGGACCGTGCGCTGCCTGGGGAAGCAATGCAAGTCTCCATAGCCTGCACAGAGCA CAATTTGAAGAGTCGGAATGGTGAGGACCGACTTCTGAGCAAGCAGAGCTCCACCGCCCCCAATGTGGTGAACGCAGCCCGGGCCAAATT CCGCACGGTCGCTATCATCGCGCGCAGCCTGGGGACGTTCACGCCTCAGCATCACATTTCTCTCAAAGAGTCCACCGCAAAGCAGACTGG CATGAAATATAGGAATCTTGGAAAATCAGGACTCAGAGTTTCTTGCTTGGGTCTTGGAACATGGGTGACATTTGGAGGTCAAATTTCAGA TGAGGTTGCTGAACGGCTGATGACCATCGCCTATGAAAGTGGTGTTAACCTCTTTGATACTGCCGAAGTCTATGCTGCTGGAAAGGCTGA AGTGATTCTGGGGAGCATCATCAAGAAGAAAGGCTGGAGGAGGTCCAGTCTGGTCATAACAACCAAACTCTACTGGGGTGGAAAAGCTGA AACAGAAAGAGGGCTGTCAAGAAAGCATATTATTGAAGGATTGAAGGGCTCCCTCCAGAGGCTGCAGCTCGAGTATGTGGATGTGGTCTT TGCAAATCGACCGGACAGTAACACTCCCATGGAAGAAATTGTCCGAGCCATGACACATGTGATAAACCAAGGCATGGCGATGTACTGGGG CACCTCGAGATGGAGTGCTATGGAGATCATGGAAGCCTATTCTGTAGCAAGACAGTTCAATATGATCCCACCGGTCTGTGAACAAGCTGA GTACCATCTTTTCCAGAGAGAGAAAGTGGAGGTCCAGCTGCCAGAGCTCTACCACAAAATAGGTGAACTGCTCCTGTTTTCCAGTGTGAT TGTCCTGCTCCAGGTGGTATGCAGCTGCCCGGACAAGTGTTACTGTCAGTCATCTACAAATTTTGTAGACTGCAGCCAGCAGGGTCTGGC CGAAATCCCTTCCCATTTACCTCCTCAGACTCGAACGCTGCATTTACAAGATAATCAGATACACCATCTTCCTGCTTTTGCATTTAGGTC AGTGCCATGGCTCATGACCTTAAACTTGTCCAACAATTCCCTTTCAAATCTGGCCCCTGGAGCTTTCCATGGGCTTCAGCACTTGCAGGT TTTAAATCTAACCCAGAATTCACTCCTTTCCCTGGAAAGCAGACTTTTCCATTCCCTCCCTCAGCTGAGGGAGCTTGATTTGTCATCAAA CAACATAAGCCACCTTCCCACATCCTTGGGAGAGACTTGGGAGAACCTAACTATACTTGCGGTTCAACAAAACCAGCTTCAGCAGCTTGA TCGAGCGCTCCTGGAATCCATGCCCAGTGTGAGGCTTTTACTTCTCAAGGACAACCTCTGGAAATGCAATTGCCACTTGCTCGGTCTTAA ACTCTGGCTGGAGAAATTTGTCTATAAAGGGGGACTAACAGACGGCATCATCTGTGAATCACCAGACACCTGGAAGGGAAAGGACCTCCT TAGGATCCCTCATGAGCTGTACCAGCCCTGCCCTCTTCCTGCTCCTGATCCAGTGTCCTCGCAGGCTCAGTGGCCCGGCTCTGCCCACGG TGTGGTCCTGAGGCCTCCTGAGAACCACAACGCGGGGGAGCGAGAACTCTTGGAGTGCGAGCTCAAACCCAAGCCAAGGCCGGCCAACCT GCGTCATGCCATTGCCACTGTCATCATCACTGGCGTTGTGTGTGGGATTGTGTGTCTCATGATGTTGGCAGCTGCCATCTATGGCTGCAC CTATGCGGCAATCACAGCCCAGTACCATGGGGGACCCTTGGCTCAAACCAATGATCCTGGGAAGGTGGAAGAAAAAGAGCGATTTGACAG CTCACCAGCCTGAGAGCTTTTGTCTCAAATAGGATTGGTCATTGCAGGCCAGAAGATAGTGTCTGAGTAGGGCTGATGTGTTTCCTGTTA GTCTGATTTTGCTTTTGCCAAAAGACAAAAAAAAAAAAAAAAAAAAAAAGAAAAAAAAAAGATGTGTCTGTAAAAAATATTTCTGGGGAG >41346_41346_1_KCNAB1-LRTM1_KCNAB1_chr3_156233009_ENST00000302490_LRTM1_chr3_54959242_ENST00000273286_length(amino acids)=630AA_BP=287 MPVFWGSERDRALPGEAMQVSIACTEHNLKSRNGEDRLLSKQSSTAPNVVNAARAKFRTVAIIARSLGTFTPQHHISLKESTAKQTGMKY RNLGKSGLRVSCLGLGTWVTFGGQISDEVAERLMTIAYESGVNLFDTAEVYAAGKAEVILGSIIKKKGWRRSSLVITTKLYWGGKAETER GLSRKHIIEGLKGSLQRLQLEYVDVVFANRPDSNTPMEEIVRAMTHVINQGMAMYWGTSRWSAMEIMEAYSVARQFNMIPPVCEQAEYHL FQREKVEVQLPELYHKIGELLLFSSVIVLLQVVCSCPDKCYCQSSTNFVDCSQQGLAEIPSHLPPQTRTLHLQDNQIHHLPAFAFRSVPW LMTLNLSNNSLSNLAPGAFHGLQHLQVLNLTQNSLLSLESRLFHSLPQLRELDLSSNNISHLPTSLGETWENLTILAVQQNQLQQLDRAL LESMPSVRLLLLKDNLWKCNCHLLGLKLWLEKFVYKGGLTDGIICESPDTWKGKDLLRIPHELYQPCPLPAPDPVSSQAQWPGSAHGVVL RPPENHNAGERELLECELKPKPRPANLRHAIATVIITGVVCGIVCLMMLAAAIYGCTYAAITAQYHGGPLAQTNDPGKVEEKERFDSSPA -------------------------------------------------------------- >41346_41346_2_KCNAB1-LRTM1_KCNAB1_chr3_156233009_ENST00000389634_LRTM1_chr3_54959242_ENST00000273286_length(transcript)=1977nt_BP=724nt ATGCAAGTCTCCATAGCCTGCACAGAGCACAATTTGAAGAGTCGGAATGGTGAGGACCGACTTCTGAGCAAGCAGAGCTCCACCGCCCCC AATGTGGTGAACGCAGCCCGGGCCAAATTCCGCACGGTCGCTATCATCGCGCGCAGCCTGGGGACGTTCACGCCTCAGCATCACATTTCT CTCAAAGAGTCCACCGCAAAGCAGACTGGCATGAAATATAGGAATCTTGGAAAATCAGGACTCAGAGTTTCTTGCTTGGGTCTTGGAACA TGGGTGACATTTGGAGGTCAAATTTCAGATGAGGTTGCTGAACGGCTGATGACCATCGCCTATGAAAGTGGTGTTAACCTCTTTGATACT GCCGAAGTCTATGCTGCTGGAAAGGCTGAAGTGATTCTGGGGAGCATCATCAAGAAGAAAGGCTGGAGGAGGTCCAGTCTGGTCATAACA ACCAAACTCTACTGGGGTGGAAAAGCTGAAACAGAAAGAGGGCTGTCAAGAAAGCATATTATTGAAGAAATTGTCCGAGCCATGACACAT GTGATAAACCAAGGCATGGCGATGTACTGGGGCACCTCGAGATGGAGTGCTATGGAGATCATGGAAGCCTATTCTGTAGCAAGACAGTTC AATATGATCCCACCGGTCTGTGAACAAGCTGAGTACCATCTTTTCCAGAGAGAGAAAGTGGAGGTCCAGCTGCCAGAGCTCTACCACAAA ATAGGTGAACTGCTCCTGTTTTCCAGTGTGATTGTCCTGCTCCAGGTGGTATGCAGCTGCCCGGACAAGTGTTACTGTCAGTCATCTACA AATTTTGTAGACTGCAGCCAGCAGGGTCTGGCCGAAATCCCTTCCCATTTACCTCCTCAGACTCGAACGCTGCATTTACAAGATAATCAG ATACACCATCTTCCTGCTTTTGCATTTAGGTCAGTGCCATGGCTCATGACCTTAAACTTGTCCAACAATTCCCTTTCAAATCTGGCCCCT GGAGCTTTCCATGGGCTTCAGCACTTGCAGGTTTTAAATCTAACCCAGAATTCACTCCTTTCCCTGGAAAGCAGACTTTTCCATTCCCTC CCTCAGCTGAGGGAGCTTGATTTGTCATCAAACAACATAAGCCACCTTCCCACATCCTTGGGAGAGACTTGGGAGAACCTAACTATACTT GCGGTTCAACAAAACCAGCTTCAGCAGCTTGATCGAGCGCTCCTGGAATCCATGCCCAGTGTGAGGCTTTTACTTCTCAAGGACAACCTC TGGAAATGCAATTGCCACTTGCTCGGTCTTAAACTCTGGCTGGAGAAATTTGTCTATAAAGGGGGACTAACAGACGGCATCATCTGTGAA TCACCAGACACCTGGAAGGGAAAGGACCTCCTTAGGATCCCTCATGAGCTGTACCAGCCCTGCCCTCTTCCTGCTCCTGATCCAGTGTCC TCGCAGGCTCAGTGGCCCGGCTCTGCCCACGGTGTGGTCCTGAGGCCTCCTGAGAACCACAACGCGGGGGAGCGAGAACTCTTGGAGTGC GAGCTCAAACCCAAGCCAAGGCCGGCCAACCTGCGTCATGCCATTGCCACTGTCATCATCACTGGCGTTGTGTGTGGGATTGTGTGTCTC ATGATGTTGGCAGCTGCCATCTATGGCTGCACCTATGCGGCAATCACAGCCCAGTACCATGGGGGACCCTTGGCTCAAACCAATGATCCT GGGAAGGTGGAAGAAAAAGAGCGATTTGACAGCTCACCAGCCTGAGAGCTTTTGTCTCAAATAGGATTGGTCATTGCAGGCCAGAAGATA GTGTCTGAGTAGGGCTGATGTGTTTCCTGTTAGTCTGATTTTGCTTTTGCCAAAAGACAAAAAAAAAAAAAAAAAAAAAAAGAAAAAAAA >41346_41346_2_KCNAB1-LRTM1_KCNAB1_chr3_156233009_ENST00000389634_LRTM1_chr3_54959242_ENST00000273286_length(amino acids)=584AA_BP=241 MQVSIACTEHNLKSRNGEDRLLSKQSSTAPNVVNAARAKFRTVAIIARSLGTFTPQHHISLKESTAKQTGMKYRNLGKSGLRVSCLGLGT WVTFGGQISDEVAERLMTIAYESGVNLFDTAEVYAAGKAEVILGSIIKKKGWRRSSLVITTKLYWGGKAETERGLSRKHIIEEIVRAMTH VINQGMAMYWGTSRWSAMEIMEAYSVARQFNMIPPVCEQAEYHLFQREKVEVQLPELYHKIGELLLFSSVIVLLQVVCSCPDKCYCQSST NFVDCSQQGLAEIPSHLPPQTRTLHLQDNQIHHLPAFAFRSVPWLMTLNLSNNSLSNLAPGAFHGLQHLQVLNLTQNSLLSLESRLFHSL PQLRELDLSSNNISHLPTSLGETWENLTILAVQQNQLQQLDRALLESMPSVRLLLLKDNLWKCNCHLLGLKLWLEKFVYKGGLTDGIICE SPDTWKGKDLLRIPHELYQPCPLPAPDPVSSQAQWPGSAHGVVLRPPENHNAGERELLECELKPKPRPANLRHAIATVIITGVVCGIVCL -------------------------------------------------------------- >41346_41346_3_KCNAB1-LRTM1_KCNAB1_chr3_156233009_ENST00000389636_LRTM1_chr3_54959242_ENST00000273286_length(transcript)=2065nt_BP=812nt AGTGACTTCCAGTCTTCTCTGAAAGATCTCCACGATGCTGGCAGCCCGGACAGGGGCAGCGGGGAGTCAGATCTCAGAGGAGAACACCAA GTTAAGGAGACAGTCTGGGTTTTCTGTAGCAGGGAAAGACAAATCTCCCAAGAAAGCCTCAGAAAACGCTAAAGACAGCAGCCTTAGTCC CTCAGGGGAAAGCCAGCTCAGGGCGCGTCAACTGGCTCTGCTGCGCGAAGTGGAGATGAACTGGTACCTAAAGCTCTGCGACCTGTCCAG CGAGCACACCACCGTCTGCACCACAGGCATGCCGCACAGGAATCTTGGAAAATCAGGACTCAGAGTTTCTTGCTTGGGTCTTGGAACATG GGTGACATTTGGAGGTCAAATTTCAGATGAGGTTGCTGAACGGCTGATGACCATCGCCTATGAAAGTGGTGTTAACCTCTTTGATACTGC CGAAGTCTATGCTGCTGGAAAGGCTGAAGTGATTCTGGGGAGCATCATCAAGAAGAAAGGCTGGAGGAGGTCCAGTCTGGTCATAACAAC CAAACTCTACTGGGGTGGAAAAGCTGAAACAGAAAGAGGGCTGTCAAGAAAGCATATTATTGAAGAAATTGTCCGAGCCATGACACATGT GATAAACCAAGGCATGGCGATGTACTGGGGCACCTCGAGATGGAGTGCTATGGAGATCATGGAAGCCTATTCTGTAGCAAGACAGTTCAA TATGATCCCACCGGTCTGTGAACAAGCTGAGTACCATCTTTTCCAGAGAGAGAAAGTGGAGGTCCAGCTGCCAGAGCTCTACCACAAAAT AGGTGAACTGCTCCTGTTTTCCAGTGTGATTGTCCTGCTCCAGGTGGTATGCAGCTGCCCGGACAAGTGTTACTGTCAGTCATCTACAAA TTTTGTAGACTGCAGCCAGCAGGGTCTGGCCGAAATCCCTTCCCATTTACCTCCTCAGACTCGAACGCTGCATTTACAAGATAATCAGAT ACACCATCTTCCTGCTTTTGCATTTAGGTCAGTGCCATGGCTCATGACCTTAAACTTGTCCAACAATTCCCTTTCAAATCTGGCCCCTGG AGCTTTCCATGGGCTTCAGCACTTGCAGGTTTTAAATCTAACCCAGAATTCACTCCTTTCCCTGGAAAGCAGACTTTTCCATTCCCTCCC TCAGCTGAGGGAGCTTGATTTGTCATCAAACAACATAAGCCACCTTCCCACATCCTTGGGAGAGACTTGGGAGAACCTAACTATACTTGC GGTTCAACAAAACCAGCTTCAGCAGCTTGATCGAGCGCTCCTGGAATCCATGCCCAGTGTGAGGCTTTTACTTCTCAAGGACAACCTCTG GAAATGCAATTGCCACTTGCTCGGTCTTAAACTCTGGCTGGAGAAATTTGTCTATAAAGGGGGACTAACAGACGGCATCATCTGTGAATC ACCAGACACCTGGAAGGGAAAGGACCTCCTTAGGATCCCTCATGAGCTGTACCAGCCCTGCCCTCTTCCTGCTCCTGATCCAGTGTCCTC GCAGGCTCAGTGGCCCGGCTCTGCCCACGGTGTGGTCCTGAGGCCTCCTGAGAACCACAACGCGGGGGAGCGAGAACTCTTGGAGTGCGA GCTCAAACCCAAGCCAAGGCCGGCCAACCTGCGTCATGCCATTGCCACTGTCATCATCACTGGCGTTGTGTGTGGGATTGTGTGTCTCAT GATGTTGGCAGCTGCCATCTATGGCTGCACCTATGCGGCAATCACAGCCCAGTACCATGGGGGACCCTTGGCTCAAACCAATGATCCTGG GAAGGTGGAAGAAAAAGAGCGATTTGACAGCTCACCAGCCTGAGAGCTTTTGTCTCAAATAGGATTGGTCATTGCAGGCCAGAAGATAGT GTCTGAGTAGGGCTGATGTGTTTCCTGTTAGTCTGATTTTGCTTTTGCCAAAAGACAAAAAAAAAAAAAAAAAAAAAAAGAAAAAAAAAA >41346_41346_3_KCNAB1-LRTM1_KCNAB1_chr3_156233009_ENST00000389636_LRTM1_chr3_54959242_ENST00000273286_length(amino acids)=602AA_BP=259 MLAARTGAAGSQISEENTKLRRQSGFSVAGKDKSPKKASENAKDSSLSPSGESQLRARQLALLREVEMNWYLKLCDLSSEHTTVCTTGMP HRNLGKSGLRVSCLGLGTWVTFGGQISDEVAERLMTIAYESGVNLFDTAEVYAAGKAEVILGSIIKKKGWRRSSLVITTKLYWGGKAETE RGLSRKHIIEEIVRAMTHVINQGMAMYWGTSRWSAMEIMEAYSVARQFNMIPPVCEQAEYHLFQREKVEVQLPELYHKIGELLLFSSVIV LLQVVCSCPDKCYCQSSTNFVDCSQQGLAEIPSHLPPQTRTLHLQDNQIHHLPAFAFRSVPWLMTLNLSNNSLSNLAPGAFHGLQHLQVL NLTQNSLLSLESRLFHSLPQLRELDLSSNNISHLPTSLGETWENLTILAVQQNQLQQLDRALLESMPSVRLLLLKDNLWKCNCHLLGLKL WLEKFVYKGGLTDGIICESPDTWKGKDLLRIPHELYQPCPLPAPDPVSSQAQWPGSAHGVVLRPPENHNAGERELLECELKPKPRPANLR -------------------------------------------------------------- >41346_41346_4_KCNAB1-LRTM1_KCNAB1_chr3_156233009_ENST00000471742_LRTM1_chr3_54959242_ENST00000273286_length(transcript)=2302nt_BP=1049nt ATGCACAGGCCTGGGCAGGGACACACAACCAACAGCTCTGTCGGGAACTTAGGAGCCTGCTCGCCTACAAAAATGATAAGAGAGATCTAA AGAAAACAAGCTAAACCTTTTGAGGGACATACTGAAGCCAGATAACCCAAGGTATTCACAGCAAGATACAGTGAGTCTTAAAGTTAAGCA CCGTGCAATTAGCTTTGCTTCCTTGGGTTTTTGAAACATGCATCTGTATAAACCTGCCTGTGCAGACATCCCGAGCCCCAAGCTGGGTCT GCCAAAATCCAGTGAATCGGCTCTAAAATGTAGATGGCACCTAGCAGTGACCAAGACTCAGCCTCAGGCGGCCTGCAAACCTGTGAGGCC CAGTGGAGCAGCCGAACAGAAATATGTGGAAAAGTTTCTACGTGTTCATGGAATTTCGTTGCAGGAAACCACCAGAGCAGAGACGGGCAT GGCATACAGGAATCTTGGAAAATCAGGACTCAGAGTTTCTTGCTTGGGTCTTGGAACATGGGTGACATTTGGAGGTCAAATTTCAGATGA GGTTGCTGAACGGCTGATGACCATCGCCTATGAAAGTGGTGTTAACCTCTTTGATACTGCCGAAGTCTATGCTGCTGGAAAGGCTGAAGT GATTCTGGGGAGCATCATCAAGAAGAAAGGCTGGAGGAGGTCCAGTCTGGTCATAACAACCAAACTCTACTGGGGTGGAAAAGCTGAAAC AGAAAGAGGGCTGTCAAGAAAGCATATTATTGAAGGATTGAAGGGCTCCCTCCAGAGGCTGCAGCTCGAGTATGTGGATGTGGTCTTTGC AAATCGACCGGACAGTAACACTCCCATGGAAGAAATTGTCCGAGCCATGACACATGTGATAAACCAAGGCATGGCGATGTACTGGGGCAC CTCGAGATGGAGTGCTATGGAGATCATGGAAGCCTATTCTGTAGCAAGACAGTTCAATATGATCCCACCGGTCTGTGAACAAGCTGAGTA CCATCTTTTCCAGAGAGAGAAAGTGGAGGTCCAGCTGCCAGAGCTCTACCACAAAATAGGTGAACTGCTCCTGTTTTCCAGTGTGATTGT CCTGCTCCAGGTGGTATGCAGCTGCCCGGACAAGTGTTACTGTCAGTCATCTACAAATTTTGTAGACTGCAGCCAGCAGGGTCTGGCCGA AATCCCTTCCCATTTACCTCCTCAGACTCGAACGCTGCATTTACAAGATAATCAGATACACCATCTTCCTGCTTTTGCATTTAGGTCAGT GCCATGGCTCATGACCTTAAACTTGTCCAACAATTCCCTTTCAAATCTGGCCCCTGGAGCTTTCCATGGGCTTCAGCACTTGCAGGTTTT AAATCTAACCCAGAATTCACTCCTTTCCCTGGAAAGCAGACTTTTCCATTCCCTCCCTCAGCTGAGGGAGCTTGATTTGTCATCAAACAA CATAAGCCACCTTCCCACATCCTTGGGAGAGACTTGGGAGAACCTAACTATACTTGCGGTTCAACAAAACCAGCTTCAGCAGCTTGATCG AGCGCTCCTGGAATCCATGCCCAGTGTGAGGCTTTTACTTCTCAAGGACAACCTCTGGAAATGCAATTGCCACTTGCTCGGTCTTAAACT CTGGCTGGAGAAATTTGTCTATAAAGGGGGACTAACAGACGGCATCATCTGTGAATCACCAGACACCTGGAAGGGAAAGGACCTCCTTAG GATCCCTCATGAGCTGTACCAGCCCTGCCCTCTTCCTGCTCCTGATCCAGTGTCCTCGCAGGCTCAGTGGCCCGGCTCTGCCCACGGTGT GGTCCTGAGGCCTCCTGAGAACCACAACGCGGGGGAGCGAGAACTCTTGGAGTGCGAGCTCAAACCCAAGCCAAGGCCGGCCAACCTGCG TCATGCCATTGCCACTGTCATCATCACTGGCGTTGTGTGTGGGATTGTGTGTCTCATGATGTTGGCAGCTGCCATCTATGGCTGCACCTA TGCGGCAATCACAGCCCAGTACCATGGGGGACCCTTGGCTCAAACCAATGATCCTGGGAAGGTGGAAGAAAAAGAGCGATTTGACAGCTC ACCAGCCTGAGAGCTTTTGTCTCAAATAGGATTGGTCATTGCAGGCCAGAAGATAGTGTCTGAGTAGGGCTGATGTGTTTCCTGTTAGTC TGATTTTGCTTTTGCCAAAAGACAAAAAAAAAAAAAAAAAAAAAAAGAAAAAAAAAAGATGTGTCTGTAAAAAATATTTCTGGGGAGGAA >41346_41346_4_KCNAB1-LRTM1_KCNAB1_chr3_156233009_ENST00000471742_LRTM1_chr3_54959242_ENST00000273286_length(amino acids)=620AA_BP=277 MHLYKPACADIPSPKLGLPKSSESALKCRWHLAVTKTQPQAACKPVRPSGAAEQKYVEKFLRVHGISLQETTRAETGMAYRNLGKSGLRV SCLGLGTWVTFGGQISDEVAERLMTIAYESGVNLFDTAEVYAAGKAEVILGSIIKKKGWRRSSLVITTKLYWGGKAETERGLSRKHIIEG LKGSLQRLQLEYVDVVFANRPDSNTPMEEIVRAMTHVINQGMAMYWGTSRWSAMEIMEAYSVARQFNMIPPVCEQAEYHLFQREKVEVQL PELYHKIGELLLFSSVIVLLQVVCSCPDKCYCQSSTNFVDCSQQGLAEIPSHLPPQTRTLHLQDNQIHHLPAFAFRSVPWLMTLNLSNNS LSNLAPGAFHGLQHLQVLNLTQNSLLSLESRLFHSLPQLRELDLSSNNISHLPTSLGETWENLTILAVQQNQLQQLDRALLESMPSVRLL LLKDNLWKCNCHLLGLKLWLEKFVYKGGLTDGIICESPDTWKGKDLLRIPHELYQPCPLPAPDPVSSQAQWPGSAHGVVLRPPENHNAGE -------------------------------------------------------------- >41346_41346_5_KCNAB1-LRTM1_KCNAB1_chr3_156233009_ENST00000490337_LRTM1_chr3_54959242_ENST00000273286_length(transcript)=2182nt_BP=929nt AGAAAAATCAATTCAGATTACTTTGATGACAGTGACTTCCAGTCTTCTCTGAAAGATCTCCACGATGCTGGCAGCCCGGACAGGGGCAGC GGGGAGTCAGATCTCAGAGGAGAACACCAAGTTAAGGAGACAGTCTGGGTTTTCTGTAGCAGGGAAAGACAAATCTCCCAAGAAAGCCTC AGAAAACGCTAAAGACAGCAGCCTTAGTCCCTCAGGGGAAAGCCAGCTCAGGGCGCGTCAACTGGCTCTGCTGCGCGAAGTGGAGATGAA CTGGTACCTAAAGCTCTGCGACCTGTCCAGCGAGCACACCACCGTCTGCACCACAGGCATGCCGCACAGGAATCTTGGAAAATCAGGACT CAGAGTTTCTTGCTTGGGTCTTGGAACATGGGTGACATTTGGAGGTCAAATTTCAGATGAGGTTGCTGAACGGCTGATGACCATCGCCTA TGAAAGTGGTGTTAACCTCTTTGATACTGCCGAAGTCTATGCTGCTGGAAAGGCTGAAGTGATTCTGGGGAGCATCATCAAGAAGAAAGG CTGGAGGAGGTCCAGTCTGGTCATAACAACCAAACTCTACTGGGGTGGAAAAGCTGAAACAGAAAGAGGGCTGTCAAGAAAGCATATTAT TGAAGGATTGAAGGGCTCCCTCCAGAGGCTGCAGCTCGAGTATGTGGATGTGGTCTTTGCAAATCGACCGGACAGTAACACTCCCATGGA AGAAATTGTCCGAGCCATGACACATGTGATAAACCAAGGCATGGCGATGTACTGGGGCACCTCGAGATGGAGTGCTATGGAGATCATGGA AGCCTATTCTGTAGCAAGACAGTTCAATATGATCCCACCGGTCTGTGAACAAGCTGAGTACCATCTTTTCCAGAGAGAGAAAGTGGAGGT CCAGCTGCCAGAGCTCTACCACAAAATAGGTGAACTGCTCCTGTTTTCCAGTGTGATTGTCCTGCTCCAGGTGGTATGCAGCTGCCCGGA CAAGTGTTACTGTCAGTCATCTACAAATTTTGTAGACTGCAGCCAGCAGGGTCTGGCCGAAATCCCTTCCCATTTACCTCCTCAGACTCG AACGCTGCATTTACAAGATAATCAGATACACCATCTTCCTGCTTTTGCATTTAGGTCAGTGCCATGGCTCATGACCTTAAACTTGTCCAA CAATTCCCTTTCAAATCTGGCCCCTGGAGCTTTCCATGGGCTTCAGCACTTGCAGGTTTTAAATCTAACCCAGAATTCACTCCTTTCCCT GGAAAGCAGACTTTTCCATTCCCTCCCTCAGCTGAGGGAGCTTGATTTGTCATCAAACAACATAAGCCACCTTCCCACATCCTTGGGAGA GACTTGGGAGAACCTAACTATACTTGCGGTTCAACAAAACCAGCTTCAGCAGCTTGATCGAGCGCTCCTGGAATCCATGCCCAGTGTGAG GCTTTTACTTCTCAAGGACAACCTCTGGAAATGCAATTGCCACTTGCTCGGTCTTAAACTCTGGCTGGAGAAATTTGTCTATAAAGGGGG ACTAACAGACGGCATCATCTGTGAATCACCAGACACCTGGAAGGGAAAGGACCTCCTTAGGATCCCTCATGAGCTGTACCAGCCCTGCCC TCTTCCTGCTCCTGATCCAGTGTCCTCGCAGGCTCAGTGGCCCGGCTCTGCCCACGGTGTGGTCCTGAGGCCTCCTGAGAACCACAACGC GGGGGAGCGAGAACTCTTGGAGTGCGAGCTCAAACCCAAGCCAAGGCCGGCCAACCTGCGTCATGCCATTGCCACTGTCATCATCACTGG CGTTGTGTGTGGGATTGTGTGTCTCATGATGTTGGCAGCTGCCATCTATGGCTGCACCTATGCGGCAATCACAGCCCAGTACCATGGGGG ACCCTTGGCTCAAACCAATGATCCTGGGAAGGTGGAAGAAAAAGAGCGATTTGACAGCTCACCAGCCTGAGAGCTTTTGTCTCAAATAGG ATTGGTCATTGCAGGCCAGAAGATAGTGTCTGAGTAGGGCTGATGTGTTTCCTGTTAGTCTGATTTTGCTTTTGCCAAAAGACAAAAAAA AAAAAAAAAAAAAAAAGAAAAAAAAAAGATGTGTCTGTAAAAAATATTTCTGGGGAGGAATTTATGTTGCGATTTTTAAAATACTATAGG >41346_41346_5_KCNAB1-LRTM1_KCNAB1_chr3_156233009_ENST00000490337_LRTM1_chr3_54959242_ENST00000273286_length(amino acids)=631AA_BP=288 MLAARTGAAGSQISEENTKLRRQSGFSVAGKDKSPKKASENAKDSSLSPSGESQLRARQLALLREVEMNWYLKLCDLSSEHTTVCTTGMP HRNLGKSGLRVSCLGLGTWVTFGGQISDEVAERLMTIAYESGVNLFDTAEVYAAGKAEVILGSIIKKKGWRRSSLVITTKLYWGGKAETE RGLSRKHIIEGLKGSLQRLQLEYVDVVFANRPDSNTPMEEIVRAMTHVINQGMAMYWGTSRWSAMEIMEAYSVARQFNMIPPVCEQAEYH LFQREKVEVQLPELYHKIGELLLFSSVIVLLQVVCSCPDKCYCQSSTNFVDCSQQGLAEIPSHLPPQTRTLHLQDNQIHHLPAFAFRSVP WLMTLNLSNNSLSNLAPGAFHGLQHLQVLNLTQNSLLSLESRLFHSLPQLRELDLSSNNISHLPTSLGETWENLTILAVQQNQLQQLDRA LLESMPSVRLLLLKDNLWKCNCHLLGLKLWLEKFVYKGGLTDGIICESPDTWKGKDLLRIPHELYQPCPLPAPDPVSSQAQWPGSAHGVV LRPPENHNAGERELLECELKPKPRPANLRHAIATVIITGVVCGIVCLMMLAAAIYGCTYAAITAQYHGGPLAQTNDPGKVEEKERFDSSP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for KCNAB1-LRTM1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for KCNAB1-LRTM1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for KCNAB1-LRTM1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
