
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:KCNMA1-DOCK1 (FusionGDB2 ID:41484) |
Fusion Gene Summary for KCNMA1-DOCK1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: KCNMA1-DOCK1 | Fusion gene ID: 41484 | Hgene | Tgene | Gene symbol | KCNMA1 | DOCK1 | Gene ID | 3778 | 1793 |
| Gene name | potassium calcium-activated channel subfamily M alpha 1 | dedicator of cytokinesis 1 | |
| Synonyms | BKTM|CADEDS|IEG16|KCa1.1|LIWAS|MaxiK|PNKD3|SAKCA|SLO|SLO-ALPHA|SLO1|bA205K10.1|hSlo|mSLO1 | DOCK180|ced5 | |
| Cytomap | 10q22.3 | 10q26.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | calcium-activated potassium channel subunit alpha-1uncharacterized proteinBK channel alpha subunitBKCA alpha subunitbig potassium channel alpha subunitcalcium-activated potassium channel, subfamily M subunit alpha-1k(VCA)alphamaxi-K channel HSLOpo | dedicator of cytokinesis protein 1180 kDa protein downstream of CRKDOwnstream of CrK | |
| Modification date | 20200315 | 20200327 | |
| UniProtAcc | Q12791 | Q5JSL3 | |
| Ensembl transtripts involved in fusion gene | ENST00000286627, ENST00000286628, ENST00000354353, ENST00000372440, ENST00000372443, ENST00000404771, ENST00000404857, ENST00000406533, ENST00000480683, ENST00000481070, ENST00000484507, | ENST00000484400, ENST00000280333, | |
| Fusion gene scores | * DoF score | 20 X 17 X 10=3400 | 16 X 15 X 8=1920 |
| # samples | 25 | 16 | |
| ** MAII score | log2(25/3400*10)=-3.76553474636298 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(16/1920*10)=-3.58496250072116 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: KCNMA1 [Title/Abstract] AND DOCK1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | KCNMA1(78880731)-DOCK1(129249596), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | KCNMA1 | GO:0001666 | response to hypoxia | 15528406 |
| Hgene | KCNMA1 | GO:0006813 | potassium ion transport | 7573516|7877450|11245614|12388065|17706472|18458941 |
| Hgene | KCNMA1 | GO:0006970 | response to osmotic stress | 10840032|12388065 |
| Hgene | KCNMA1 | GO:0030007 | cellular potassium ion homeostasis | 11245614 |
| Hgene | KCNMA1 | GO:0034465 | response to carbon monoxide | 15528406 |
| Hgene | KCNMA1 | GO:0042391 | regulation of membrane potential | 7877450|7993625 |
| Hgene | KCNMA1 | GO:0045794 | negative regulation of cell volume | 12388065 |
| Hgene | KCNMA1 | GO:0051592 | response to calcium ion | 12388065|18458941 |
| Hgene | KCNMA1 | GO:0060073 | micturition | 11641143 |
| Hgene | KCNMA1 | GO:0060083 | smooth muscle contraction involved in micturition | 11641143 |
 Fusion gene breakpoints across KCNMA1 (5'-gene) Fusion gene breakpoints across KCNMA1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
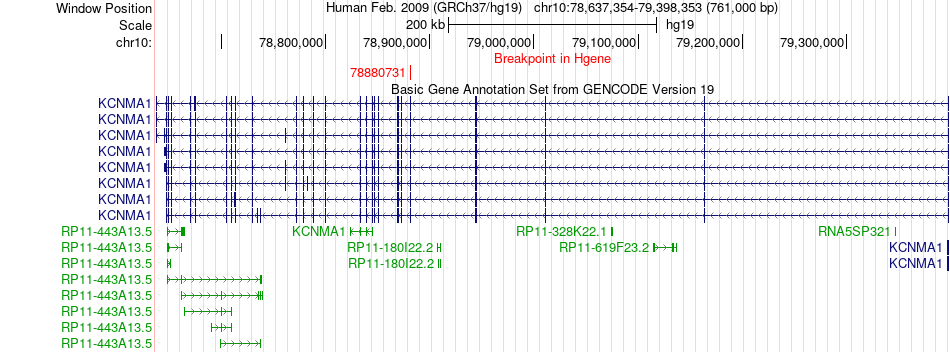 |
 Fusion gene breakpoints across DOCK1 (3'-gene) Fusion gene breakpoints across DOCK1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
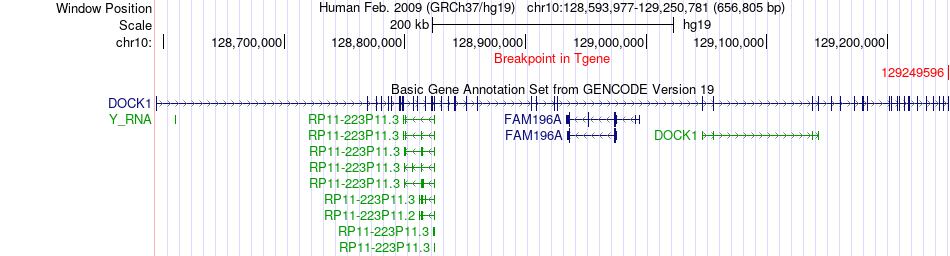 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LIHC | TCGA-CC-A7IJ-01A | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + |
Top |
Fusion Gene ORF analysis for KCNMA1-DOCK1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000286627 | ENST00000484400 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + |
| 5CDS-intron | ENST00000286628 | ENST00000484400 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + |
| 5CDS-intron | ENST00000354353 | ENST00000484400 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + |
| 5CDS-intron | ENST00000372440 | ENST00000484400 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + |
| 5CDS-intron | ENST00000372443 | ENST00000484400 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + |
| 5CDS-intron | ENST00000404771 | ENST00000484400 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + |
| 5CDS-intron | ENST00000404857 | ENST00000484400 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + |
| 5CDS-intron | ENST00000406533 | ENST00000484400 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + |
| In-frame | ENST00000286627 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + |
| In-frame | ENST00000286628 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + |
| In-frame | ENST00000354353 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + |
| In-frame | ENST00000372440 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + |
| In-frame | ENST00000372443 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + |
| In-frame | ENST00000404771 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + |
| In-frame | ENST00000404857 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + |
| In-frame | ENST00000406533 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + |
| intron-3CDS | ENST00000480683 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + |
| intron-3CDS | ENST00000481070 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + |
| intron-3CDS | ENST00000484507 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + |
| intron-intron | ENST00000480683 | ENST00000484400 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + |
| intron-intron | ENST00000481070 | ENST00000484400 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + |
| intron-intron | ENST00000484507 | ENST00000484400 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000404771 | KCNMA1 | chr10 | 78880731 | - | ENST00000280333 | DOCK1 | chr10 | 129249596 | + | 2186 | 1000 | 62 | 1267 | 401 |
| ENST00000372440 | KCNMA1 | chr10 | 78880731 | - | ENST00000280333 | DOCK1 | chr10 | 129249596 | + | 2421 | 1235 | 297 | 1502 | 401 |
| ENST00000372443 | KCNMA1 | chr10 | 78880731 | - | ENST00000280333 | DOCK1 | chr10 | 129249596 | + | 2797 | 1611 | 673 | 1878 | 401 |
| ENST00000286628 | KCNMA1 | chr10 | 78880731 | - | ENST00000280333 | DOCK1 | chr10 | 129249596 | + | 2070 | 884 | 0 | 1151 | 383 |
| ENST00000286627 | KCNMA1 | chr10 | 78880731 | - | ENST00000280333 | DOCK1 | chr10 | 129249596 | + | 3023 | 1837 | 899 | 2104 | 401 |
| ENST00000354353 | KCNMA1 | chr10 | 78880731 | - | ENST00000280333 | DOCK1 | chr10 | 129249596 | + | 2070 | 884 | 0 | 1151 | 383 |
| ENST00000404857 | KCNMA1 | chr10 | 78880731 | - | ENST00000280333 | DOCK1 | chr10 | 129249596 | + | 2070 | 884 | 0 | 1151 | 383 |
| ENST00000406533 | KCNMA1 | chr10 | 78880731 | - | ENST00000280333 | DOCK1 | chr10 | 129249596 | + | 2070 | 884 | 0 | 1151 | 383 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000404771 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + | 0.035057057 | 0.96494293 |
| ENST00000372440 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + | 0.044519797 | 0.9554802 |
| ENST00000372443 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + | 0.04783887 | 0.95216113 |
| ENST00000286628 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + | 0.03677205 | 0.963228 |
| ENST00000286627 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + | 0.055738155 | 0.94426185 |
| ENST00000354353 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + | 0.03677205 | 0.963228 |
| ENST00000404857 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + | 0.03677205 | 0.963228 |
| ENST00000406533 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + | 0.03677205 | 0.963228 |
Top |
Fusion Genomic Features for KCNMA1-DOCK1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| KCNMA1 | chr10 | 78880730 | - | DOCK1 | chr10 | 129249595 | + | 0.0006194 | 0.9993806 |
| KCNMA1 | chr10 | 78880730 | - | DOCK1 | chr10 | 129249595 | + | 0.0006194 | 0.9993806 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
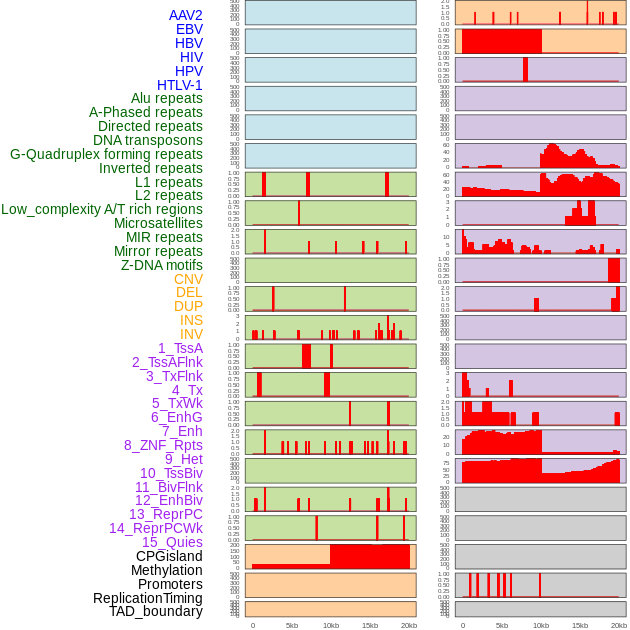 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
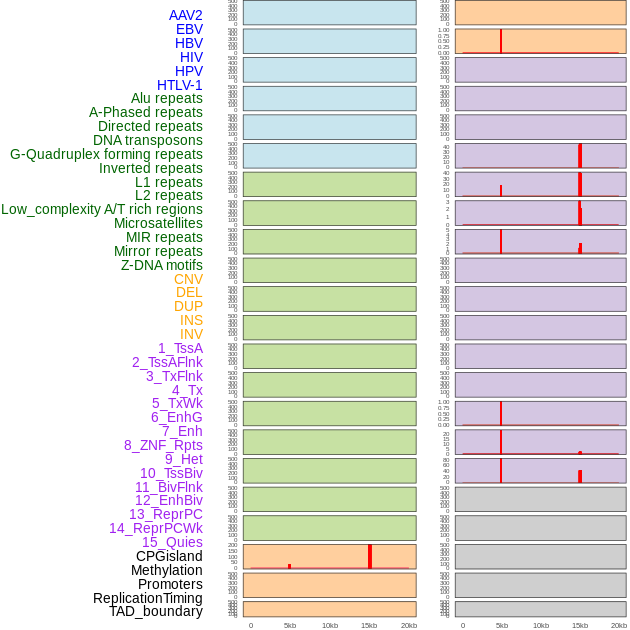 |
Top |
Fusion Protein Features for KCNMA1-DOCK1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr10:78880731/chr10:129249596) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| KCNMA1 | DOCK1 |
| FUNCTION: Potassium channel activated by both membrane depolarization or increase in cytosolic Ca(2+) that mediates export of K(+) (PubMed:29330545, PubMed:31152168). It is also activated by the concentration of cytosolic Mg(2+). Its activation dampens the excitatory events that elevate the cytosolic Ca(2+) concentration and/or depolarize the cell membrane. It therefore contributes to repolarization of the membrane potential. Plays a key role in controlling excitability in a number of systems, such as regulation of the contraction of smooth muscle, the tuning of hair cells in the cochlea, regulation of transmitter release, and innate immunity. In smooth muscles, its activation by high level of Ca(2+), caused by ryanodine receptors in the sarcoplasmic reticulum, regulates the membrane potential. In cochlea cells, its number and kinetic properties partly determine the characteristic frequency of each hair cell and thereby helps to establish a tonotopic map. Kinetics of KCNMA1 channels are determined by alternative splicing, phosphorylation status and its combination with modulating beta subunits. Highly sensitive to both iberiotoxin (IbTx) and charybdotoxin (CTX). {ECO:0000269|PubMed:29330545, ECO:0000269|PubMed:31152168}. | FUNCTION: Guanine nucleotide-exchange factor (GEF) that activates CDC42 by exchanging bound GDP for free GTP. Required for marginal zone (MZ) B-cell development, is associated with early bone marrow B-cell development, MZ B-cell formation, MZ B-cell number and marginal metallophilic macrophages morphology. Facilitates filopodia formation through the activation of CDC42. {ECO:0000250|UniProtKB:A2AF47}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 13_20 | 294 | 1179.0 | Compositional bias | Note=Poly-Gly |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 39_60 | 294 | 1179.0 | Compositional bias | Note=Poly-Ser |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 4_10 | 294 | 1179.0 | Compositional bias | Note=Poly-Gly |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 13_20 | 294 | 1237.0 | Compositional bias | Note=Poly-Gly |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 39_60 | 294 | 1237.0 | Compositional bias | Note=Poly-Ser |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 4_10 | 294 | 1237.0 | Compositional bias | Note=Poly-Gly |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 13_20 | 294 | 1220.0 | Compositional bias | Note=Poly-Gly |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 39_60 | 294 | 1220.0 | Compositional bias | Note=Poly-Ser |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 4_10 | 294 | 1220.0 | Compositional bias | Note=Poly-Gly |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 108_178 | 294 | 1179.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 1_86 | 294 | 1179.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 200_214 | 294 | 1179.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 236_239 | 294 | 1179.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 261_264 | 294 | 1179.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 108_178 | 294 | 1237.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 1_86 | 294 | 1237.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 200_214 | 294 | 1237.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 236_239 | 294 | 1237.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 261_264 | 294 | 1237.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 108_178 | 294 | 1220.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 1_86 | 294 | 1220.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 200_214 | 294 | 1220.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 236_239 | 294 | 1220.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 261_264 | 294 | 1220.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 179_199 | 294 | 1179.0 | Transmembrane | Helical%3B Name%3DSegment S1 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 215_235 | 294 | 1179.0 | Transmembrane | Helical%3B Name%3DSegment S2 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 240_260 | 294 | 1179.0 | Transmembrane | Helical%3B Name%3DSegment S3 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 265_285 | 294 | 1179.0 | Transmembrane | Helical%3B Name%3DSegment S4 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 87_107 | 294 | 1179.0 | Transmembrane | Helical%3B Name%3DSegment S0 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 179_199 | 294 | 1237.0 | Transmembrane | Helical%3B Name%3DSegment S1 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 215_235 | 294 | 1237.0 | Transmembrane | Helical%3B Name%3DSegment S2 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 240_260 | 294 | 1237.0 | Transmembrane | Helical%3B Name%3DSegment S3 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 265_285 | 294 | 1237.0 | Transmembrane | Helical%3B Name%3DSegment S4 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 87_107 | 294 | 1237.0 | Transmembrane | Helical%3B Name%3DSegment S0 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 179_199 | 294 | 1220.0 | Transmembrane | Helical%3B Name%3DSegment S1 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 215_235 | 294 | 1220.0 | Transmembrane | Helical%3B Name%3DSegment S2 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 240_260 | 294 | 1220.0 | Transmembrane | Helical%3B Name%3DSegment S3 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 265_285 | 294 | 1220.0 | Transmembrane | Helical%3B Name%3DSegment S4 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 87_107 | 294 | 1220.0 | Transmembrane | Helical%3B Name%3DSegment S0 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 13_20 | 0 | 169.0 | Compositional bias | Note=Poly-Gly |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 39_60 | 0 | 169.0 | Compositional bias | Note=Poly-Ser |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 4_10 | 0 | 169.0 | Compositional bias | Note=Poly-Gly |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 415_558 | 294 | 1179.0 | Domain | Note=RCK N-terminal |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 415_558 | 294 | 1237.0 | Domain | Note=RCK N-terminal |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 415_558 | 294 | 1220.0 | Domain | Note=RCK N-terminal |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 415_558 | 0 | 169.0 | Domain | Note=RCK N-terminal |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 336_358 | 294 | 1179.0 | Intramembrane | Pore-forming%3B Name%3DP region |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 336_358 | 294 | 1237.0 | Intramembrane | Pore-forming%3B Name%3DP region |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 336_358 | 294 | 1220.0 | Intramembrane | Pore-forming%3B Name%3DP region |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 336_358 | 0 | 169.0 | Intramembrane | Pore-forming%3B Name%3DP region |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 1003_1025 | 294 | 1179.0 | Motif | Note=Calcium bowl |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 352_355 | 294 | 1179.0 | Motif | Note=Selectivity for potassium |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 1003_1025 | 294 | 1237.0 | Motif | Note=Calcium bowl |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 352_355 | 294 | 1237.0 | Motif | Note=Selectivity for potassium |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 1003_1025 | 294 | 1220.0 | Motif | Note=Calcium bowl |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 352_355 | 294 | 1220.0 | Motif | Note=Selectivity for potassium |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 1003_1025 | 0 | 169.0 | Motif | Note=Calcium bowl |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 352_355 | 0 | 169.0 | Motif | Note=Selectivity for potassium |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 1032_1052 | 294 | 1179.0 | Region | Note=Segment S10 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 556_576 | 294 | 1179.0 | Region | Note=Segment S7 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 613_633 | 294 | 1179.0 | Region | Note=Segment S8 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 677_681 | 294 | 1179.0 | Region | Note=Heme-binding motif |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 837_857 | 294 | 1179.0 | Region | Note=Segment S9 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 1032_1052 | 294 | 1237.0 | Region | Note=Segment S10 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 556_576 | 294 | 1237.0 | Region | Note=Segment S7 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 613_633 | 294 | 1237.0 | Region | Note=Segment S8 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 677_681 | 294 | 1237.0 | Region | Note=Heme-binding motif |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 837_857 | 294 | 1237.0 | Region | Note=Segment S9 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 1032_1052 | 294 | 1220.0 | Region | Note=Segment S10 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 556_576 | 294 | 1220.0 | Region | Note=Segment S7 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 613_633 | 294 | 1220.0 | Region | Note=Segment S8 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 677_681 | 294 | 1220.0 | Region | Note=Heme-binding motif |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 837_857 | 294 | 1220.0 | Region | Note=Segment S9 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 1032_1052 | 0 | 169.0 | Region | Note=Segment S10 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 556_576 | 0 | 169.0 | Region | Note=Segment S7 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 613_633 | 0 | 169.0 | Region | Note=Segment S8 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 677_681 | 0 | 169.0 | Region | Note=Heme-binding motif |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 837_857 | 0 | 169.0 | Region | Note=Segment S9 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 286_300 | 294 | 1179.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 322_335 | 294 | 1179.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 359_367 | 294 | 1179.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 389_1236 | 294 | 1179.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 286_300 | 294 | 1237.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 322_335 | 294 | 1237.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 359_367 | 294 | 1237.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 389_1236 | 294 | 1237.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 286_300 | 294 | 1220.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 322_335 | 294 | 1220.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 359_367 | 294 | 1220.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 389_1236 | 294 | 1220.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 108_178 | 0 | 169.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 1_86 | 0 | 169.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 200_214 | 0 | 169.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 236_239 | 0 | 169.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 261_264 | 0 | 169.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 286_300 | 0 | 169.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 322_335 | 0 | 169.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 359_367 | 0 | 169.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 389_1236 | 0 | 169.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 301_321 | 294 | 1179.0 | Transmembrane | Helical%3B Name%3DSegment S5 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 368_388 | 294 | 1179.0 | Transmembrane | Helical%3B Name%3DSegment S6 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 301_321 | 294 | 1237.0 | Transmembrane | Helical%3B Name%3DSegment S5 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 368_388 | 294 | 1237.0 | Transmembrane | Helical%3B Name%3DSegment S6 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 301_321 | 294 | 1220.0 | Transmembrane | Helical%3B Name%3DSegment S5 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 368_388 | 294 | 1220.0 | Transmembrane | Helical%3B Name%3DSegment S6 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 179_199 | 0 | 169.0 | Transmembrane | Helical%3B Name%3DSegment S1 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 215_235 | 0 | 169.0 | Transmembrane | Helical%3B Name%3DSegment S2 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 240_260 | 0 | 169.0 | Transmembrane | Helical%3B Name%3DSegment S3 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 265_285 | 0 | 169.0 | Transmembrane | Helical%3B Name%3DSegment S4 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 301_321 | 0 | 169.0 | Transmembrane | Helical%3B Name%3DSegment S5 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 368_388 | 0 | 169.0 | Transmembrane | Helical%3B Name%3DSegment S6 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 87_107 | 0 | 169.0 | Transmembrane | Helical%3B Name%3DSegment S0 |
| Tgene | DOCK1 | chr10:78880731 | chr10:129249596 | ENST00000280333 | 50 | 52 | 1207_1617 | 1834 | 1866.0 | Domain | DOCKER | |
| Tgene | DOCK1 | chr10:78880731 | chr10:129249596 | ENST00000280333 | 50 | 52 | 425_609 | 1834 | 1866.0 | Domain | C2 DOCK-type | |
| Tgene | DOCK1 | chr10:78880731 | chr10:129249596 | ENST00000280333 | 50 | 52 | 9_70 | 1834 | 1866.0 | Domain | SH3 | |
| Tgene | DOCK1 | chr10:78880731 | chr10:129249596 | ENST00000280333 | 50 | 52 | 1687_1695 | 1834 | 1866.0 | Region | Phosphoinositide-binding |
Top |
Fusion Gene Sequence for KCNMA1-DOCK1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >41484_41484_1_KCNMA1-DOCK1_KCNMA1_chr10_78880731_ENST00000286627_DOCK1_chr10_129249596_ENST00000280333_length(transcript)=3023nt_BP=1837nt CCGGGCCGCTGCCGAGGGGCCCTGCCCACCCCTGCCCAGCCCGGAGCCCAGGCGAGCCAGCGCGGCGAGCCTCCTCCCCTCCCGCCTGGG TCGTCCTGAGTCCGCGCCCGCTCCAGTTGCTGCAGCTGCTGCTGCTGGCGGCTGCGGGGAGCGCGGGGCGCGCGCCGAAGAGGCTTGGGC GGCGAGGACGGGGACGAGGAGGAGTCGGAGGCGGCAAGGAGGAAGAGCGAGCGGAGGCCGAGGGGCTCAGCGGCGTGGCGACAGGACTAG GGGGTCCCGAGAACATGGGCTAGCAGGCGGCCACGGGCCGGCGGCCGGACGCGGTGAACCTGCCGCGGCCGGGCGCGCACTGGCGCGGCG AGCCGGAGCCGAGCGCAGCCGGGGGAGGAGGGCTGGCGCAGGGGCGCCCGGGCGGAGGGGGCCGATTATCCATTATTAACGAGTTACCAC CTAATAAGCGGAGAGCGGCTCGTTAGCCGCGCGGGAGGCGCTCGGGGAAGGGGGTCACCGGCCGAGCGCAGCGCACACCCGTGGCGCACG GCGGGCGGCGCGGCGCCGAGGACACCCGGGACCGCGCGCGGAGCCGGGGCGGGCCCGGGGGCGGCGCAGACCCCGGGCGCGGGCGGCCCG CGGCGCCCCCTGGCTCTCGGCGCTCGGGGCCAGCGCAGCCCGGGGCGAGCGGGGCGCGGGCCGGGGAGGGGGAGCTGCCCGCGCACTCCA TCCCCCTCCCCCCCGGGCGGGCGGCGGCGGCTCGCTGTATATATCTCGGCGCACCCCGCCAGGTCGCGCACAGCGCCCCGAGCCCAGGCG CCTCCCCGCCCCCCTCCCGCGCTCCGCGGCGGCGGCGGCGGCGGCAGCAGTAGCAGCAATATGGCTGTTGATGGGTGTTTGGGGTGGCGC TGGCGGCGGGAGGAGCTCCCCCGAGCCCCTGCGCCGGCTGCCCGTTGCTAGCTATGGCAAATGGTGGCGGCGGCGGCGGCGGCAGCAGCG GCGGCGGCGGCGGCGGCGGAGGCAGCAGTCTTAGAATGAGTAGCAATATCCACGCGAACCATCTCAGCCTAGACGCGTCCTCCTCCTCCT CCTCCTCCTCTTCCTCTTCTTCTTCTTCCTCCTCCTCTTCCTCCTCGTCCTCGGTCCACGAGCCCAAGATGGATGCGCTCATCATCCCGG TGACCATGGAGGTGCCGTGCGACAGCCGGGGCCAACGCATGTGGTGGGCTTTCCTGGCCTCCTCCATGGTGACTTTCTTCGGGGGCCTCT TCATCATCTTGCTCTGGCGGACGCTCAAGTACCTGTGGACCGTGTGCTGCCACTGCGGGGGCAAGACGAAGGAGGCCCAGAAGATTAACA ATGGCTCAAGCCAGGCGGATGGCACTCTCAAACCAGTGGATGAAAAAGAGGAGGCAGTGGCCGCCGAGGTCGGCTGGATGACCTCCGTGA AGGACTGGGCGGGGGTGATGATATCCGCCCAGACACTGACTGGCAGAGTCCTGGTTGTCTTAGTCTTTGCTCTCAGCATCGGTGCACTTG TAATATACTTCATAGATTCATCAAACCCAATAGAATCCTGCCAGAATTTCTACAAAGATTTCACATTACAGATCGACATGGCTTTCAACG TGTTCTTCCTTCTCTACTTCGGCTTGCGGTTTATTGCAGCCAACGATAAATTGTGGTTCTGGCTGGAAGTGAACTCTGTAGTGGATTTCT TCACGGTGCCCCCCGTGTTTGTGTCTGTGTACTTAAACAGAAGTTGGCTTGGTTTGAGATTTTTAAGAGCTCTGAGACTGATACAGTTTT CAGAAATTTTGCAGTTTCTGAATATTCTTAAAACAAGCATCTGCCACCTCCACTGCCCAGCAAAACTCCGCCTCCTCCCCCTCCAAAGAC AACTCGCAAGCAGGCATCGGTGGACTCCGGGATCGTGCAGTGACGTCGCAAGCCTCTCTGGAAAGAGTGTGCTGCCCCTCCCCATCTCCA TGCCCTCTCCTTCTGTGTCCCCTGAGTCTGCTGTTTACCTCATTGGGCCTGTGATGTTAACATTTCGTGCGACTGCTTTTTCTTCAAAGG AGTTCAGTTCTCACCATGGAGTGAGTGGCCTTTAGCGTCATGGAGCAAGGTGGGTCTGGGAGGTAGATATGGGTCCGGGATGTGCTATCG TAGTTATCAGAGTTGGGGGCCTCTGAGTGTGTCTGGCTCTGAGAGAGTCTGAGTCTTGCCCAAACATTCTTTCTTTTTGTGCCAAATGAC TTGCATTTGCAAAGAGCTCAATTGCTCTGAGCTCAGCCAAGTAGGAGAGGCTAGGCCATCACTCTTAGGAAGCTGTGTAGTGATGATGTA TAAGAATCCTCCTCACTGTCATGGGATGTTGTATCCAGCCCCTCCTTGTTCCAGCCGGTGGTGTGACTTCGTTGGTTGAGGTGTGTCTCC AACCTACATCAGACCATGAAGTTCAACCCCTCCAGGGAAGCTCCTGATTTCCCCTGCATAATTGAAAATAGGATATTTCTCAGCTATTGA ACAGTTACTAATTTATGGGGTGGAAACAGCATTAAGAATACTGAATCAAATGGAAAAACAAATGAATACAGGAAGATAAGTGTTCGTTCT TTTCTGAAAAAGAGTATGTGTACCACAAGAGCTGGTTTTAATTGGGTGAATTGTTTTTGTCCTCATTCTGTACAGAAATTTGTATATATG ATGGTTCTTAGAACTTGTTTTAATTTTTGTGGTCCTTCCGTTTATTATAATAGGCGTCCACCAATGATTATCCATATGTGTTCTTAATTT TTAACTGCTGGAAGTGTTAAAACACACACACACACACACACACATTTTTTTTTTGAGAACTCCAAAGTCCTGAAAATTTTGGTGGACAAT GATTTTTAAAAAACTAACTTTGTATAACCTAATATTTGTATTCTCTCATCTATATTTTTTTATTCACTATAATCATGATACTCTTCTAAT >41484_41484_1_KCNMA1-DOCK1_KCNMA1_chr10_78880731_ENST00000286627_DOCK1_chr10_129249596_ENST00000280333_length(amino acids)=401AA_BP=312 MAAGGAPPSPCAGCPLLAMANGGGGGGGSSGGGGGGGGSSLRMSSNIHANHLSLDASSSSSSSSSSSSSSSSSSSSSSVHEPKMDALIIP VTMEVPCDSRGQRMWWAFLASSMVTFFGGLFIILLWRTLKYLWTVCCHCGGKTKEAQKINNGSSQADGTLKPVDEKEEAVAAEVGWMTSV KDWAGVMISAQTLTGRVLVVLVFALSIGALVIYFIDSSNPIESCQNFYKDFTLQIDMAFNVFFLLYFGLRFIAANDKLWFWLEVNSVVDF FTVPPVFVSVYLNRSWLGLRFLRALRLIQFSEILQFLNILKTSICHLHCPAKLRLLPLQRQLASRHRWTPGSCSDVASLSGKSVLPLPIS -------------------------------------------------------------- >41484_41484_2_KCNMA1-DOCK1_KCNMA1_chr10_78880731_ENST00000286628_DOCK1_chr10_129249596_ENST00000280333_length(transcript)=2070nt_BP=884nt ATGGCAAATGGTGGCGGCGGCGGCGGCGGCAGCAGCGGCGGCGGCGGCGGCGGCGGAGGCAGCAGTCTTAGAATGAGTAGCAATATCCAC GCGAACCATCTCAGCCTAGACGCGTCCTCCTCCTCCTCCTCCTCCTCTTCCTCTTCTTCTTCTTCCTCCTCCTCTTCCTCCTCGTCCTCG GTCCACGAGCCCAAGATGGATGCGCTCATCATCCCGGTGACCATGGAGGTGCCGTGCGACAGCCGGGGCCAACGCATGTGGTGGGCTTTC CTGGCCTCCTCCATGGTGACTTTCTTCGGGGGCCTCTTCATCATCTTGCTCTGGCGGACGCTCAAGTACCTGTGGACCGTGTGCTGCCAC TGCGGGGGCAAGACGAAGGAGGCCCAGAAGATTAACAATGGCTCAAGCCAGGCGGATGGCACTCTCAAACCAGTGGATGAAAAAGAGGAG GCAGTGGCCGCCGAGGTCGGCTGGATGACCTCCGTGAAGGACTGGGCGGGGGTGATGATATCCGCCCAGACACTGACTGGCAGAGTCCTG GTTGTCTTAGTCTTTGCTCTCAGCATCGGTGCACTTGTAATATACTTCATAGATTCATCAAACCCAATAGAATCCTGCCAGAATTTCTAC AAAGATTTCACATTACAGATCGACATGGCTTTCAACGTGTTCTTCCTTCTCTACTTCGGCTTGCGGTTTATTGCAGCCAACGATAAATTG TGGTTCTGGCTGGAAGTGAACTCTGTAGTGGATTTCTTCACGGTGCCCCCCGTGTTTGTGTCTGTGTACTTAAACAGAAGTTGGCTTGGT TTGAGATTTTTAAGAGCTCTGAGACTGATACAGTTTTCAGAAATTTTGCAGTTTCTGAATATTCTTAAAACAAGCATCTGCCACCTCCAC TGCCCAGCAAAACTCCGCCTCCTCCCCCTCCAAAGACAACTCGCAAGCAGGCATCGGTGGACTCCGGGATCGTGCAGTGACGTCGCAAGC CTCTCTGGAAAGAGTGTGCTGCCCCTCCCCATCTCCATGCCCTCTCCTTCTGTGTCCCCTGAGTCTGCTGTTTACCTCATTGGGCCTGTG ATGTTAACATTTCGTGCGACTGCTTTTTCTTCAAAGGAGTTCAGTTCTCACCATGGAGTGAGTGGCCTTTAGCGTCATGGAGCAAGGTGG GTCTGGGAGGTAGATATGGGTCCGGGATGTGCTATCGTAGTTATCAGAGTTGGGGGCCTCTGAGTGTGTCTGGCTCTGAGAGAGTCTGAG TCTTGCCCAAACATTCTTTCTTTTTGTGCCAAATGACTTGCATTTGCAAAGAGCTCAATTGCTCTGAGCTCAGCCAAGTAGGAGAGGCTA GGCCATCACTCTTAGGAAGCTGTGTAGTGATGATGTATAAGAATCCTCCTCACTGTCATGGGATGTTGTATCCAGCCCCTCCTTGTTCCA GCCGGTGGTGTGACTTCGTTGGTTGAGGTGTGTCTCCAACCTACATCAGACCATGAAGTTCAACCCCTCCAGGGAAGCTCCTGATTTCCC CTGCATAATTGAAAATAGGATATTTCTCAGCTATTGAACAGTTACTAATTTATGGGGTGGAAACAGCATTAAGAATACTGAATCAAATGG AAAAACAAATGAATACAGGAAGATAAGTGTTCGTTCTTTTCTGAAAAAGAGTATGTGTACCACAAGAGCTGGTTTTAATTGGGTGAATTG TTTTTGTCCTCATTCTGTACAGAAATTTGTATATATGATGGTTCTTAGAACTTGTTTTAATTTTTGTGGTCCTTCCGTTTATTATAATAG GCGTCCACCAATGATTATCCATATGTGTTCTTAATTTTTAACTGCTGGAAGTGTTAAAACACACACACACACACACACACATTTTTTTTT TGAGAACTCCAAAGTCCTGAAAATTTTGGTGGACAATGATTTTTAAAAAACTAACTTTGTATAACCTAATATTTGTATTCTCTCATCTAT ATTTTTTTATTCACTATAATCATGATACTCTTCTAATAGACTTAAAAGTTAATCATTGACAACGAAAAATAAAATGTTATTTTAAAAGAC >41484_41484_2_KCNMA1-DOCK1_KCNMA1_chr10_78880731_ENST00000286628_DOCK1_chr10_129249596_ENST00000280333_length(amino acids)=383AA_BP=294 MANGGGGGGGSSGGGGGGGGSSLRMSSNIHANHLSLDASSSSSSSSSSSSSSSSSSSSSSVHEPKMDALIIPVTMEVPCDSRGQRMWWAF LASSMVTFFGGLFIILLWRTLKYLWTVCCHCGGKTKEAQKINNGSSQADGTLKPVDEKEEAVAAEVGWMTSVKDWAGVMISAQTLTGRVL VVLVFALSIGALVIYFIDSSNPIESCQNFYKDFTLQIDMAFNVFFLLYFGLRFIAANDKLWFWLEVNSVVDFFTVPPVFVSVYLNRSWLG LRFLRALRLIQFSEILQFLNILKTSICHLHCPAKLRLLPLQRQLASRHRWTPGSCSDVASLSGKSVLPLPISMPSPSVSPESAVYLIGPV -------------------------------------------------------------- >41484_41484_3_KCNMA1-DOCK1_KCNMA1_chr10_78880731_ENST00000354353_DOCK1_chr10_129249596_ENST00000280333_length(transcript)=2070nt_BP=884nt ATGGCAAATGGTGGCGGCGGCGGCGGCGGCAGCAGCGGCGGCGGCGGCGGCGGCGGAGGCAGCAGTCTTAGAATGAGTAGCAATATCCAC GCGAACCATCTCAGCCTAGACGCGTCCTCCTCCTCCTCCTCCTCCTCTTCCTCTTCTTCTTCTTCCTCCTCCTCTTCCTCCTCGTCCTCG GTCCACGAGCCCAAGATGGATGCGCTCATCATCCCGGTGACCATGGAGGTGCCGTGCGACAGCCGGGGCCAACGCATGTGGTGGGCTTTC CTGGCCTCCTCCATGGTGACTTTCTTCGGGGGCCTCTTCATCATCTTGCTCTGGCGGACGCTCAAGTACCTGTGGACCGTGTGCTGCCAC TGCGGGGGCAAGACGAAGGAGGCCCAGAAGATTAACAATGGCTCAAGCCAGGCGGATGGCACTCTCAAACCAGTGGATGAAAAAGAGGAG GCAGTGGCCGCCGAGGTCGGCTGGATGACCTCCGTGAAGGACTGGGCGGGGGTGATGATATCCGCCCAGACACTGACTGGCAGAGTCCTG GTTGTCTTAGTCTTTGCTCTCAGCATCGGTGCACTTGTAATATACTTCATAGATTCATCAAACCCAATAGAATCCTGCCAGAATTTCTAC AAAGATTTCACATTACAGATCGACATGGCTTTCAACGTGTTCTTCCTTCTCTACTTCGGCTTGCGGTTTATTGCAGCCAACGATAAATTG TGGTTCTGGCTGGAAGTGAACTCTGTAGTGGATTTCTTCACGGTGCCCCCCGTGTTTGTGTCTGTGTACTTAAACAGAAGTTGGCTTGGT TTGAGATTTTTAAGAGCTCTGAGACTGATACAGTTTTCAGAAATTTTGCAGTTTCTGAATATTCTTAAAACAAGCATCTGCCACCTCCAC TGCCCAGCAAAACTCCGCCTCCTCCCCCTCCAAAGACAACTCGCAAGCAGGCATCGGTGGACTCCGGGATCGTGCAGTGACGTCGCAAGC CTCTCTGGAAAGAGTGTGCTGCCCCTCCCCATCTCCATGCCCTCTCCTTCTGTGTCCCCTGAGTCTGCTGTTTACCTCATTGGGCCTGTG ATGTTAACATTTCGTGCGACTGCTTTTTCTTCAAAGGAGTTCAGTTCTCACCATGGAGTGAGTGGCCTTTAGCGTCATGGAGCAAGGTGG GTCTGGGAGGTAGATATGGGTCCGGGATGTGCTATCGTAGTTATCAGAGTTGGGGGCCTCTGAGTGTGTCTGGCTCTGAGAGAGTCTGAG TCTTGCCCAAACATTCTTTCTTTTTGTGCCAAATGACTTGCATTTGCAAAGAGCTCAATTGCTCTGAGCTCAGCCAAGTAGGAGAGGCTA GGCCATCACTCTTAGGAAGCTGTGTAGTGATGATGTATAAGAATCCTCCTCACTGTCATGGGATGTTGTATCCAGCCCCTCCTTGTTCCA GCCGGTGGTGTGACTTCGTTGGTTGAGGTGTGTCTCCAACCTACATCAGACCATGAAGTTCAACCCCTCCAGGGAAGCTCCTGATTTCCC CTGCATAATTGAAAATAGGATATTTCTCAGCTATTGAACAGTTACTAATTTATGGGGTGGAAACAGCATTAAGAATACTGAATCAAATGG AAAAACAAATGAATACAGGAAGATAAGTGTTCGTTCTTTTCTGAAAAAGAGTATGTGTACCACAAGAGCTGGTTTTAATTGGGTGAATTG TTTTTGTCCTCATTCTGTACAGAAATTTGTATATATGATGGTTCTTAGAACTTGTTTTAATTTTTGTGGTCCTTCCGTTTATTATAATAG GCGTCCACCAATGATTATCCATATGTGTTCTTAATTTTTAACTGCTGGAAGTGTTAAAACACACACACACACACACACACATTTTTTTTT TGAGAACTCCAAAGTCCTGAAAATTTTGGTGGACAATGATTTTTAAAAAACTAACTTTGTATAACCTAATATTTGTATTCTCTCATCTAT ATTTTTTTATTCACTATAATCATGATACTCTTCTAATAGACTTAAAAGTTAATCATTGACAACGAAAAATAAAATGTTATTTTAAAAGAC >41484_41484_3_KCNMA1-DOCK1_KCNMA1_chr10_78880731_ENST00000354353_DOCK1_chr10_129249596_ENST00000280333_length(amino acids)=383AA_BP=294 MANGGGGGGGSSGGGGGGGGSSLRMSSNIHANHLSLDASSSSSSSSSSSSSSSSSSSSSSVHEPKMDALIIPVTMEVPCDSRGQRMWWAF LASSMVTFFGGLFIILLWRTLKYLWTVCCHCGGKTKEAQKINNGSSQADGTLKPVDEKEEAVAAEVGWMTSVKDWAGVMISAQTLTGRVL VVLVFALSIGALVIYFIDSSNPIESCQNFYKDFTLQIDMAFNVFFLLYFGLRFIAANDKLWFWLEVNSVVDFFTVPPVFVSVYLNRSWLG LRFLRALRLIQFSEILQFLNILKTSICHLHCPAKLRLLPLQRQLASRHRWTPGSCSDVASLSGKSVLPLPISMPSPSVSPESAVYLIGPV -------------------------------------------------------------- >41484_41484_4_KCNMA1-DOCK1_KCNMA1_chr10_78880731_ENST00000372440_DOCK1_chr10_129249596_ENST00000280333_length(transcript)=2421nt_BP=1235nt GGCGCAGACCCCGGGCGCGGGCGGCCCGCGGCGCCCCCTGGCTCTCGGCGCTCGGGGCCAGCGCAGCCCGGGGCGAGCGGGGCGCGGGCC GGGGAGGGGGAGCTGCCCGCGCACTCCATCCCCCTCCCCCCCGGGCGGGCGGCGGCGGCTCGCTGTATATATCTCGGCGCACCCCGCCAG GTCGCGCACAGCGCCCCGAGCCCAGGCGCCTCCCCGCCCCCCTCCCGCGCTCCGCGGCGGCGGCGGCGGCGGCAGCAGTAGCAGCAATAT GGCTGTTGATGGGTGTTTGGGGTGGCGCTGGCGGCGGGAGGAGCTCCCCCGAGCCCCTGCGCCGGCTGCCCGTTGCTAGCTATGGCAAAT GGTGGCGGCGGCGGCGGCGGCAGCAGCGGCGGCGGCGGCGGCGGCGGAGGCAGCAGTCTTAGAATGAGTAGCAATATCCACGCGAACCAT CTCAGCCTAGACGCGTCCTCCTCCTCCTCCTCCTCCTCTTCCTCTTCTTCTTCTTCCTCCTCCTCTTCCTCCTCGTCCTCGGTCCACGAG CCCAAGATGGATGCGCTCATCATCCCGGTGACCATGGAGGTGCCGTGCGACAGCCGGGGCCAACGCATGTGGTGGGCTTTCCTGGCCTCC TCCATGGTGACTTTCTTCGGGGGCCTCTTCATCATCTTGCTCTGGCGGACGCTCAAGTACCTGTGGACCGTGTGCTGCCACTGCGGGGGC AAGACGAAGGAGGCCCAGAAGATTAACAATGGCTCAAGCCAGGCGGATGGCACTCTCAAACCAGTGGATGAAAAAGAGGAGGCAGTGGCC GCCGAGGTCGGCTGGATGACCTCCGTGAAGGACTGGGCGGGGGTGATGATATCCGCCCAGACACTGACTGGCAGAGTCCTGGTTGTCTTA GTCTTTGCTCTCAGCATCGGTGCACTTGTAATATACTTCATAGATTCATCAAACCCAATAGAATCCTGCCAGAATTTCTACAAAGATTTC ACATTACAGATCGACATGGCTTTCAACGTGTTCTTCCTTCTCTACTTCGGCTTGCGGTTTATTGCAGCCAACGATAAATTGTGGTTCTGG CTGGAAGTGAACTCTGTAGTGGATTTCTTCACGGTGCCCCCCGTGTTTGTGTCTGTGTACTTAAACAGAAGTTGGCTTGGTTTGAGATTT TTAAGAGCTCTGAGACTGATACAGTTTTCAGAAATTTTGCAGTTTCTGAATATTCTTAAAACAAGCATCTGCCACCTCCACTGCCCAGCA AAACTCCGCCTCCTCCCCCTCCAAAGACAACTCGCAAGCAGGCATCGGTGGACTCCGGGATCGTGCAGTGACGTCGCAAGCCTCTCTGGA AAGAGTGTGCTGCCCCTCCCCATCTCCATGCCCTCTCCTTCTGTGTCCCCTGAGTCTGCTGTTTACCTCATTGGGCCTGTGATGTTAACA TTTCGTGCGACTGCTTTTTCTTCAAAGGAGTTCAGTTCTCACCATGGAGTGAGTGGCCTTTAGCGTCATGGAGCAAGGTGGGTCTGGGAG GTAGATATGGGTCCGGGATGTGCTATCGTAGTTATCAGAGTTGGGGGCCTCTGAGTGTGTCTGGCTCTGAGAGAGTCTGAGTCTTGCCCA AACATTCTTTCTTTTTGTGCCAAATGACTTGCATTTGCAAAGAGCTCAATTGCTCTGAGCTCAGCCAAGTAGGAGAGGCTAGGCCATCAC TCTTAGGAAGCTGTGTAGTGATGATGTATAAGAATCCTCCTCACTGTCATGGGATGTTGTATCCAGCCCCTCCTTGTTCCAGCCGGTGGT GTGACTTCGTTGGTTGAGGTGTGTCTCCAACCTACATCAGACCATGAAGTTCAACCCCTCCAGGGAAGCTCCTGATTTCCCCTGCATAAT TGAAAATAGGATATTTCTCAGCTATTGAACAGTTACTAATTTATGGGGTGGAAACAGCATTAAGAATACTGAATCAAATGGAAAAACAAA TGAATACAGGAAGATAAGTGTTCGTTCTTTTCTGAAAAAGAGTATGTGTACCACAAGAGCTGGTTTTAATTGGGTGAATTGTTTTTGTCC TCATTCTGTACAGAAATTTGTATATATGATGGTTCTTAGAACTTGTTTTAATTTTTGTGGTCCTTCCGTTTATTATAATAGGCGTCCACC AATGATTATCCATATGTGTTCTTAATTTTTAACTGCTGGAAGTGTTAAAACACACACACACACACACACACATTTTTTTTTTGAGAACTC CAAAGTCCTGAAAATTTTGGTGGACAATGATTTTTAAAAAACTAACTTTGTATAACCTAATATTTGTATTCTCTCATCTATATTTTTTTA >41484_41484_4_KCNMA1-DOCK1_KCNMA1_chr10_78880731_ENST00000372440_DOCK1_chr10_129249596_ENST00000280333_length(amino acids)=401AA_BP=312 MAAGGAPPSPCAGCPLLAMANGGGGGGGSSGGGGGGGGSSLRMSSNIHANHLSLDASSSSSSSSSSSSSSSSSSSSSSVHEPKMDALIIP VTMEVPCDSRGQRMWWAFLASSMVTFFGGLFIILLWRTLKYLWTVCCHCGGKTKEAQKINNGSSQADGTLKPVDEKEEAVAAEVGWMTSV KDWAGVMISAQTLTGRVLVVLVFALSIGALVIYFIDSSNPIESCQNFYKDFTLQIDMAFNVFFLLYFGLRFIAANDKLWFWLEVNSVVDF FTVPPVFVSVYLNRSWLGLRFLRALRLIQFSEILQFLNILKTSICHLHCPAKLRLLPLQRQLASRHRWTPGSCSDVASLSGKSVLPLPIS -------------------------------------------------------------- >41484_41484_5_KCNMA1-DOCK1_KCNMA1_chr10_78880731_ENST00000372443_DOCK1_chr10_129249596_ENST00000280333_length(transcript)=2797nt_BP=1611nt GCGAGCGGAGGCCGAGGGGCTCAGCGGCGTGGCGACAGGACTAGGGGGTCCCGAGAACATGGGCTAGCAGGCGGCCACGGGCCGGCGGCC GGACGCGGTGAACCTGCCGCGGCCGGGCGCGCACTGGCGCGGCGAGCCGGAGCCGAGCGCAGCCGGGGGAGGAGGGCTGGCGCAGGGGCG CCCGGGCGGAGGGGGCCGATTATCCATTATTAACGAGTTACCACCTAATAAGCGGAGAGCGGCTCGTTAGCCGCGCGGGAGGCGCTCGGG GAAGGGGGTCACCGGCCGAGCGCAGCGCACACCCGTGGCGCACGGCGGGCGGCGCGGCGCCGAGGACACCCGGGACCGCGCGCGGAGCCG GGGCGGGCCCGGGGGCGGCGCAGACCCCGGGCGCGGGCGGCCCGCGGCGCCCCCTGGCTCTCGGCGCTCGGGGCCAGCGCAGCCCGGGGC GAGCGGGGCGCGGGCCGGGGAGGGGGAGCTGCCCGCGCACTCCATCCCCCTCCCCCCCGGGCGGGCGGCGGCGGCTCGCTGTATATATCT CGGCGCACCCCGCCAGGTCGCGCACAGCGCCCCGAGCCCAGGCGCCTCCCCGCCCCCCTCCCGCGCTCCGCGGCGGCGGCGGCGGCGGCA GCAGTAGCAGCAATATGGCTGTTGATGGGTGTTTGGGGTGGCGCTGGCGGCGGGAGGAGCTCCCCCGAGCCCCTGCGCCGGCTGCCCGTT GCTAGCTATGGCAAATGGTGGCGGCGGCGGCGGCGGCAGCAGCGGCGGCGGCGGCGGCGGCGGAGGCAGCAGTCTTAGAATGAGTAGCAA TATCCACGCGAACCATCTCAGCCTAGACGCGTCCTCCTCCTCCTCCTCCTCCTCTTCCTCTTCTTCTTCTTCCTCCTCCTCTTCCTCCTC GTCCTCGGTCCACGAGCCCAAGATGGATGCGCTCATCATCCCGGTGACCATGGAGGTGCCGTGCGACAGCCGGGGCCAACGCATGTGGTG GGCTTTCCTGGCCTCCTCCATGGTGACTTTCTTCGGGGGCCTCTTCATCATCTTGCTCTGGCGGACGCTCAAGTACCTGTGGACCGTGTG CTGCCACTGCGGGGGCAAGACGAAGGAGGCCCAGAAGATTAACAATGGCTCAAGCCAGGCGGATGGCACTCTCAAACCAGTGGATGAAAA AGAGGAGGCAGTGGCCGCCGAGGTCGGCTGGATGACCTCCGTGAAGGACTGGGCGGGGGTGATGATATCCGCCCAGACACTGACTGGCAG AGTCCTGGTTGTCTTAGTCTTTGCTCTCAGCATCGGTGCACTTGTAATATACTTCATAGATTCATCAAACCCAATAGAATCCTGCCAGAA TTTCTACAAAGATTTCACATTACAGATCGACATGGCTTTCAACGTGTTCTTCCTTCTCTACTTCGGCTTGCGGTTTATTGCAGCCAACGA TAAATTGTGGTTCTGGCTGGAAGTGAACTCTGTAGTGGATTTCTTCACGGTGCCCCCCGTGTTTGTGTCTGTGTACTTAAACAGAAGTTG GCTTGGTTTGAGATTTTTAAGAGCTCTGAGACTGATACAGTTTTCAGAAATTTTGCAGTTTCTGAATATTCTTAAAACAAGCATCTGCCA CCTCCACTGCCCAGCAAAACTCCGCCTCCTCCCCCTCCAAAGACAACTCGCAAGCAGGCATCGGTGGACTCCGGGATCGTGCAGTGACGT CGCAAGCCTCTCTGGAAAGAGTGTGCTGCCCCTCCCCATCTCCATGCCCTCTCCTTCTGTGTCCCCTGAGTCTGCTGTTTACCTCATTGG GCCTGTGATGTTAACATTTCGTGCGACTGCTTTTTCTTCAAAGGAGTTCAGTTCTCACCATGGAGTGAGTGGCCTTTAGCGTCATGGAGC AAGGTGGGTCTGGGAGGTAGATATGGGTCCGGGATGTGCTATCGTAGTTATCAGAGTTGGGGGCCTCTGAGTGTGTCTGGCTCTGAGAGA GTCTGAGTCTTGCCCAAACATTCTTTCTTTTTGTGCCAAATGACTTGCATTTGCAAAGAGCTCAATTGCTCTGAGCTCAGCCAAGTAGGA GAGGCTAGGCCATCACTCTTAGGAAGCTGTGTAGTGATGATGTATAAGAATCCTCCTCACTGTCATGGGATGTTGTATCCAGCCCCTCCT TGTTCCAGCCGGTGGTGTGACTTCGTTGGTTGAGGTGTGTCTCCAACCTACATCAGACCATGAAGTTCAACCCCTCCAGGGAAGCTCCTG ATTTCCCCTGCATAATTGAAAATAGGATATTTCTCAGCTATTGAACAGTTACTAATTTATGGGGTGGAAACAGCATTAAGAATACTGAAT CAAATGGAAAAACAAATGAATACAGGAAGATAAGTGTTCGTTCTTTTCTGAAAAAGAGTATGTGTACCACAAGAGCTGGTTTTAATTGGG TGAATTGTTTTTGTCCTCATTCTGTACAGAAATTTGTATATATGATGGTTCTTAGAACTTGTTTTAATTTTTGTGGTCCTTCCGTTTATT ATAATAGGCGTCCACCAATGATTATCCATATGTGTTCTTAATTTTTAACTGCTGGAAGTGTTAAAACACACACACACACACACACACATT TTTTTTTTGAGAACTCCAAAGTCCTGAAAATTTTGGTGGACAATGATTTTTAAAAAACTAACTTTGTATAACCTAATATTTGTATTCTCT CATCTATATTTTTTTATTCACTATAATCATGATACTCTTCTAATAGACTTAAAAGTTAATCATTGACAACGAAAAATAAAATGTTATTTT >41484_41484_5_KCNMA1-DOCK1_KCNMA1_chr10_78880731_ENST00000372443_DOCK1_chr10_129249596_ENST00000280333_length(amino acids)=401AA_BP=312 MAAGGAPPSPCAGCPLLAMANGGGGGGGSSGGGGGGGGSSLRMSSNIHANHLSLDASSSSSSSSSSSSSSSSSSSSSSVHEPKMDALIIP VTMEVPCDSRGQRMWWAFLASSMVTFFGGLFIILLWRTLKYLWTVCCHCGGKTKEAQKINNGSSQADGTLKPVDEKEEAVAAEVGWMTSV KDWAGVMISAQTLTGRVLVVLVFALSIGALVIYFIDSSNPIESCQNFYKDFTLQIDMAFNVFFLLYFGLRFIAANDKLWFWLEVNSVVDF FTVPPVFVSVYLNRSWLGLRFLRALRLIQFSEILQFLNILKTSICHLHCPAKLRLLPLQRQLASRHRWTPGSCSDVASLSGKSVLPLPIS -------------------------------------------------------------- >41484_41484_6_KCNMA1-DOCK1_KCNMA1_chr10_78880731_ENST00000404771_DOCK1_chr10_129249596_ENST00000280333_length(transcript)=2186nt_BP=1000nt GGCGGCGGCGGCGGCGGCAGCAGTAGCAGCAATATGGCTGTTGATGGGTGTTTGGGGTGGCGCTGGCGGCGGGAGGAGCTCCCCCGAGCC CCTGCGCCGGCTGCCCGTTGCTAGCTATGGCAAATGGTGGCGGCGGCGGCGGCGGCAGCAGCGGCGGCGGCGGCGGCGGCGGAGGCAGCA GTCTTAGAATGAGTAGCAATATCCACGCGAACCATCTCAGCCTAGACGCGTCCTCCTCCTCCTCCTCCTCCTCTTCCTCTTCTTCTTCTT CCTCCTCCTCTTCCTCCTCGTCCTCGGTCCACGAGCCCAAGATGGATGCGCTCATCATCCCGGTGACCATGGAGGTGCCGTGCGACAGCC GGGGCCAACGCATGTGGTGGGCTTTCCTGGCCTCCTCCATGGTGACTTTCTTCGGGGGCCTCTTCATCATCTTGCTCTGGCGGACGCTCA AGTACCTGTGGACCGTGTGCTGCCACTGCGGGGGCAAGACGAAGGAGGCCCAGAAGATTAACAATGGCTCAAGCCAGGCGGATGGCACTC TCAAACCAGTGGATGAAAAAGAGGAGGCAGTGGCCGCCGAGGTCGGCTGGATGACCTCCGTGAAGGACTGGGCGGGGGTGATGATATCCG CCCAGACACTGACTGGCAGAGTCCTGGTTGTCTTAGTCTTTGCTCTCAGCATCGGTGCACTTGTAATATACTTCATAGATTCATCAAACC CAATAGAATCCTGCCAGAATTTCTACAAAGATTTCACATTACAGATCGACATGGCTTTCAACGTGTTCTTCCTTCTCTACTTCGGCTTGC GGTTTATTGCAGCCAACGATAAATTGTGGTTCTGGCTGGAAGTGAACTCTGTAGTGGATTTCTTCACGGTGCCCCCCGTGTTTGTGTCTG TGTACTTAAACAGAAGTTGGCTTGGTTTGAGATTTTTAAGAGCTCTGAGACTGATACAGTTTTCAGAAATTTTGCAGTTTCTGAATATTC TTAAAACAAGCATCTGCCACCTCCACTGCCCAGCAAAACTCCGCCTCCTCCCCCTCCAAAGACAACTCGCAAGCAGGCATCGGTGGACTC CGGGATCGTGCAGTGACGTCGCAAGCCTCTCTGGAAAGAGTGTGCTGCCCCTCCCCATCTCCATGCCCTCTCCTTCTGTGTCCCCTGAGT CTGCTGTTTACCTCATTGGGCCTGTGATGTTAACATTTCGTGCGACTGCTTTTTCTTCAAAGGAGTTCAGTTCTCACCATGGAGTGAGTG GCCTTTAGCGTCATGGAGCAAGGTGGGTCTGGGAGGTAGATATGGGTCCGGGATGTGCTATCGTAGTTATCAGAGTTGGGGGCCTCTGAG TGTGTCTGGCTCTGAGAGAGTCTGAGTCTTGCCCAAACATTCTTTCTTTTTGTGCCAAATGACTTGCATTTGCAAAGAGCTCAATTGCTC TGAGCTCAGCCAAGTAGGAGAGGCTAGGCCATCACTCTTAGGAAGCTGTGTAGTGATGATGTATAAGAATCCTCCTCACTGTCATGGGAT GTTGTATCCAGCCCCTCCTTGTTCCAGCCGGTGGTGTGACTTCGTTGGTTGAGGTGTGTCTCCAACCTACATCAGACCATGAAGTTCAAC CCCTCCAGGGAAGCTCCTGATTTCCCCTGCATAATTGAAAATAGGATATTTCTCAGCTATTGAACAGTTACTAATTTATGGGGTGGAAAC AGCATTAAGAATACTGAATCAAATGGAAAAACAAATGAATACAGGAAGATAAGTGTTCGTTCTTTTCTGAAAAAGAGTATGTGTACCACA AGAGCTGGTTTTAATTGGGTGAATTGTTTTTGTCCTCATTCTGTACAGAAATTTGTATATATGATGGTTCTTAGAACTTGTTTTAATTTT TGTGGTCCTTCCGTTTATTATAATAGGCGTCCACCAATGATTATCCATATGTGTTCTTAATTTTTAACTGCTGGAAGTGTTAAAACACAC ACACACACACACACACATTTTTTTTTTGAGAACTCCAAAGTCCTGAAAATTTTGGTGGACAATGATTTTTAAAAAACTAACTTTGTATAA CCTAATATTTGTATTCTCTCATCTATATTTTTTTATTCACTATAATCATGATACTCTTCTAATAGACTTAAAAGTTAATCATTGACAACG >41484_41484_6_KCNMA1-DOCK1_KCNMA1_chr10_78880731_ENST00000404771_DOCK1_chr10_129249596_ENST00000280333_length(amino acids)=401AA_BP=312 MAAGGAPPSPCAGCPLLAMANGGGGGGGSSGGGGGGGGSSLRMSSNIHANHLSLDASSSSSSSSSSSSSSSSSSSSSSVHEPKMDALIIP VTMEVPCDSRGQRMWWAFLASSMVTFFGGLFIILLWRTLKYLWTVCCHCGGKTKEAQKINNGSSQADGTLKPVDEKEEAVAAEVGWMTSV KDWAGVMISAQTLTGRVLVVLVFALSIGALVIYFIDSSNPIESCQNFYKDFTLQIDMAFNVFFLLYFGLRFIAANDKLWFWLEVNSVVDF FTVPPVFVSVYLNRSWLGLRFLRALRLIQFSEILQFLNILKTSICHLHCPAKLRLLPLQRQLASRHRWTPGSCSDVASLSGKSVLPLPIS -------------------------------------------------------------- >41484_41484_7_KCNMA1-DOCK1_KCNMA1_chr10_78880731_ENST00000404857_DOCK1_chr10_129249596_ENST00000280333_length(transcript)=2070nt_BP=884nt ATGGCAAATGGTGGCGGCGGCGGCGGCGGCAGCAGCGGCGGCGGCGGCGGCGGCGGAGGCAGCAGTCTTAGAATGAGTAGCAATATCCAC GCGAACCATCTCAGCCTAGACGCGTCCTCCTCCTCCTCCTCCTCCTCTTCCTCTTCTTCTTCTTCCTCCTCCTCTTCCTCCTCGTCCTCG GTCCACGAGCCCAAGATGGATGCGCTCATCATCCCGGTGACCATGGAGGTGCCGTGCGACAGCCGGGGCCAACGCATGTGGTGGGCTTTC CTGGCCTCCTCCATGGTGACTTTCTTCGGGGGCCTCTTCATCATCTTGCTCTGGCGGACGCTCAAGTACCTGTGGACCGTGTGCTGCCAC TGCGGGGGCAAGACGAAGGAGGCCCAGAAGATTAACAATGGCTCAAGCCAGGCGGATGGCACTCTCAAACCAGTGGATGAAAAAGAGGAG GCAGTGGCCGCCGAGGTCGGCTGGATGACCTCCGTGAAGGACTGGGCGGGGGTGATGATATCCGCCCAGACACTGACTGGCAGAGTCCTG GTTGTCTTAGTCTTTGCTCTCAGCATCGGTGCACTTGTAATATACTTCATAGATTCATCAAACCCAATAGAATCCTGCCAGAATTTCTAC AAAGATTTCACATTACAGATCGACATGGCTTTCAACGTGTTCTTCCTTCTCTACTTCGGCTTGCGGTTTATTGCAGCCAACGATAAATTG TGGTTCTGGCTGGAAGTGAACTCTGTAGTGGATTTCTTCACGGTGCCCCCCGTGTTTGTGTCTGTGTACTTAAACAGAAGTTGGCTTGGT TTGAGATTTTTAAGAGCTCTGAGACTGATACAGTTTTCAGAAATTTTGCAGTTTCTGAATATTCTTAAAACAAGCATCTGCCACCTCCAC TGCCCAGCAAAACTCCGCCTCCTCCCCCTCCAAAGACAACTCGCAAGCAGGCATCGGTGGACTCCGGGATCGTGCAGTGACGTCGCAAGC CTCTCTGGAAAGAGTGTGCTGCCCCTCCCCATCTCCATGCCCTCTCCTTCTGTGTCCCCTGAGTCTGCTGTTTACCTCATTGGGCCTGTG ATGTTAACATTTCGTGCGACTGCTTTTTCTTCAAAGGAGTTCAGTTCTCACCATGGAGTGAGTGGCCTTTAGCGTCATGGAGCAAGGTGG GTCTGGGAGGTAGATATGGGTCCGGGATGTGCTATCGTAGTTATCAGAGTTGGGGGCCTCTGAGTGTGTCTGGCTCTGAGAGAGTCTGAG TCTTGCCCAAACATTCTTTCTTTTTGTGCCAAATGACTTGCATTTGCAAAGAGCTCAATTGCTCTGAGCTCAGCCAAGTAGGAGAGGCTA GGCCATCACTCTTAGGAAGCTGTGTAGTGATGATGTATAAGAATCCTCCTCACTGTCATGGGATGTTGTATCCAGCCCCTCCTTGTTCCA GCCGGTGGTGTGACTTCGTTGGTTGAGGTGTGTCTCCAACCTACATCAGACCATGAAGTTCAACCCCTCCAGGGAAGCTCCTGATTTCCC CTGCATAATTGAAAATAGGATATTTCTCAGCTATTGAACAGTTACTAATTTATGGGGTGGAAACAGCATTAAGAATACTGAATCAAATGG AAAAACAAATGAATACAGGAAGATAAGTGTTCGTTCTTTTCTGAAAAAGAGTATGTGTACCACAAGAGCTGGTTTTAATTGGGTGAATTG TTTTTGTCCTCATTCTGTACAGAAATTTGTATATATGATGGTTCTTAGAACTTGTTTTAATTTTTGTGGTCCTTCCGTTTATTATAATAG GCGTCCACCAATGATTATCCATATGTGTTCTTAATTTTTAACTGCTGGAAGTGTTAAAACACACACACACACACACACACATTTTTTTTT TGAGAACTCCAAAGTCCTGAAAATTTTGGTGGACAATGATTTTTAAAAAACTAACTTTGTATAACCTAATATTTGTATTCTCTCATCTAT ATTTTTTTATTCACTATAATCATGATACTCTTCTAATAGACTTAAAAGTTAATCATTGACAACGAAAAATAAAATGTTATTTTAAAAGAC >41484_41484_7_KCNMA1-DOCK1_KCNMA1_chr10_78880731_ENST00000404857_DOCK1_chr10_129249596_ENST00000280333_length(amino acids)=383AA_BP=294 MANGGGGGGGSSGGGGGGGGSSLRMSSNIHANHLSLDASSSSSSSSSSSSSSSSSSSSSSVHEPKMDALIIPVTMEVPCDSRGQRMWWAF LASSMVTFFGGLFIILLWRTLKYLWTVCCHCGGKTKEAQKINNGSSQADGTLKPVDEKEEAVAAEVGWMTSVKDWAGVMISAQTLTGRVL VVLVFALSIGALVIYFIDSSNPIESCQNFYKDFTLQIDMAFNVFFLLYFGLRFIAANDKLWFWLEVNSVVDFFTVPPVFVSVYLNRSWLG LRFLRALRLIQFSEILQFLNILKTSICHLHCPAKLRLLPLQRQLASRHRWTPGSCSDVASLSGKSVLPLPISMPSPSVSPESAVYLIGPV -------------------------------------------------------------- >41484_41484_8_KCNMA1-DOCK1_KCNMA1_chr10_78880731_ENST00000406533_DOCK1_chr10_129249596_ENST00000280333_length(transcript)=2070nt_BP=884nt ATGGCAAATGGTGGCGGCGGCGGCGGCGGCAGCAGCGGCGGCGGCGGCGGCGGCGGAGGCAGCAGTCTTAGAATGAGTAGCAATATCCAC GCGAACCATCTCAGCCTAGACGCGTCCTCCTCCTCCTCCTCCTCCTCTTCCTCTTCTTCTTCTTCCTCCTCCTCTTCCTCCTCGTCCTCG GTCCACGAGCCCAAGATGGATGCGCTCATCATCCCGGTGACCATGGAGGTGCCGTGCGACAGCCGGGGCCAACGCATGTGGTGGGCTTTC CTGGCCTCCTCCATGGTGACTTTCTTCGGGGGCCTCTTCATCATCTTGCTCTGGCGGACGCTCAAGTACCTGTGGACCGTGTGCTGCCAC TGCGGGGGCAAGACGAAGGAGGCCCAGAAGATTAACAATGGCTCAAGCCAGGCGGATGGCACTCTCAAACCAGTGGATGAAAAAGAGGAG GCAGTGGCCGCCGAGGTCGGCTGGATGACCTCCGTGAAGGACTGGGCGGGGGTGATGATATCCGCCCAGACACTGACTGGCAGAGTCCTG GTTGTCTTAGTCTTTGCTCTCAGCATCGGTGCACTTGTAATATACTTCATAGATTCATCAAACCCAATAGAATCCTGCCAGAATTTCTAC AAAGATTTCACATTACAGATCGACATGGCTTTCAACGTGTTCTTCCTTCTCTACTTCGGCTTGCGGTTTATTGCAGCCAACGATAAATTG TGGTTCTGGCTGGAAGTGAACTCTGTAGTGGATTTCTTCACGGTGCCCCCCGTGTTTGTGTCTGTGTACTTAAACAGAAGTTGGCTTGGT TTGAGATTTTTAAGAGCTCTGAGACTGATACAGTTTTCAGAAATTTTGCAGTTTCTGAATATTCTTAAAACAAGCATCTGCCACCTCCAC TGCCCAGCAAAACTCCGCCTCCTCCCCCTCCAAAGACAACTCGCAAGCAGGCATCGGTGGACTCCGGGATCGTGCAGTGACGTCGCAAGC CTCTCTGGAAAGAGTGTGCTGCCCCTCCCCATCTCCATGCCCTCTCCTTCTGTGTCCCCTGAGTCTGCTGTTTACCTCATTGGGCCTGTG ATGTTAACATTTCGTGCGACTGCTTTTTCTTCAAAGGAGTTCAGTTCTCACCATGGAGTGAGTGGCCTTTAGCGTCATGGAGCAAGGTGG GTCTGGGAGGTAGATATGGGTCCGGGATGTGCTATCGTAGTTATCAGAGTTGGGGGCCTCTGAGTGTGTCTGGCTCTGAGAGAGTCTGAG TCTTGCCCAAACATTCTTTCTTTTTGTGCCAAATGACTTGCATTTGCAAAGAGCTCAATTGCTCTGAGCTCAGCCAAGTAGGAGAGGCTA GGCCATCACTCTTAGGAAGCTGTGTAGTGATGATGTATAAGAATCCTCCTCACTGTCATGGGATGTTGTATCCAGCCCCTCCTTGTTCCA GCCGGTGGTGTGACTTCGTTGGTTGAGGTGTGTCTCCAACCTACATCAGACCATGAAGTTCAACCCCTCCAGGGAAGCTCCTGATTTCCC CTGCATAATTGAAAATAGGATATTTCTCAGCTATTGAACAGTTACTAATTTATGGGGTGGAAACAGCATTAAGAATACTGAATCAAATGG AAAAACAAATGAATACAGGAAGATAAGTGTTCGTTCTTTTCTGAAAAAGAGTATGTGTACCACAAGAGCTGGTTTTAATTGGGTGAATTG TTTTTGTCCTCATTCTGTACAGAAATTTGTATATATGATGGTTCTTAGAACTTGTTTTAATTTTTGTGGTCCTTCCGTTTATTATAATAG GCGTCCACCAATGATTATCCATATGTGTTCTTAATTTTTAACTGCTGGAAGTGTTAAAACACACACACACACACACACACATTTTTTTTT TGAGAACTCCAAAGTCCTGAAAATTTTGGTGGACAATGATTTTTAAAAAACTAACTTTGTATAACCTAATATTTGTATTCTCTCATCTAT ATTTTTTTATTCACTATAATCATGATACTCTTCTAATAGACTTAAAAGTTAATCATTGACAACGAAAAATAAAATGTTATTTTAAAAGAC >41484_41484_8_KCNMA1-DOCK1_KCNMA1_chr10_78880731_ENST00000406533_DOCK1_chr10_129249596_ENST00000280333_length(amino acids)=383AA_BP=294 MANGGGGGGGSSGGGGGGGGSSLRMSSNIHANHLSLDASSSSSSSSSSSSSSSSSSSSSSVHEPKMDALIIPVTMEVPCDSRGQRMWWAF LASSMVTFFGGLFIILLWRTLKYLWTVCCHCGGKTKEAQKINNGSSQADGTLKPVDEKEEAVAAEVGWMTSVKDWAGVMISAQTLTGRVL VVLVFALSIGALVIYFIDSSNPIESCQNFYKDFTLQIDMAFNVFFLLYFGLRFIAANDKLWFWLEVNSVVDFFTVPPVFVSVYLNRSWLG LRFLRALRLIQFSEILQFLNILKTSICHLHCPAKLRLLPLQRQLASRHRWTPGSCSDVASLSGKSVLPLPISMPSPSVSPESAVYLIGPV -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for KCNMA1-DOCK1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | DOCK1 | chr10:78880731 | chr10:129249596 | ENST00000280333 | 50 | 52 | 1837_1852 | 1834.0 | 1866.0 | NCK2 (minor) |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Tgene | DOCK1 | chr10:78880731 | chr10:129249596 | ENST00000280333 | 50 | 52 | 1793_1819 | 1834.0 | 1866.0 | NCK2 second and third SH3 domain (minor) | |
| Tgene | DOCK1 | chr10:78880731 | chr10:129249596 | ENST00000280333 | 50 | 52 | 1820_1836 | 1834.0 | 1866.0 | NCK2 third SH3 domain (major) |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for KCNMA1-DOCK1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for KCNMA1-DOCK1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
