
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:KCNQ3-NDRG1 (FusionGDB2 ID:41565) |
Fusion Gene Summary for KCNQ3-NDRG1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: KCNQ3-NDRG1 | Fusion gene ID: 41565 | Hgene | Tgene | Gene symbol | KCNQ3 | NDRG1 | Gene ID | 3786 | 10397 |
| Gene name | potassium voltage-gated channel subfamily Q member 3 | N-myc downstream regulated 1 | |
| Synonyms | BFNC2|EBN2|KV7.3 | CAP43|CMT4D|DRG-1|DRG1|GC4|HMSNL|NDR1|NMSL|PROXY1|RIT42|RTP|TARG1|TDD5 | |
| Cytomap | 8q24.22 | 8q24.22 | |
| Type of gene | protein-coding | protein-coding | |
| Description | potassium voltage-gated channel subfamily KQT member 3potassium channel subunit alpha KvLQT3potassium channel, voltage gated KQT-like subfamily Q, member 3potassium channel, voltage-gated, subfamily Q, member 3potassium voltage-gated channel, KQT-like | protein NDRG1N-myc downstream-regulated gene 1 proteindifferentiation-related gene 1 proteinnickel-specific induction protein Cap43protein regulated by oxygen-1reducing agents and tunicamycin-responsive protein | |
| Modification date | 20200313 | 20200328 | |
| UniProtAcc | . | Q92597 | |
| Ensembl transtripts involved in fusion gene | ENST00000388996, ENST00000519445, ENST00000521134, | ENST00000522476, ENST00000414097, ENST00000518066, ENST00000518176, ENST00000521414, ENST00000537882, ENST00000323851, ENST00000354944, | |
| Fusion gene scores | * DoF score | 13 X 10 X 5=650 | 24 X 17 X 10=4080 |
| # samples | 14 | 26 | |
| ** MAII score | log2(14/650*10)=-2.21501289097085 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(26/4080*10)=-3.9719856238304 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: KCNQ3 [Title/Abstract] AND NDRG1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | KCNQ3(133492393)-NDRG1(134276895), # samples:1 KCNQ3(133310284)-NDRG1(134266781), # samples:1 KCNQ3(133310371)-NDRG1(134266781), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | KCNQ3 | GO:0071805 | potassium ion transmembrane transport | 11159685|27564677 |
 Fusion gene breakpoints across KCNQ3 (5'-gene) Fusion gene breakpoints across KCNQ3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
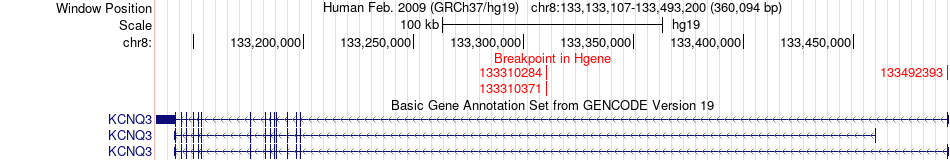 |
 Fusion gene breakpoints across NDRG1 (3'-gene) Fusion gene breakpoints across NDRG1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
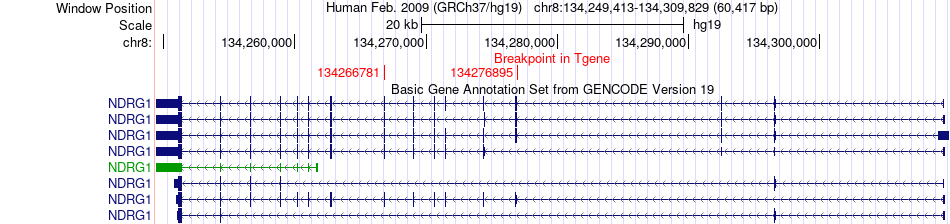 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-AC-A7VB-01A | KCNQ3 | chr8 | 133310284 | - | NDRG1 | chr8 | 134266781 | + |
| ChimerDB4 | BRCA | TCGA-AC-A7VB-01A | KCNQ3 | chr8 | 133310371 | - | NDRG1 | chr8 | 134266781 | + |
| ChimerDB4 | OV | TCGA-23-1023 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - |
Top |
Fusion Gene ORF analysis for KCNQ3-NDRG1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000388996 | ENST00000522476 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - |
| 5CDS-5UTR | ENST00000519445 | ENST00000522476 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - |
| 5CDS-intron | ENST00000388996 | ENST00000414097 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - |
| 5CDS-intron | ENST00000388996 | ENST00000518066 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - |
| 5CDS-intron | ENST00000388996 | ENST00000518176 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - |
| 5CDS-intron | ENST00000388996 | ENST00000521414 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - |
| 5CDS-intron | ENST00000388996 | ENST00000537882 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - |
| 5CDS-intron | ENST00000519445 | ENST00000414097 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - |
| 5CDS-intron | ENST00000519445 | ENST00000518066 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - |
| 5CDS-intron | ENST00000519445 | ENST00000518176 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - |
| 5CDS-intron | ENST00000519445 | ENST00000521414 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - |
| 5CDS-intron | ENST00000519445 | ENST00000537882 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - |
| In-frame | ENST00000388996 | ENST00000323851 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - |
| In-frame | ENST00000388996 | ENST00000354944 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - |
| In-frame | ENST00000519445 | ENST00000323851 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - |
| In-frame | ENST00000519445 | ENST00000354944 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - |
| intron-3CDS | ENST00000521134 | ENST00000323851 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - |
| intron-3CDS | ENST00000521134 | ENST00000354944 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - |
| intron-5UTR | ENST00000521134 | ENST00000522476 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - |
| intron-intron | ENST00000388996 | ENST00000323851 | KCNQ3 | chr8 | 133310284 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000388996 | ENST00000323851 | KCNQ3 | chr8 | 133310371 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000388996 | ENST00000354944 | KCNQ3 | chr8 | 133310284 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000388996 | ENST00000354944 | KCNQ3 | chr8 | 133310371 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000388996 | ENST00000414097 | KCNQ3 | chr8 | 133310284 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000388996 | ENST00000414097 | KCNQ3 | chr8 | 133310371 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000388996 | ENST00000518066 | KCNQ3 | chr8 | 133310284 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000388996 | ENST00000518066 | KCNQ3 | chr8 | 133310371 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000388996 | ENST00000518176 | KCNQ3 | chr8 | 133310284 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000388996 | ENST00000518176 | KCNQ3 | chr8 | 133310371 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000388996 | ENST00000521414 | KCNQ3 | chr8 | 133310284 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000388996 | ENST00000521414 | KCNQ3 | chr8 | 133310371 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000388996 | ENST00000522476 | KCNQ3 | chr8 | 133310284 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000388996 | ENST00000522476 | KCNQ3 | chr8 | 133310371 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000388996 | ENST00000537882 | KCNQ3 | chr8 | 133310284 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000388996 | ENST00000537882 | KCNQ3 | chr8 | 133310371 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000519445 | ENST00000323851 | KCNQ3 | chr8 | 133310284 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000519445 | ENST00000323851 | KCNQ3 | chr8 | 133310371 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000519445 | ENST00000354944 | KCNQ3 | chr8 | 133310284 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000519445 | ENST00000354944 | KCNQ3 | chr8 | 133310371 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000519445 | ENST00000414097 | KCNQ3 | chr8 | 133310284 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000519445 | ENST00000414097 | KCNQ3 | chr8 | 133310371 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000519445 | ENST00000518066 | KCNQ3 | chr8 | 133310284 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000519445 | ENST00000518066 | KCNQ3 | chr8 | 133310371 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000519445 | ENST00000518176 | KCNQ3 | chr8 | 133310284 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000519445 | ENST00000518176 | KCNQ3 | chr8 | 133310371 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000519445 | ENST00000521414 | KCNQ3 | chr8 | 133310284 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000519445 | ENST00000521414 | KCNQ3 | chr8 | 133310371 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000519445 | ENST00000522476 | KCNQ3 | chr8 | 133310284 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000519445 | ENST00000522476 | KCNQ3 | chr8 | 133310371 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000519445 | ENST00000537882 | KCNQ3 | chr8 | 133310284 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000519445 | ENST00000537882 | KCNQ3 | chr8 | 133310371 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000521134 | ENST00000323851 | KCNQ3 | chr8 | 133310284 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000521134 | ENST00000323851 | KCNQ3 | chr8 | 133310371 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000521134 | ENST00000354944 | KCNQ3 | chr8 | 133310284 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000521134 | ENST00000354944 | KCNQ3 | chr8 | 133310371 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000521134 | ENST00000414097 | KCNQ3 | chr8 | 133310284 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000521134 | ENST00000414097 | KCNQ3 | chr8 | 133310371 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000521134 | ENST00000414097 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - |
| intron-intron | ENST00000521134 | ENST00000518066 | KCNQ3 | chr8 | 133310284 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000521134 | ENST00000518066 | KCNQ3 | chr8 | 133310371 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000521134 | ENST00000518066 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - |
| intron-intron | ENST00000521134 | ENST00000518176 | KCNQ3 | chr8 | 133310284 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000521134 | ENST00000518176 | KCNQ3 | chr8 | 133310371 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000521134 | ENST00000518176 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - |
| intron-intron | ENST00000521134 | ENST00000521414 | KCNQ3 | chr8 | 133310284 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000521134 | ENST00000521414 | KCNQ3 | chr8 | 133310371 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000521134 | ENST00000521414 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - |
| intron-intron | ENST00000521134 | ENST00000522476 | KCNQ3 | chr8 | 133310284 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000521134 | ENST00000522476 | KCNQ3 | chr8 | 133310371 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000521134 | ENST00000537882 | KCNQ3 | chr8 | 133310284 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000521134 | ENST00000537882 | KCNQ3 | chr8 | 133310371 | - | NDRG1 | chr8 | 134266781 | + |
| intron-intron | ENST00000521134 | ENST00000537882 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000388996 | KCNQ3 | chr8 | 133492393 | - | ENST00000323851 | NDRG1 | chr8 | 134276895 | - | 3600 | 807 | 780 | 1892 | 370 |
| ENST00000388996 | KCNQ3 | chr8 | 133492393 | - | ENST00000354944 | NDRG1 | chr8 | 134276895 | - | 3389 | 807 | 780 | 1682 | 300 |
| ENST00000519445 | KCNQ3 | chr8 | 133492393 | - | ENST00000323851 | NDRG1 | chr8 | 134276895 | - | 3225 | 432 | 405 | 1517 | 370 |
| ENST00000519445 | KCNQ3 | chr8 | 133492393 | - | ENST00000354944 | NDRG1 | chr8 | 134276895 | - | 3014 | 432 | 405 | 1307 | 300 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000388996 | ENST00000323851 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - | 0.004114726 | 0.9958853 |
| ENST00000388996 | ENST00000354944 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - | 0.03660723 | 0.9633928 |
| ENST00000519445 | ENST00000323851 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - | 0.003552263 | 0.9964477 |
| ENST00000519445 | ENST00000354944 | KCNQ3 | chr8 | 133492393 | - | NDRG1 | chr8 | 134276895 | - | 0.033578716 | 0.96642125 |
Top |
Fusion Genomic Features for KCNQ3-NDRG1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
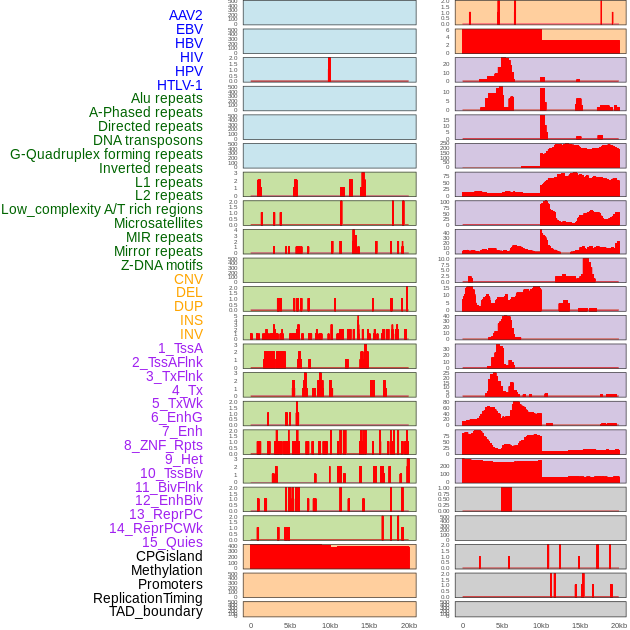 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for KCNQ3-NDRG1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr8:133492393/chr8:134276895) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | NDRG1 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Stress-responsive protein involved in hormone responses, cell growth, and differentiation. Acts as a tumor suppressor in many cell types. Necessary but not sufficient for p53/TP53-mediated caspase activation and apoptosis. Has a role in cell trafficking, notably of the Schwann cell, and is necessary for the maintenance and development of the peripheral nerve myelin sheath. Required for vesicular recycling of CDH1 and TF. May also function in lipid trafficking. Protects cells from spindle disruption damage. Functions in p53/TP53-dependent mitotic spindle checkpoint. Regulates microtubule dynamics and maintains euploidy. {ECO:0000269|PubMed:15247272, ECO:0000269|PubMed:15377670, ECO:0000269|PubMed:17786215, ECO:0000269|PubMed:9766676}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000388996 | - | 1 | 15 | 13_24 | 128 | 873.0 | Compositional bias | Note=Poly-Gly |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000388996 | - | 1 | 15 | 1_121 | 128 | 873.0 | Topological domain | Cytoplasmic |
| Tgene | NDRG1 | chr8:133492393 | chr8:134276895 | ENST00000323851 | 2 | 16 | 339_368 | 33 | 395.0 | Region | Note=3 X 10 AA tandem repeats of G-T-R-S-R-S-H-T-S-E | |
| Tgene | NDRG1 | chr8:133492393 | chr8:134276895 | ENST00000414097 | 2 | 16 | 339_368 | 33 | 395.0 | Region | Note=3 X 10 AA tandem repeats of G-T-R-S-R-S-H-T-S-E | |
| Tgene | NDRG1 | chr8:133492393 | chr8:134276895 | ENST00000522476 | 0 | 14 | 339_368 | 0 | 329.0 | Region | Note=3 X 10 AA tandem repeats of G-T-R-S-R-S-H-T-S-E | |
| Tgene | NDRG1 | chr8:133492393 | chr8:134276895 | ENST00000537882 | 0 | 15 | 339_368 | 0 | 314.0 | Region | Note=3 X 10 AA tandem repeats of G-T-R-S-R-S-H-T-S-E | |
| Tgene | NDRG1 | chr8:133492393 | chr8:134276895 | ENST00000323851 | 2 | 16 | 339_348 | 33 | 395.0 | Repeat | Note=1 | |
| Tgene | NDRG1 | chr8:133492393 | chr8:134276895 | ENST00000323851 | 2 | 16 | 349_358 | 33 | 395.0 | Repeat | Note=2 | |
| Tgene | NDRG1 | chr8:133492393 | chr8:134276895 | ENST00000323851 | 2 | 16 | 359_368 | 33 | 395.0 | Repeat | Note=3 | |
| Tgene | NDRG1 | chr8:133492393 | chr8:134276895 | ENST00000414097 | 2 | 16 | 339_348 | 33 | 395.0 | Repeat | Note=1 | |
| Tgene | NDRG1 | chr8:133492393 | chr8:134276895 | ENST00000414097 | 2 | 16 | 349_358 | 33 | 395.0 | Repeat | Note=2 | |
| Tgene | NDRG1 | chr8:133492393 | chr8:134276895 | ENST00000414097 | 2 | 16 | 359_368 | 33 | 395.0 | Repeat | Note=3 | |
| Tgene | NDRG1 | chr8:133492393 | chr8:134276895 | ENST00000522476 | 0 | 14 | 339_348 | 0 | 329.0 | Repeat | Note=1 | |
| Tgene | NDRG1 | chr8:133492393 | chr8:134276895 | ENST00000522476 | 0 | 14 | 349_358 | 0 | 329.0 | Repeat | Note=2 | |
| Tgene | NDRG1 | chr8:133492393 | chr8:134276895 | ENST00000522476 | 0 | 14 | 359_368 | 0 | 329.0 | Repeat | Note=3 | |
| Tgene | NDRG1 | chr8:133492393 | chr8:134276895 | ENST00000537882 | 0 | 15 | 339_348 | 0 | 314.0 | Repeat | Note=1 | |
| Tgene | NDRG1 | chr8:133492393 | chr8:134276895 | ENST00000537882 | 0 | 15 | 349_358 | 0 | 314.0 | Repeat | Note=2 | |
| Tgene | NDRG1 | chr8:133492393 | chr8:134276895 | ENST00000537882 | 0 | 15 | 359_368 | 0 | 314.0 | Repeat | Note=3 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000521134 | - | 1 | 15 | 13_24 | 0 | 753.0 | Compositional bias | Note=Poly-Gly |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000388996 | - | 1 | 15 | 304_324 | 128 | 873.0 | Intramembrane | Pore-forming%3B Name%3DSegment H5 |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000521134 | - | 1 | 15 | 304_324 | 0 | 753.0 | Intramembrane | Pore-forming%3B Name%3DSegment H5 |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000388996 | - | 1 | 15 | 316_321 | 128 | 873.0 | Motif | Selectivity filter |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000521134 | - | 1 | 15 | 316_321 | 0 | 753.0 | Motif | Selectivity filter |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000388996 | - | 1 | 15 | 143_152 | 128 | 873.0 | Topological domain | Extracellular |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000388996 | - | 1 | 15 | 174_196 | 128 | 873.0 | Topological domain | Cytoplasmic |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000388996 | - | 1 | 15 | 218_225 | 128 | 873.0 | Topological domain | Extracellular |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000388996 | - | 1 | 15 | 248_261 | 128 | 873.0 | Topological domain | Cytoplasmic |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000388996 | - | 1 | 15 | 283_303 | 128 | 873.0 | Topological domain | Extracellular |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000388996 | - | 1 | 15 | 325_330 | 128 | 873.0 | Topological domain | Extracellular |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000388996 | - | 1 | 15 | 352_872 | 128 | 873.0 | Topological domain | Cytoplasmic |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000521134 | - | 1 | 15 | 143_152 | 0 | 753.0 | Topological domain | Extracellular |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000521134 | - | 1 | 15 | 174_196 | 0 | 753.0 | Topological domain | Cytoplasmic |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000521134 | - | 1 | 15 | 1_121 | 0 | 753.0 | Topological domain | Cytoplasmic |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000521134 | - | 1 | 15 | 218_225 | 0 | 753.0 | Topological domain | Extracellular |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000521134 | - | 1 | 15 | 248_261 | 0 | 753.0 | Topological domain | Cytoplasmic |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000521134 | - | 1 | 15 | 283_303 | 0 | 753.0 | Topological domain | Extracellular |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000521134 | - | 1 | 15 | 325_330 | 0 | 753.0 | Topological domain | Extracellular |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000521134 | - | 1 | 15 | 352_872 | 0 | 753.0 | Topological domain | Cytoplasmic |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000388996 | - | 1 | 15 | 122_142 | 128 | 873.0 | Transmembrane | Helical%3B Name%3DSegment S1 |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000388996 | - | 1 | 15 | 153_173 | 128 | 873.0 | Transmembrane | Helical%3B Name%3DSegment S2 |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000388996 | - | 1 | 15 | 197_217 | 128 | 873.0 | Transmembrane | Helical%3B Name%3DSegment S3 |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000388996 | - | 1 | 15 | 226_247 | 128 | 873.0 | Transmembrane | Helical%3B Voltage-sensor%3B Name%3DSegment S4 |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000388996 | - | 1 | 15 | 262_282 | 128 | 873.0 | Transmembrane | Helical%3B Name%3DSegment S5 |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000388996 | - | 1 | 15 | 331_351 | 128 | 873.0 | Transmembrane | Helical%3B Name%3DSegment S6 |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000521134 | - | 1 | 15 | 122_142 | 0 | 753.0 | Transmembrane | Helical%3B Name%3DSegment S1 |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000521134 | - | 1 | 15 | 153_173 | 0 | 753.0 | Transmembrane | Helical%3B Name%3DSegment S2 |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000521134 | - | 1 | 15 | 197_217 | 0 | 753.0 | Transmembrane | Helical%3B Name%3DSegment S3 |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000521134 | - | 1 | 15 | 226_247 | 0 | 753.0 | Transmembrane | Helical%3B Voltage-sensor%3B Name%3DSegment S4 |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000521134 | - | 1 | 15 | 262_282 | 0 | 753.0 | Transmembrane | Helical%3B Name%3DSegment S5 |
| Hgene | KCNQ3 | chr8:133492393 | chr8:134276895 | ENST00000521134 | - | 1 | 15 | 331_351 | 0 | 753.0 | Transmembrane | Helical%3B Name%3DSegment S6 |
Top |
Fusion Gene Sequence for KCNQ3-NDRG1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >41565_41565_1_KCNQ3-NDRG1_KCNQ3_chr8_133492393_ENST00000388996_NDRG1_chr8_134276895_ENST00000323851_length(transcript)=3600nt_BP=807nt GGCTGGCACCGGGCTTGTAAATAAACTGCGATGCTGTCTTGGGCTCGCCGGCCTGGGTGCCGCCGCCGCCGCCGCCGCCCCGGAGCGCCG GTTCCAGGCTCCCGCCGCCGCGCGCCCTAGCGCCTCCGCGCCAGCCTCGCGCGCCCTCCCAGCTTCTCCGCAGCCCCCGGCCCGCGACCC TCGGGGTCCTGCGGCTGACCCGCGCGGGATGTGACCCCCTGACCCCCTGCCTGGCCTCCCCTGCCCCCCAGGGGGCCCGCCTTTGCCTGC TTTTGGGGGGGGGTGGGGAGGGGCGCGCGGATCATGGCATTGGAGTTCCCGGGCTTGCAGCCGCCGCCGCCGCCTCGTTCACGCACCCCG AGCGCCCCTTCTTCCCAGAGCAGCAGCGGAGAAGGCGAAGCGGCCGGCGGGGGCGAGGCAGATGGGGCTCAAGGCGCGCAGGGCGGCGGG GGCGGCTGGCGGCGGCGGCGACGGGGGCGGCGGAGGCGGCGGGGCGGCTAACCCAGCCGGAGGGGACGCGGCGGCGGCCGGCGACGAGGA GCGGAAAGTGGGGCTGGCGCCCGGCGACGTGGAGCAAGTCACCTTGGCGCTCGGGGCCGGAGCCGACAAAGACGGGACCCTGCTGCTGGA GGGCGGCGGCCGCGACGAGGGGCAGCGGAGGACCCCGCAGGGCATCGGGCTCCTGGCCAAGACCCCGCTGAGCCGCCCAGTCAAGAGAAA CAACGCCAAGTACCGGCGCATCCAAACTTTGATCTACGACGCCCTGGAGAGACCGCGGGGCTGGGCGCTGCTTTACCACGCGTTGGTGAG CAGGACATCGAGACTTTACATGGCTCTGTTCACGTCACGCTGTGTGGGACTCCCAAGGGAAACCGGCCTGTCATCCTCACCTACCATGAC ATCGGCATGAACCACAAAACCTGCTACAACCCCCTCTTCAACTACGAGGACATGCAGGAGATCACCCAGCACTTTGCCGTCTGCCACGTG GACGCCCCTGGCCAGCAGGACGGCGCAGCCTCCTTCCCCGCAGGGTACATGTACCCCTCCATGGATCAGCTGGCTGAAATGCTTCCTGGA GTCCTTCAACAGTTTGGGCTGAAAAGCATTATTGGCATGGGAACAGGAGCAGGCGCCTACATCCTAACTCGATTTGCTCTAAACAACCCT GAGATGGTGGAGGGCCTTGTCCTTATCAACGTGAACCCTTGTGCGGAAGGCTGGATGGACTGGGCCGCCTCCAAGATCTCAGGATGGACC CAAGCTCTGCCGGACATGGTGGTGTCCCACCTTTTTGGGAAGGAAGAAATGCAGAGTAACGTGGAAGTGGTCCACACCTACCGCCAGCAC ATTGTGAATGACATGAACCCCGGCAACCTGCACCTGTTCATCAATGCCTACAACAGCCGGCGCGACCTGGAGATTGAGCGACCAATGCCG GGAACCCACACAGTCACCCTGCAGTGCCCTGCTCTGTTGGTGGTTGGGGACAGCTCGCCTGCAGTGGATGCCGTGGTGGAGTGCAACTCA AAATTGGACCCAACAAAGACCACTCTCCTCAAGATGGCGGACTGTGGCGGCCTCCCGCAGATCTCCCAGCCGGCCAAGCTCGCTGAGGCC TTCAAGTACTTCGTGCAGGGCATGGGATACATGCCCTCGGCTAGCATGACCCGCCTGATGCGGTCCCGCACAGCCTCTGGTTCCAGCGTC ACTTCTCTGGATGGCACCCGCAGCCGCTCCCACACCAGCGAGGGCACCCGAAGCCGCTCCCACACCAGCGAGGGCACCCGCAGCCGCTCG CACACCAGCGAGGGGGCCCACCTGGACATCACCCCCAACTCGGGTGCTGCTGGGAACAGCGCCGGGCCCAAGTCCATGGAGGTCTCCTGC TAGGCGGCCTGCCCAGCTGCCGCCCCCGGACTCTGATCTCTGTAGTGGCCCCCTCCTCCCCGGCCCCTTTTCGCCCCCTGCCTGCCATAC TGCGCCTAACTCGGTATTAATCCAAAGCTTATTTTGTAAGAGTGAGCTCTGGTGGAGACAAATGAGGTCTATTACGTGGGTGCCCTCTCC AAAGGCGGGGTGGCGGTGGACCAAAGGAAGGAAGCAAGCATCTCCGCATCGCATCCTCTTCCATTAACCAGTGGCCGGTTGCCACTCTCC TCCCCTCCCTCAGAGACACCAAACTGCCAAAAACAAGACGCGTAGCAGCACACACTTCACAAAGCCAAGCCTAGGCCGCCCTGAGCATCC TGGTTCAAACGGGTGCCTGGTCAGAAGGCCAGCCGCCCACTTCCCGTTTCCTCTTTAACTGAGGAGAAGCTGATCCAGTTTCCGGAAACA AAATCCTTTTCTCATTTGGGGAGGGGGGTAATAGTGACATGCAGGCACCTCTTTTAAACAGGCAAAACAGGAAGGGGGAAAAGGTGGGAT TCATGTCGAGGCTAGAGGCATTTGGAACAACAAATCTACGTAGTTAACTTGAAGAAACCGATTTTTAAAGTTGGTGCATCTAGAAAGCTT TGAATGCAGAAGCAAACAAGCTTGATTTTTCTAGCATCCTCTTAATGTGCAGCAAAAGCAGGCGACAAAATCTCCTGGCTTTACAGACAA AAATATTTCAGCAAACGTTGGGCATCATGGTTTTTGAAGGCTTTAGTTCTGCTTTCTGCCTCTCCTCCACAGCCCCAACCTCCCACCCCT GATACATGAGCCAGTGATTATTCTTGTTCAGGGAGAAGATCATTTAGATTTGTTTTGCATTCCTTAGAATGGAGGGCAACATTCCACAGC TGCCCTGGCTGTGATGAGTGTCCTTGCAGGGGCCGGAGTAGGAGCACTGGGGTGGGGGTGGAATTGGGGTTACTCGATGTAAGGGATTCC TTGTTGTTGTGTTGAGATCCAGTGCAGTTGTGATTTCTGTGGATCCCAGCTTGGTTCCAGGAATTTTGTGTGATTGGCTTAAATCCAGTT TTCAATCTTCGACAGCTGGGCTGGAACGTGAACTCAGTAGCTGAACCTGTCTGACCCGGTCACGTTCTTGGATCCTCAGAACTCTTTGCT CTTGTCGGGGTGGGGGTGGGAACTCACGTGGGGAGCGGTGGCTGAGAAAATGTAAGGATTCTGGAATACATATTCCATGGGACTTTCCTT CCCTCTCCTGCTTCCTCTTTTCCTGCTCCCTAACCTTTCGCCGAATGGGGCAGCACCACTGACGTTTCTGGGCGGCCAGTGCGGCTGCCA GGTTCCTGTACTACTGCCTTGTACTTTTCATTTTGGCTCACCGTGGATTTTCTCATAGGAAGTTTGGTCAGAGTGAATTGAATATTGTAA GTCAGCCACTGGGACCCGAGGATTTCTGGGACCCCGCAGTTGGGAGGAGGAAGTAGTCCAGCCTTCCAGGTGGCGTGAGAGGCAATGACT CGTTACCTGCCGCCCATCACCTTGGAGGCCTTCCCTGGCCTTGAGTAGAAAAGTCGGGGATCGGGGCAAGAGAGGCTGAGTACGGATGGG AAACTATTGTGCACAAGTCTTTCCAGAGGAGTTTCTTAATGAGATATTTGTATTTATTTCCAGACCAATAAATTTGTAACTTTGCAGCGG >41565_41565_1_KCNQ3-NDRG1_KCNQ3_chr8_133492393_ENST00000388996_NDRG1_chr8_134276895_ENST00000323851_length(amino acids)=370AA_BP=9 MGAALPRVGEQDIETLHGSVHVTLCGTPKGNRPVILTYHDIGMNHKTCYNPLFNYEDMQEITQHFAVCHVDAPGQQDGAASFPAGYMYPS MDQLAEMLPGVLQQFGLKSIIGMGTGAGAYILTRFALNNPEMVEGLVLINVNPCAEGWMDWAASKISGWTQALPDMVVSHLFGKEEMQSN VEVVHTYRQHIVNDMNPGNLHLFINAYNSRRDLEIERPMPGTHTVTLQCPALLVVGDSSPAVDAVVECNSKLDPTKTTLLKMADCGGLPQ ISQPAKLAEAFKYFVQGMGYMPSASMTRLMRSRTASGSSVTSLDGTRSRSHTSEGTRSRSHTSEGTRSRSHTSEGAHLDITPNSGAAGNS -------------------------------------------------------------- >41565_41565_2_KCNQ3-NDRG1_KCNQ3_chr8_133492393_ENST00000388996_NDRG1_chr8_134276895_ENST00000354944_length(transcript)=3389nt_BP=807nt GGCTGGCACCGGGCTTGTAAATAAACTGCGATGCTGTCTTGGGCTCGCCGGCCTGGGTGCCGCCGCCGCCGCCGCCGCCCCGGAGCGCCG GTTCCAGGCTCCCGCCGCCGCGCGCCCTAGCGCCTCCGCGCCAGCCTCGCGCGCCCTCCCAGCTTCTCCGCAGCCCCCGGCCCGCGACCC TCGGGGTCCTGCGGCTGACCCGCGCGGGATGTGACCCCCTGACCCCCTGCCTGGCCTCCCCTGCCCCCCAGGGGGCCCGCCTTTGCCTGC TTTTGGGGGGGGGTGGGGAGGGGCGCGCGGATCATGGCATTGGAGTTCCCGGGCTTGCAGCCGCCGCCGCCGCCTCGTTCACGCACCCCG AGCGCCCCTTCTTCCCAGAGCAGCAGCGGAGAAGGCGAAGCGGCCGGCGGGGGCGAGGCAGATGGGGCTCAAGGCGCGCAGGGCGGCGGG GGCGGCTGGCGGCGGCGGCGACGGGGGCGGCGGAGGCGGCGGGGCGGCTAACCCAGCCGGAGGGGACGCGGCGGCGGCCGGCGACGAGGA GCGGAAAGTGGGGCTGGCGCCCGGCGACGTGGAGCAAGTCACCTTGGCGCTCGGGGCCGGAGCCGACAAAGACGGGACCCTGCTGCTGGA GGGCGGCGGCCGCGACGAGGGGCAGCGGAGGACCCCGCAGGGCATCGGGCTCCTGGCCAAGACCCCGCTGAGCCGCCCAGTCAAGAGAAA CAACGCCAAGTACCGGCGCATCCAAACTTTGATCTACGACGCCCTGGAGAGACCGCGGGGCTGGGCGCTGCTTTACCACGCGTTGGTGAG CAGGACATCGAGACTTTACATGGCTCTGTTCACGTCACGCTGTGTGGGACTCCCAAGGGAAACCGGCCTGTCATCCTCACCTACCATGAC ATCGGCATGAACCACAAAACCTGCGCCTACATCCTAACTCGATTTGCTCTAAACAACCCTGAGATGGTGGAGGGCCTTGTCCTTATCAAC GTGAACCCTTGTGCGGAAGGCTGGATGGACTGGGCCGCCTCCAAGATCTCAGGATGGACCCAAGCTCTGCCGGACATGGTGGTGTCCCAC CTTTTTGGGAAGGAAGAAATGCAGAGTAACGTGGAAGTGGTCCACACCTACCGCCAGCACATTGTGAATGACATGAACCCCGGCAACCTG CACCTGTTCATCAATGCCTACAACAGCCGGCGCGACCTGGAGATTGAGCGACCAATGCCGGGAACCCACACAGTCACCCTGCAGTGCCCT GCTCTGTTGGTGGTTGGGGACAGCTCGCCTGCAGTGGATGCCGTGGTGGAGTGCAACTCAAAATTGGACCCAACAAAGACCACTCTCCTC AAGATGGCGGACTGTGGCGGCCTCCCGCAGATCTCCCAGCCGGCCAAGCTCGCTGAGGCCTTCAAGTACTTCGTGCAGGGCATGGGATAC ATGCCCTCGGCTAGCATGACCCGCCTGATGCGGTCCCGCACAGCCTCTGGTTCCAGCGTCACTTCTCTGGATGGCACCCGCAGCCGCTCC CACACCAGCGAGGGCACCCGAAGCCGCTCCCACACCAGCGAGGGCACCCGCAGCCGCTCGCACACCAGCGAGGGGGCCCACCTGGACATC ACCCCCAACTCGGGTGCTGCTGGGAACAGCGCCGGGCCCAAGTCCATGGAGGTCTCCTGCTAGGCGGCCTGCCCAGCTGCCGCCCCCGGA CTCTGATCTCTGTAGTGGCCCCCTCCTCCCCGGCCCCTTTTCGCCCCCTGCCTGCCATACTGCGCCTAACTCGGTATTAATCCAAAGCTT ATTTTGTAAGAGTGAGCTCTGGTGGAGACAAATGAGGTCTATTACGTGGGTGCCCTCTCCAAAGGCGGGGTGGCGGTGGACCAAAGGAAG GAAGCAAGCATCTCCGCATCGCATCCTCTTCCATTAACCAGTGGCCGGTTGCCACTCTCCTCCCCTCCCTCAGAGACACCAAACTGCCAA AAACAAGACGCGTAGCAGCACACACTTCACAAAGCCAAGCCTAGGCCGCCCTGAGCATCCTGGTTCAAACGGGTGCCTGGTCAGAAGGCC AGCCGCCCACTTCCCGTTTCCTCTTTAACTGAGGAGAAGCTGATCCAGTTTCCGGAAACAAAATCCTTTTCTCATTTGGGGAGGGGGGTA ATAGTGACATGCAGGCACCTCTTTTAAACAGGCAAAACAGGAAGGGGGAAAAGGTGGGATTCATGTCGAGGCTAGAGGCATTTGGAACAA CAAATCTACGTAGTTAACTTGAAGAAACCGATTTTTAAAGTTGGTGCATCTAGAAAGCTTTGAATGCAGAAGCAAACAAGCTTGATTTTT CTAGCATCCTCTTAATGTGCAGCAAAAGCAGGCGACAAAATCTCCTGGCTTTACAGACAAAAATATTTCAGCAAACGTTGGGCATCATGG TTTTTGAAGGCTTTAGTTCTGCTTTCTGCCTCTCCTCCACAGCCCCAACCTCCCACCCCTGATACATGAGCCAGTGATTATTCTTGTTCA GGGAGAAGATCATTTAGATTTGTTTTGCATTCCTTAGAATGGAGGGCAACATTCCACAGCTGCCCTGGCTGTGATGAGTGTCCTTGCAGG GGCCGGAGTAGGAGCACTGGGGTGGGGGTGGAATTGGGGTTACTCGATGTAAGGGATTCCTTGTTGTTGTGTTGAGATCCAGTGCAGTTG TGATTTCTGTGGATCCCAGCTTGGTTCCAGGAATTTTGTGTGATTGGCTTAAATCCAGTTTTCAATCTTCGACAGCTGGGCTGGAACGTG AACTCAGTAGCTGAACCTGTCTGACCCGGTCACGTTCTTGGATCCTCAGAACTCTTTGCTCTTGTCGGGGTGGGGGTGGGAACTCACGTG GGGAGCGGTGGCTGAGAAAATGTAAGGATTCTGGAATACATATTCCATGGGACTTTCCTTCCCTCTCCTGCTTCCTCTTTTCCTGCTCCC TAACCTTTCGCCGAATGGGGCAGCACCACTGACGTTTCTGGGCGGCCAGTGCGGCTGCCAGGTTCCTGTACTACTGCCTTGTACTTTTCA TTTTGGCTCACCGTGGATTTTCTCATAGGAAGTTTGGTCAGAGTGAATTGAATATTGTAAGTCAGCCACTGGGACCCGAGGATTTCTGGG ACCCCGCAGTTGGGAGGAGGAAGTAGTCCAGCCTTCCAGGTGGCGTGAGAGGCAATGACTCGTTACCTGCCGCCCATCACCTTGGAGGCC TTCCCTGGCCTTGAGTAGAAAAGTCGGGGATCGGGGCAAGAGAGGCTGAGTACGGATGGGAAACTATTGTGCACAAGTCTTTCCAGAGGA >41565_41565_2_KCNQ3-NDRG1_KCNQ3_chr8_133492393_ENST00000388996_NDRG1_chr8_134276895_ENST00000354944_length(amino acids)=300AA_BP=9 MGAALPRVGEQDIETLHGSVHVTLCGTPKGNRPVILTYHDIGMNHKTCAYILTRFALNNPEMVEGLVLINVNPCAEGWMDWAASKISGWT QALPDMVVSHLFGKEEMQSNVEVVHTYRQHIVNDMNPGNLHLFINAYNSRRDLEIERPMPGTHTVTLQCPALLVVGDSSPAVDAVVECNS KLDPTKTTLLKMADCGGLPQISQPAKLAEAFKYFVQGMGYMPSASMTRLMRSRTASGSSVTSLDGTRSRSHTSEGTRSRSHTSEGTRSRS -------------------------------------------------------------- >41565_41565_3_KCNQ3-NDRG1_KCNQ3_chr8_133492393_ENST00000519445_NDRG1_chr8_134276895_ENST00000323851_length(transcript)=3225nt_BP=432nt CAGAGCAGCAGCGGAGAAGGCGAAGCGGCCGGCGGGGGCGAGGCAGATGGGGCTCAAGGCGCGCAGGGCGGCGGGGGCGGCTGGCGGCGG CGGCGACGGGGGCGGCGGAGGCGGCGGGGCGGCTAACCCAGCCGGAGGGGACGCGGCGGCGGCCGGCGACGAGGAGCGGAAAGTGGGGCT GGCGCCCGGCGACGTGGAGCAAGTCACCTTGGCGCTCGGGGCCGGAGCCGACAAAGACGGGACCCTGCTGCTGGAGGGCGGCGGCCGCGA CGAGGGGCAGCGGAGGACCCCGCAGGGCATCGGGCTCCTGGCCAAGACCCCGCTGAGCCGCCCAGTCAAGAGAAACAACGCCAAGTACCG GCGCATCCAAACTTTGATCTACGACGCCCTGGAGAGACCGCGGGGCTGGGCGCTGCTTTACCACGCGTTGGTGAGCAGGACATCGAGACT TTACATGGCTCTGTTCACGTCACGCTGTGTGGGACTCCCAAGGGAAACCGGCCTGTCATCCTCACCTACCATGACATCGGCATGAACCAC AAAACCTGCTACAACCCCCTCTTCAACTACGAGGACATGCAGGAGATCACCCAGCACTTTGCCGTCTGCCACGTGGACGCCCCTGGCCAG CAGGACGGCGCAGCCTCCTTCCCCGCAGGGTACATGTACCCCTCCATGGATCAGCTGGCTGAAATGCTTCCTGGAGTCCTTCAACAGTTT GGGCTGAAAAGCATTATTGGCATGGGAACAGGAGCAGGCGCCTACATCCTAACTCGATTTGCTCTAAACAACCCTGAGATGGTGGAGGGC CTTGTCCTTATCAACGTGAACCCTTGTGCGGAAGGCTGGATGGACTGGGCCGCCTCCAAGATCTCAGGATGGACCCAAGCTCTGCCGGAC ATGGTGGTGTCCCACCTTTTTGGGAAGGAAGAAATGCAGAGTAACGTGGAAGTGGTCCACACCTACCGCCAGCACATTGTGAATGACATG AACCCCGGCAACCTGCACCTGTTCATCAATGCCTACAACAGCCGGCGCGACCTGGAGATTGAGCGACCAATGCCGGGAACCCACACAGTC ACCCTGCAGTGCCCTGCTCTGTTGGTGGTTGGGGACAGCTCGCCTGCAGTGGATGCCGTGGTGGAGTGCAACTCAAAATTGGACCCAACA AAGACCACTCTCCTCAAGATGGCGGACTGTGGCGGCCTCCCGCAGATCTCCCAGCCGGCCAAGCTCGCTGAGGCCTTCAAGTACTTCGTG CAGGGCATGGGATACATGCCCTCGGCTAGCATGACCCGCCTGATGCGGTCCCGCACAGCCTCTGGTTCCAGCGTCACTTCTCTGGATGGC ACCCGCAGCCGCTCCCACACCAGCGAGGGCACCCGAAGCCGCTCCCACACCAGCGAGGGCACCCGCAGCCGCTCGCACACCAGCGAGGGG GCCCACCTGGACATCACCCCCAACTCGGGTGCTGCTGGGAACAGCGCCGGGCCCAAGTCCATGGAGGTCTCCTGCTAGGCGGCCTGCCCA GCTGCCGCCCCCGGACTCTGATCTCTGTAGTGGCCCCCTCCTCCCCGGCCCCTTTTCGCCCCCTGCCTGCCATACTGCGCCTAACTCGGT ATTAATCCAAAGCTTATTTTGTAAGAGTGAGCTCTGGTGGAGACAAATGAGGTCTATTACGTGGGTGCCCTCTCCAAAGGCGGGGTGGCG GTGGACCAAAGGAAGGAAGCAAGCATCTCCGCATCGCATCCTCTTCCATTAACCAGTGGCCGGTTGCCACTCTCCTCCCCTCCCTCAGAG ACACCAAACTGCCAAAAACAAGACGCGTAGCAGCACACACTTCACAAAGCCAAGCCTAGGCCGCCCTGAGCATCCTGGTTCAAACGGGTG CCTGGTCAGAAGGCCAGCCGCCCACTTCCCGTTTCCTCTTTAACTGAGGAGAAGCTGATCCAGTTTCCGGAAACAAAATCCTTTTCTCAT TTGGGGAGGGGGGTAATAGTGACATGCAGGCACCTCTTTTAAACAGGCAAAACAGGAAGGGGGAAAAGGTGGGATTCATGTCGAGGCTAG AGGCATTTGGAACAACAAATCTACGTAGTTAACTTGAAGAAACCGATTTTTAAAGTTGGTGCATCTAGAAAGCTTTGAATGCAGAAGCAA ACAAGCTTGATTTTTCTAGCATCCTCTTAATGTGCAGCAAAAGCAGGCGACAAAATCTCCTGGCTTTACAGACAAAAATATTTCAGCAAA CGTTGGGCATCATGGTTTTTGAAGGCTTTAGTTCTGCTTTCTGCCTCTCCTCCACAGCCCCAACCTCCCACCCCTGATACATGAGCCAGT GATTATTCTTGTTCAGGGAGAAGATCATTTAGATTTGTTTTGCATTCCTTAGAATGGAGGGCAACATTCCACAGCTGCCCTGGCTGTGAT GAGTGTCCTTGCAGGGGCCGGAGTAGGAGCACTGGGGTGGGGGTGGAATTGGGGTTACTCGATGTAAGGGATTCCTTGTTGTTGTGTTGA GATCCAGTGCAGTTGTGATTTCTGTGGATCCCAGCTTGGTTCCAGGAATTTTGTGTGATTGGCTTAAATCCAGTTTTCAATCTTCGACAG CTGGGCTGGAACGTGAACTCAGTAGCTGAACCTGTCTGACCCGGTCACGTTCTTGGATCCTCAGAACTCTTTGCTCTTGTCGGGGTGGGG GTGGGAACTCACGTGGGGAGCGGTGGCTGAGAAAATGTAAGGATTCTGGAATACATATTCCATGGGACTTTCCTTCCCTCTCCTGCTTCC TCTTTTCCTGCTCCCTAACCTTTCGCCGAATGGGGCAGCACCACTGACGTTTCTGGGCGGCCAGTGCGGCTGCCAGGTTCCTGTACTACT GCCTTGTACTTTTCATTTTGGCTCACCGTGGATTTTCTCATAGGAAGTTTGGTCAGAGTGAATTGAATATTGTAAGTCAGCCACTGGGAC CCGAGGATTTCTGGGACCCCGCAGTTGGGAGGAGGAAGTAGTCCAGCCTTCCAGGTGGCGTGAGAGGCAATGACTCGTTACCTGCCGCCC ATCACCTTGGAGGCCTTCCCTGGCCTTGAGTAGAAAAGTCGGGGATCGGGGCAAGAGAGGCTGAGTACGGATGGGAAACTATTGTGCACA >41565_41565_3_KCNQ3-NDRG1_KCNQ3_chr8_133492393_ENST00000519445_NDRG1_chr8_134276895_ENST00000323851_length(amino acids)=370AA_BP=9 MGAALPRVGEQDIETLHGSVHVTLCGTPKGNRPVILTYHDIGMNHKTCYNPLFNYEDMQEITQHFAVCHVDAPGQQDGAASFPAGYMYPS MDQLAEMLPGVLQQFGLKSIIGMGTGAGAYILTRFALNNPEMVEGLVLINVNPCAEGWMDWAASKISGWTQALPDMVVSHLFGKEEMQSN VEVVHTYRQHIVNDMNPGNLHLFINAYNSRRDLEIERPMPGTHTVTLQCPALLVVGDSSPAVDAVVECNSKLDPTKTTLLKMADCGGLPQ ISQPAKLAEAFKYFVQGMGYMPSASMTRLMRSRTASGSSVTSLDGTRSRSHTSEGTRSRSHTSEGTRSRSHTSEGAHLDITPNSGAAGNS -------------------------------------------------------------- >41565_41565_4_KCNQ3-NDRG1_KCNQ3_chr8_133492393_ENST00000519445_NDRG1_chr8_134276895_ENST00000354944_length(transcript)=3014nt_BP=432nt CAGAGCAGCAGCGGAGAAGGCGAAGCGGCCGGCGGGGGCGAGGCAGATGGGGCTCAAGGCGCGCAGGGCGGCGGGGGCGGCTGGCGGCGG CGGCGACGGGGGCGGCGGAGGCGGCGGGGCGGCTAACCCAGCCGGAGGGGACGCGGCGGCGGCCGGCGACGAGGAGCGGAAAGTGGGGCT GGCGCCCGGCGACGTGGAGCAAGTCACCTTGGCGCTCGGGGCCGGAGCCGACAAAGACGGGACCCTGCTGCTGGAGGGCGGCGGCCGCGA CGAGGGGCAGCGGAGGACCCCGCAGGGCATCGGGCTCCTGGCCAAGACCCCGCTGAGCCGCCCAGTCAAGAGAAACAACGCCAAGTACCG GCGCATCCAAACTTTGATCTACGACGCCCTGGAGAGACCGCGGGGCTGGGCGCTGCTTTACCACGCGTTGGTGAGCAGGACATCGAGACT TTACATGGCTCTGTTCACGTCACGCTGTGTGGGACTCCCAAGGGAAACCGGCCTGTCATCCTCACCTACCATGACATCGGCATGAACCAC AAAACCTGCGCCTACATCCTAACTCGATTTGCTCTAAACAACCCTGAGATGGTGGAGGGCCTTGTCCTTATCAACGTGAACCCTTGTGCG GAAGGCTGGATGGACTGGGCCGCCTCCAAGATCTCAGGATGGACCCAAGCTCTGCCGGACATGGTGGTGTCCCACCTTTTTGGGAAGGAA GAAATGCAGAGTAACGTGGAAGTGGTCCACACCTACCGCCAGCACATTGTGAATGACATGAACCCCGGCAACCTGCACCTGTTCATCAAT GCCTACAACAGCCGGCGCGACCTGGAGATTGAGCGACCAATGCCGGGAACCCACACAGTCACCCTGCAGTGCCCTGCTCTGTTGGTGGTT GGGGACAGCTCGCCTGCAGTGGATGCCGTGGTGGAGTGCAACTCAAAATTGGACCCAACAAAGACCACTCTCCTCAAGATGGCGGACTGT GGCGGCCTCCCGCAGATCTCCCAGCCGGCCAAGCTCGCTGAGGCCTTCAAGTACTTCGTGCAGGGCATGGGATACATGCCCTCGGCTAGC ATGACCCGCCTGATGCGGTCCCGCACAGCCTCTGGTTCCAGCGTCACTTCTCTGGATGGCACCCGCAGCCGCTCCCACACCAGCGAGGGC ACCCGAAGCCGCTCCCACACCAGCGAGGGCACCCGCAGCCGCTCGCACACCAGCGAGGGGGCCCACCTGGACATCACCCCCAACTCGGGT GCTGCTGGGAACAGCGCCGGGCCCAAGTCCATGGAGGTCTCCTGCTAGGCGGCCTGCCCAGCTGCCGCCCCCGGACTCTGATCTCTGTAG TGGCCCCCTCCTCCCCGGCCCCTTTTCGCCCCCTGCCTGCCATACTGCGCCTAACTCGGTATTAATCCAAAGCTTATTTTGTAAGAGTGA GCTCTGGTGGAGACAAATGAGGTCTATTACGTGGGTGCCCTCTCCAAAGGCGGGGTGGCGGTGGACCAAAGGAAGGAAGCAAGCATCTCC GCATCGCATCCTCTTCCATTAACCAGTGGCCGGTTGCCACTCTCCTCCCCTCCCTCAGAGACACCAAACTGCCAAAAACAAGACGCGTAG CAGCACACACTTCACAAAGCCAAGCCTAGGCCGCCCTGAGCATCCTGGTTCAAACGGGTGCCTGGTCAGAAGGCCAGCCGCCCACTTCCC GTTTCCTCTTTAACTGAGGAGAAGCTGATCCAGTTTCCGGAAACAAAATCCTTTTCTCATTTGGGGAGGGGGGTAATAGTGACATGCAGG CACCTCTTTTAAACAGGCAAAACAGGAAGGGGGAAAAGGTGGGATTCATGTCGAGGCTAGAGGCATTTGGAACAACAAATCTACGTAGTT AACTTGAAGAAACCGATTTTTAAAGTTGGTGCATCTAGAAAGCTTTGAATGCAGAAGCAAACAAGCTTGATTTTTCTAGCATCCTCTTAA TGTGCAGCAAAAGCAGGCGACAAAATCTCCTGGCTTTACAGACAAAAATATTTCAGCAAACGTTGGGCATCATGGTTTTTGAAGGCTTTA GTTCTGCTTTCTGCCTCTCCTCCACAGCCCCAACCTCCCACCCCTGATACATGAGCCAGTGATTATTCTTGTTCAGGGAGAAGATCATTT AGATTTGTTTTGCATTCCTTAGAATGGAGGGCAACATTCCACAGCTGCCCTGGCTGTGATGAGTGTCCTTGCAGGGGCCGGAGTAGGAGC ACTGGGGTGGGGGTGGAATTGGGGTTACTCGATGTAAGGGATTCCTTGTTGTTGTGTTGAGATCCAGTGCAGTTGTGATTTCTGTGGATC CCAGCTTGGTTCCAGGAATTTTGTGTGATTGGCTTAAATCCAGTTTTCAATCTTCGACAGCTGGGCTGGAACGTGAACTCAGTAGCTGAA CCTGTCTGACCCGGTCACGTTCTTGGATCCTCAGAACTCTTTGCTCTTGTCGGGGTGGGGGTGGGAACTCACGTGGGGAGCGGTGGCTGA GAAAATGTAAGGATTCTGGAATACATATTCCATGGGACTTTCCTTCCCTCTCCTGCTTCCTCTTTTCCTGCTCCCTAACCTTTCGCCGAA TGGGGCAGCACCACTGACGTTTCTGGGCGGCCAGTGCGGCTGCCAGGTTCCTGTACTACTGCCTTGTACTTTTCATTTTGGCTCACCGTG GATTTTCTCATAGGAAGTTTGGTCAGAGTGAATTGAATATTGTAAGTCAGCCACTGGGACCCGAGGATTTCTGGGACCCCGCAGTTGGGA GGAGGAAGTAGTCCAGCCTTCCAGGTGGCGTGAGAGGCAATGACTCGTTACCTGCCGCCCATCACCTTGGAGGCCTTCCCTGGCCTTGAG TAGAAAAGTCGGGGATCGGGGCAAGAGAGGCTGAGTACGGATGGGAAACTATTGTGCACAAGTCTTTCCAGAGGAGTTTCTTAATGAGAT >41565_41565_4_KCNQ3-NDRG1_KCNQ3_chr8_133492393_ENST00000519445_NDRG1_chr8_134276895_ENST00000354944_length(amino acids)=300AA_BP=9 MGAALPRVGEQDIETLHGSVHVTLCGTPKGNRPVILTYHDIGMNHKTCAYILTRFALNNPEMVEGLVLINVNPCAEGWMDWAASKISGWT QALPDMVVSHLFGKEEMQSNVEVVHTYRQHIVNDMNPGNLHLFINAYNSRRDLEIERPMPGTHTVTLQCPALLVVGDSSPAVDAVVECNS KLDPTKTTLLKMADCGGLPQISQPAKLAEAFKYFVQGMGYMPSASMTRLMRSRTASGSSVTSLDGTRSRSHTSEGTRSRSHTSEGTRSRS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for KCNQ3-NDRG1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for KCNQ3-NDRG1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for KCNQ3-NDRG1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
