
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:KCTD18-SNED1 (FusionGDB2 ID:41625) |
Fusion Gene Summary for KCTD18-SNED1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: KCTD18-SNED1 | Fusion gene ID: 41625 | Hgene | Tgene | Gene symbol | KCTD18 | SNED1 | Gene ID | 130535 | 25992 |
| Gene name | potassium channel tetramerization domain containing 18 | sushi, nidogen and EGF like domains 1 | |
| Synonyms | 6530404F10Rik | IRE-BP1|SST3|Snep | |
| Cytomap | 2q33.1 | 2q37.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | BTB/POZ domain-containing protein KCTD18potassium channel tetramerisation domain containing 18 | sushi, nidogen and EGF-like domain-containing protein 1insulin responsive sequence DNA binding protein-1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000359878, ENST00000409157, ENST00000468413, | ENST00000469006, ENST00000310397, ENST00000342631, ENST00000401884, ENST00000405547, | |
| Fusion gene scores | * DoF score | 3 X 2 X 3=18 | 5 X 6 X 3=90 |
| # samples | 3 | 6 | |
| ** MAII score | log2(3/18*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(6/90*10)=-0.584962500721156 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: KCTD18 [Title/Abstract] AND SNED1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | KCTD18(201362476)-SNED1(242007179), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | KCTD18-SNED1 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across KCTD18 (5'-gene) Fusion gene breakpoints across KCTD18 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
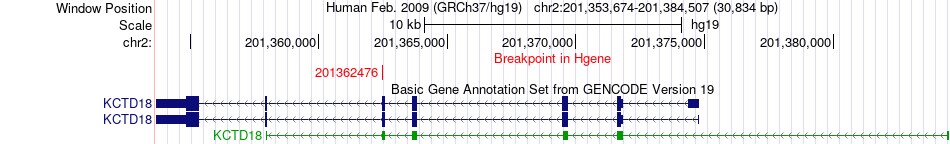 |
 Fusion gene breakpoints across SNED1 (3'-gene) Fusion gene breakpoints across SNED1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
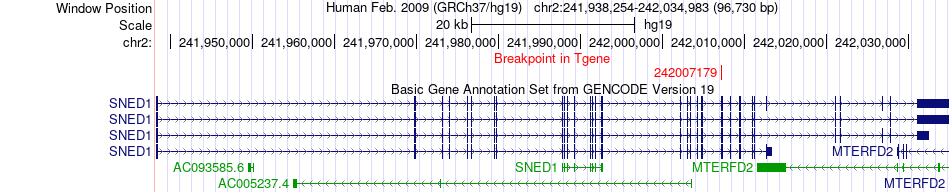 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUAD | TCGA-75-6207-01A | KCTD18 | chr2 | 201362476 | - | SNED1 | chr2 | 242007179 | + |
Top |
Fusion Gene ORF analysis for KCTD18-SNED1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000359878 | ENST00000469006 | KCTD18 | chr2 | 201362476 | - | SNED1 | chr2 | 242007179 | + |
| 5CDS-intron | ENST00000409157 | ENST00000469006 | KCTD18 | chr2 | 201362476 | - | SNED1 | chr2 | 242007179 | + |
| 5UTR-3CDS | ENST00000468413 | ENST00000310397 | KCTD18 | chr2 | 201362476 | - | SNED1 | chr2 | 242007179 | + |
| 5UTR-3CDS | ENST00000468413 | ENST00000342631 | KCTD18 | chr2 | 201362476 | - | SNED1 | chr2 | 242007179 | + |
| 5UTR-3CDS | ENST00000468413 | ENST00000401884 | KCTD18 | chr2 | 201362476 | - | SNED1 | chr2 | 242007179 | + |
| 5UTR-3CDS | ENST00000468413 | ENST00000405547 | KCTD18 | chr2 | 201362476 | - | SNED1 | chr2 | 242007179 | + |
| 5UTR-intron | ENST00000468413 | ENST00000469006 | KCTD18 | chr2 | 201362476 | - | SNED1 | chr2 | 242007179 | + |
| Frame-shift | ENST00000359878 | ENST00000310397 | KCTD18 | chr2 | 201362476 | - | SNED1 | chr2 | 242007179 | + |
| Frame-shift | ENST00000359878 | ENST00000342631 | KCTD18 | chr2 | 201362476 | - | SNED1 | chr2 | 242007179 | + |
| Frame-shift | ENST00000359878 | ENST00000401884 | KCTD18 | chr2 | 201362476 | - | SNED1 | chr2 | 242007179 | + |
| Frame-shift | ENST00000359878 | ENST00000405547 | KCTD18 | chr2 | 201362476 | - | SNED1 | chr2 | 242007179 | + |
| In-frame | ENST00000409157 | ENST00000310397 | KCTD18 | chr2 | 201362476 | - | SNED1 | chr2 | 242007179 | + |
| In-frame | ENST00000409157 | ENST00000342631 | KCTD18 | chr2 | 201362476 | - | SNED1 | chr2 | 242007179 | + |
| In-frame | ENST00000409157 | ENST00000401884 | KCTD18 | chr2 | 201362476 | - | SNED1 | chr2 | 242007179 | + |
| In-frame | ENST00000409157 | ENST00000405547 | KCTD18 | chr2 | 201362476 | - | SNED1 | chr2 | 242007179 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000409157 | KCTD18 | chr2 | 201362476 | - | ENST00000401884 | SNED1 | chr2 | 242007179 | + | 2322 | 794 | 133 | 1758 | 541 |
| ENST00000409157 | KCTD18 | chr2 | 201362476 | - | ENST00000342631 | SNED1 | chr2 | 242007179 | + | 3359 | 794 | 133 | 1926 | 597 |
| ENST00000409157 | KCTD18 | chr2 | 201362476 | - | ENST00000405547 | SNED1 | chr2 | 242007179 | + | 5770 | 794 | 133 | 1842 | 569 |
| ENST00000409157 | KCTD18 | chr2 | 201362476 | - | ENST00000310397 | SNED1 | chr2 | 242007179 | + | 5958 | 794 | 133 | 2025 | 630 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000409157 | ENST00000401884 | KCTD18 | chr2 | 201362476 | - | SNED1 | chr2 | 242007179 | + | 0.012944975 | 0.98705506 |
| ENST00000409157 | ENST00000342631 | KCTD18 | chr2 | 201362476 | - | SNED1 | chr2 | 242007179 | + | 0.005543607 | 0.9944564 |
| ENST00000409157 | ENST00000405547 | KCTD18 | chr2 | 201362476 | - | SNED1 | chr2 | 242007179 | + | 0.005826781 | 0.9941732 |
| ENST00000409157 | ENST00000310397 | KCTD18 | chr2 | 201362476 | - | SNED1 | chr2 | 242007179 | + | 0.005101784 | 0.99489826 |
Top |
Fusion Genomic Features for KCTD18-SNED1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| KCTD18 | chr2 | 201362475 | - | SNED1 | chr2 | 242007178 | + | 0.002478897 | 0.9975211 |
| KCTD18 | chr2 | 201362475 | - | SNED1 | chr2 | 242007178 | + | 0.002478897 | 0.9975211 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
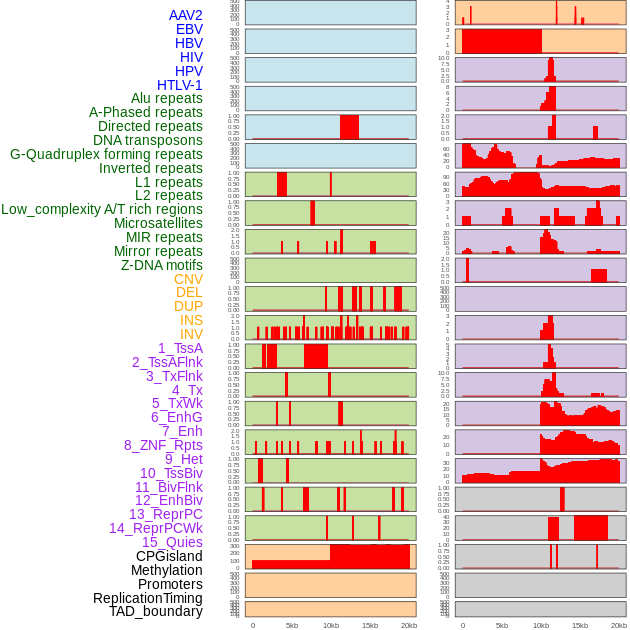 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
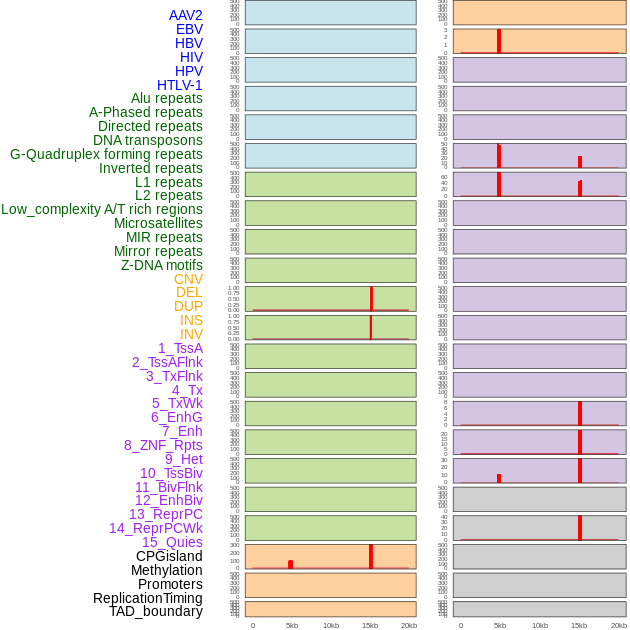 |
Top |
Fusion Protein Features for KCTD18-SNED1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:201362476/chr2:242007179) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KCTD18 | chr2:201362476 | chr2:242007179 | ENST00000359878 | - | 5 | 7 | 12_80 | 220 | 427.0 | Domain | Note=BTB |
| Hgene | KCTD18 | chr2:201362476 | chr2:242007179 | ENST00000409157 | - | 5 | 7 | 12_80 | 220 | 427.0 | Domain | Note=BTB |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000310397 | 20 | 32 | 1007_1105 | 1003 | 2653.3333333333335 | Domain | Fibronectin type-III 2 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000310397 | 20 | 32 | 1106_1200 | 1003 | 2653.3333333333335 | Domain | Fibronectin type-III 3 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000310397 | 20 | 32 | 1307_1343 | 1003 | 2653.3333333333335 | Domain | EGF-like 15 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000401884 | 20 | 27 | 1007_1105 | 1003 | 1325.0 | Domain | Fibronectin type-III 2 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000401884 | 20 | 27 | 1106_1200 | 1003 | 1325.0 | Domain | Fibronectin type-III 3 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000401884 | 20 | 27 | 1307_1343 | 1003 | 1325.0 | Domain | EGF-like 15 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000405547 | 20 | 30 | 1007_1105 | 1003 | 2590.6666666666665 | Domain | Fibronectin type-III 2 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000405547 | 20 | 30 | 1106_1200 | 1003 | 2590.6666666666665 | Domain | Fibronectin type-III 3 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000405547 | 20 | 30 | 1307_1343 | 1003 | 2590.6666666666665 | Domain | EGF-like 15 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000310397 | 20 | 32 | 103_258 | 1003 | 2653.3333333333335 | Domain | NIDO | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000310397 | 20 | 32 | 268_309 | 1003 | 2653.3333333333335 | Domain | EGF-like 1 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000310397 | 20 | 32 | 311_347 | 1003 | 2653.3333333333335 | Domain | EGF-like 2 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000310397 | 20 | 32 | 349_385 | 1003 | 2653.3333333333335 | Domain | EGF-like 3 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000310397 | 20 | 32 | 352_374 | 1003 | 2653.3333333333335 | Domain | Note=Follistatin-like 1 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000310397 | 20 | 32 | 387_423 | 1003 | 2653.3333333333335 | Domain | EGF-like 4%3B calcium-binding | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000310397 | 20 | 32 | 429_465 | 1003 | 2653.3333333333335 | Domain | EGF-like 5 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000310397 | 20 | 32 | 468_500 | 1003 | 2653.3333333333335 | Domain | EGF-like 6 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000310397 | 20 | 32 | 507_530 | 1003 | 2653.3333333333335 | Domain | Note=Follistatin-like 2 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000310397 | 20 | 32 | 541_577 | 1003 | 2653.3333333333335 | Domain | EGF-like 7 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000310397 | 20 | 32 | 580_616 | 1003 | 2653.3333333333335 | Domain | EGF-like 8 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000310397 | 20 | 32 | 619_655 | 1003 | 2653.3333333333335 | Domain | EGF-like 9 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000310397 | 20 | 32 | 657_693 | 1003 | 2653.3333333333335 | Domain | EGF-like 10 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000310397 | 20 | 32 | 696_753 | 1003 | 2653.3333333333335 | Domain | Sushi | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000310397 | 20 | 32 | 753_789 | 1003 | 2653.3333333333335 | Domain | EGF-like 11%3B calcium-binding | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000310397 | 20 | 32 | 791_827 | 1003 | 2653.3333333333335 | Domain | EGF-like 12%3B calcium-binding | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000310397 | 20 | 32 | 829_865 | 1003 | 2653.3333333333335 | Domain | EGF-like 13 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000310397 | 20 | 32 | 867_903 | 1003 | 2653.3333333333335 | Domain | EGF-like 14 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000310397 | 20 | 32 | 908_1006 | 1003 | 2653.3333333333335 | Domain | Fibronectin type-III 1 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000401884 | 20 | 27 | 103_258 | 1003 | 1325.0 | Domain | NIDO | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000401884 | 20 | 27 | 268_309 | 1003 | 1325.0 | Domain | EGF-like 1 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000401884 | 20 | 27 | 311_347 | 1003 | 1325.0 | Domain | EGF-like 2 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000401884 | 20 | 27 | 349_385 | 1003 | 1325.0 | Domain | EGF-like 3 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000401884 | 20 | 27 | 352_374 | 1003 | 1325.0 | Domain | Note=Follistatin-like 1 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000401884 | 20 | 27 | 387_423 | 1003 | 1325.0 | Domain | EGF-like 4%3B calcium-binding | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000401884 | 20 | 27 | 429_465 | 1003 | 1325.0 | Domain | EGF-like 5 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000401884 | 20 | 27 | 468_500 | 1003 | 1325.0 | Domain | EGF-like 6 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000401884 | 20 | 27 | 507_530 | 1003 | 1325.0 | Domain | Note=Follistatin-like 2 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000401884 | 20 | 27 | 541_577 | 1003 | 1325.0 | Domain | EGF-like 7 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000401884 | 20 | 27 | 580_616 | 1003 | 1325.0 | Domain | EGF-like 8 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000401884 | 20 | 27 | 619_655 | 1003 | 1325.0 | Domain | EGF-like 9 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000401884 | 20 | 27 | 657_693 | 1003 | 1325.0 | Domain | EGF-like 10 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000401884 | 20 | 27 | 696_753 | 1003 | 1325.0 | Domain | Sushi | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000401884 | 20 | 27 | 753_789 | 1003 | 1325.0 | Domain | EGF-like 11%3B calcium-binding | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000401884 | 20 | 27 | 791_827 | 1003 | 1325.0 | Domain | EGF-like 12%3B calcium-binding | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000401884 | 20 | 27 | 829_865 | 1003 | 1325.0 | Domain | EGF-like 13 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000401884 | 20 | 27 | 867_903 | 1003 | 1325.0 | Domain | EGF-like 14 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000401884 | 20 | 27 | 908_1006 | 1003 | 1325.0 | Domain | Fibronectin type-III 1 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000405547 | 20 | 30 | 103_258 | 1003 | 2590.6666666666665 | Domain | NIDO | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000405547 | 20 | 30 | 268_309 | 1003 | 2590.6666666666665 | Domain | EGF-like 1 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000405547 | 20 | 30 | 311_347 | 1003 | 2590.6666666666665 | Domain | EGF-like 2 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000405547 | 20 | 30 | 349_385 | 1003 | 2590.6666666666665 | Domain | EGF-like 3 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000405547 | 20 | 30 | 352_374 | 1003 | 2590.6666666666665 | Domain | Note=Follistatin-like 1 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000405547 | 20 | 30 | 387_423 | 1003 | 2590.6666666666665 | Domain | EGF-like 4%3B calcium-binding | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000405547 | 20 | 30 | 429_465 | 1003 | 2590.6666666666665 | Domain | EGF-like 5 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000405547 | 20 | 30 | 468_500 | 1003 | 2590.6666666666665 | Domain | EGF-like 6 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000405547 | 20 | 30 | 507_530 | 1003 | 2590.6666666666665 | Domain | Note=Follistatin-like 2 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000405547 | 20 | 30 | 541_577 | 1003 | 2590.6666666666665 | Domain | EGF-like 7 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000405547 | 20 | 30 | 580_616 | 1003 | 2590.6666666666665 | Domain | EGF-like 8 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000405547 | 20 | 30 | 619_655 | 1003 | 2590.6666666666665 | Domain | EGF-like 9 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000405547 | 20 | 30 | 657_693 | 1003 | 2590.6666666666665 | Domain | EGF-like 10 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000405547 | 20 | 30 | 696_753 | 1003 | 2590.6666666666665 | Domain | Sushi | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000405547 | 20 | 30 | 753_789 | 1003 | 2590.6666666666665 | Domain | EGF-like 11%3B calcium-binding | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000405547 | 20 | 30 | 791_827 | 1003 | 2590.6666666666665 | Domain | EGF-like 12%3B calcium-binding | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000405547 | 20 | 30 | 829_865 | 1003 | 2590.6666666666665 | Domain | EGF-like 13 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000405547 | 20 | 30 | 867_903 | 1003 | 2590.6666666666665 | Domain | EGF-like 14 | |
| Tgene | SNED1 | chr2:201362476 | chr2:242007179 | ENST00000405547 | 20 | 30 | 908_1006 | 1003 | 2590.6666666666665 | Domain | Fibronectin type-III 1 |
Top |
Fusion Gene Sequence for KCTD18-SNED1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >41625_41625_1_KCTD18-SNED1_KCTD18_chr2_201362476_ENST00000409157_SNED1_chr2_242007179_ENST00000310397_length(transcript)=5958nt_BP=794nt ACTTCCGCGTCCGCCCCGCGGCGTCCGGCGGCCGCGCCGAGGGTCTGGCGGGCCGCAGAGGGAGACCAAGACATCATTGTTTTGAAGTTT CTGAAAGTGTTGGGCGGCAAAAATTCAGGTGCCTGGGGGAGAAATGGAAGGCCACAAGGCAGAAGAAGAGGTGCTAGATGTTCTCCGACT GAACGTGGGTGGCTGTATTTACACAGCCCGGCGGGAGTCCTTGTGCCGCTTCAAGGACTCCATGTTGGCATCTATGTTCAGTGGTCGCTT TCCTCTAAAAACAGATGAGTCAGGGGCTTGTGTTATTGACCGTGATGGACGTCTATTTAAATACCTTTTGGATTACCTTCATGGAGAAGT TCAGATTCCCACAGATGAGCAAACCCGCATCGCCCTACAGGAAGAGGCTGATTACTTTGGCATCCCTTATCCATACAGCCTGTCTGACCA TTTGGCCAATGAAATGGAGACATATTCTTTAAGGTCAAATATAGAACTTAAAAAGGCTTTAACAGACTTCTGTGATTCATATGGCTTAGT CTGCAATAAACCAACAGTTTGGGTTCTCCACTATCTTAACACATCTGGTGCAAGCTGTGAGAGTAGAATTATTGGTGTATATGCCACAAA AACTGATGGAACAGACGCTATTGAAAAGCAGCTGGGAGGAAGAATTCACAGCAAAGGCATTTTTAAAAGAGAGGCGGGAAATAATGTTCA GTACATTTGGAGCTATTATTCAGTAGCAGAGTTGAAGAAAATGATGGATGCCTTTGATGCTTGGGAAGGAAAAGGACCCCGCCCTGTGGA AGGCTTCGAGGTCACCAATGTGACGGCTAGCACCATCTCAGTGCAGTGGGCCCTGCACAGGATCCGCCATGCCACCGTCAGTGGGGTCCG TGTGTCCATCCGCCACCCTGAGGCCCTCAGGGACCAGGCCACCGATGTGGACAGGAGTGTGGACAGGTTCACCTTTAGGGCCCTGCTGCC TGGGAAGAGGTACACCATCCAGCTGACCACCCTCAGTGGGCTCAGGGGAGAGGAGCACCCCACAGAGAGCCTGGCCACCGCGCCGACGCA CGTGTGGACCCGGCCCCTGCCTCCAGCAAACCTGACCGCCGCCCGAGTCACTGCCACCTCTGCCCACGTGGTCTGGGATGCCCCGACTCC AGGCAGCTTGCTGGAGGCTTATGTCATCAATGTGACCACCAGCCAGAGCACCAAGAGCCGCTATGTCCCCAACGGGAAGCTGGCGTCCTA CACGGTGCGCGACCTGCTGCCGGGACGGCGGTACCAGCTCTCTGTGATAGCAGTGCAGAGCACGGAGCTCGGGCCGCAGCACAGCGAGCC CGCCCACCTCTACATCATCACCTCCCCCAGGGATGGCGCTGACAGACGCTGGCACCAGGGAGGACACCACCCTCGGGTGCTCAAGAACAG ACCGCCCCCGGCGCGCCTGCCGGAGCTGCGCCTGCTCAATGACCACAGCGCCCCCGAGACCCCCACCCAGCCCCCCAGGTTCTCGGAGCT TGTGGACGGCAGAGGAAGAGTGAGCGCCAGGTTCGGTGGCTCACCCAGCAAAGCAGCCACCGTGAGATCACAACCCACAGCCTCGGCGCA GCTCGAGAACATGGAGGAAGCCCCCAAGCGGGTCAGCCTGGCCCTCCAGCTCCCTGAACACGGCAGCAAGGACATCGGAAACGTCCCTGG CAACTGTTCAGAAAACCCCTGTCAGAACGGAGGCACTTGTGTGCCGGGCGCAGACGCCCACAGCTGTGACTGCGGGCCAGGGTTCAAAGG CAGACGCTGCGAGCTCGCCTGTATAAAGGTGTCCCGCCCCTGCACAAGGCTGTTCTCCGAGACAAAGGCCTTTCCAGTCTGGGAGGGAGG CGTCTGTCACCACGTGTATAAAAGAGTCTACCGAGTTCACCAAGACATCTGCTTCAAAGAGAGCTGTGAAAGCACAAGCCTCAAGAAGAC CCCAAACAGGAAACAAAGTAAGAGTCAGACACTGGAGAAATCTTAAGGATTTAAGACGTTCTTGTTACACTCCACCAACCTCACGAGTTT CTAACACCCAGGAAGATGAGGTCTAAAAACTGGATGAAAAAGGACACCCTGAGAAAAGGTCCTAGCTGGAGTCAGTCCCCTCTGTGACCT CTCTCCTCAGGCCTCTAGAGGACAGATGGCCAGGCCTGTGCACACACCAGCCCACCCTGAGAGACCCCTCTGGGACCAACCACCTGTGAG TCCTGCGATGCGTTTAAGCAGCCTGTGCCCTCACCCAAGCTGCAGTTCCTGAAGGTGTAGTCTGTGTCTCTGCGGATGAGATGACAGCTC GCCATTCCCCGGAATCAGTGAGGCTGTCAGTCAGCCACGCTTCTGCAGTATGCAGAAACCTGTTCTTAGACTCCAAAGCCAGAGAAAGAA TTCTCCCTTCGAGGCCCAACAAATTGAGAAGGAACTGTGATGGACCACTTCCAAAACAGAGACGGGGGCAGGGGCTGAAGGGCAGAGACC AGGTGATGTCAGAAGGAAAGCCGGGTTGCAGACACAGCCGCCCCTGCTCTGGTCCTCCAGCGTGTTTATGACGCTCGTGCAGGTCGACGA GCCATCCTATGGACTAGTTAACACTAAGGTGGAGTTCAGACTTTTTTAGACAACGGCGCGACTGGCAGCCTTTCTCTATCAAGGGTCAGA CGGTAAACGTTTTCAGCTTTGCAGACCAGAGGTCCCTGTGGCTACAGTAGCGCAGACACAGCCACAGGCATGTCATTGAATGGCTGCGGC TATGTTCCAATAAAAACTTATTTACAATAACAGGTGGTGGCCAAATTGGCCCATGGGCCTTATTTGGTGAACCCTGTTCTATGAGATCAC CTAGGCTTCAGCCTTAAACAGTGGAAGCCATCCCCTGAATGACAAGTCACAAGGGTATCAAAGAAAGACCCCTGAATTTTCATGGAAAAA GCTATTCAGACCCCTGCTTGGAAAGCTAAGGCACACTGCCACGAAGCAGCAAGGACGCCTTACAAGTCTCAGTGCAACAGAGATGGACAC CTGGGCTGGGCTGGACAATGTTTAAGGTTCCTTTTAGTCCATGACTCAAGTGATACTGTTTTAGGCTATCAGGTAGTAAACACGATCTTA GACATCCCCATCTTTGTAAGCAGAACAGTACGGCACTTCACCACATCTGCTTCCCACCATGCTTCTAAGCAGCTGTCTTCCCCCTGCTAA TGTTACAACCAAAGCAGCCACCCCACCTCCTCTCGTGTTGAGCCTCACGACCGCTGACCCAGCTGGAAAGCCAGCGCCCTGCCGCGTCAC CCTGACTCTGCTCAGAGCCAGCATTCCAGCCACAAAGAGGGCCTCCTTCCTTTCCTCTTTCATAAAAATGTTTTTTGAAGAGTTAGAGTA TATTTTAGGCTTTTTATCTTTATTAAAATTTCATGTGCATGTGTCTGTGTATTCTGCAATTTGTCATTTTCAGAAAGAAGGAACAGGCAA TTCGCGAAGTTTCACCTGTACTCCCGAGCTGTTCCCCAGGCTCCAGACCCACTTGAGAGCAGAAGGTGGAGCTCAATGAAGGGTCTCGAG CTCAGCGAAGGGTCACTGGGTGAACTGAGAGAAACCACGTTCACAAACGCGTACTGCGGACTTCCTGCCGCCCTGGGACCTGTCACTGTT TGTCCATGTAAGCTACAGCATTACTAGCAGATGCTAAGATCGAGTGATATCACTGGAAAAGTAGGTGAATCCTACTAGGAAACTTTCTAC TCCCTACTAGGACCTCAAGCCCCTCAGCCACACAGCAAATGCTAATATGCTCCAGTGTTAGCTTAGAAGCCTTGTGTCAACAAGAACTGG CTCCTGAGTCCCAAGCTTGGTGCCACACAGCAAATGCTAATATGCTCCAGTGTTAGCTTAGAAGCCTTGTGTCAACAAGAACTGGCTCCT GAGTCCCAAGCTTGGTGCCACACAGCAAATGCTAATATGCTCCAGTGTTAGCTTAGAAGCCTTGTGTCAACAAGAACTGGCTCCTGAGTC CCAAGTGCTGTCACAGGACTTGCCCATTGGGATGTTTTCCACATTAAATATCAAGTAAAAAGACTTCCTGGTGCTCAGGAATTACAGTTC GTTCTTGAAACATTCCAAAGAGGCCACCACAGCTTTTCCCATGTGGCTTCTTTTAAAAACTCAAATGGCTTCCTTGAAAATACTCAAAGT CCACCCAAGGAAATTAGTAATAATAGAATCAGAAAACTGTCAGGAGCATAAAGATTTCTGTATCAAAATGAAAGAAGCAATCCTGAGTTG CTGAATTACCCATCTGCTAATGAAACCGGGATGGACTGATCATCTAACCAAGTGCAGACTGAGGATTCTACTTAGTCCTCCGACTGGGTA CAACAACAGCCTAGGTTCTAGGGAGGGTGGCAGTGACCGGGATGCCACAATGGAAGAGAAAATGAAAACACTGGCACAGTGAAATGTCTC ATTTCCAAACAGTTTTGCCTATGGCCAAGCAAGGCAATAAAGACAAACTCTCCCTTTTCCCCATTGCGTGTGGGCTGCCAGGTACAAGTA AGGGAATCTTTGCTGTGCCCACTGTCCTCCAGTAGAGAACCCAACAGGCAAGGGCCCCACTCAGGTATCAGCTCACCTCCTGCACCTCCC CTTAGCAGGAACTCCTTCCACTGGCAAAGGACTGCCACTGCCATCTGACTCAACTGTGGCGATGTGGACGGAGTCACCGAGCTGCTTTTC TTTTGCAAAACAAAAGTCTTTTTCTTTGCAGTCACGCTGTAAGACGAGGCTGCTGGAGAAAACAAAAGCACCTAGATTTCAGTGCTGAAT CCCCACAATTGCATGCAGCTCACACCTACCAGGGGTATTCCAGTGCATAGGGGAAAGGAACCCGGCTGAAAAACCAGCTCCTTATTTTTC TTTTAAATAAAATAATACGATCCTAAGTCCATTTACCATCTGAAGTTGTCACGAGTGAACAGTCACATTACTGTTGTGGACCAGGCCTTA GATGAGTTTCTCAGGCTCAGCACTGACATTCTGGGCCGGATCATCCTCTCTTGTGGGACCATCCTGTGCACTGCAGGATGTTTCACAGCA CCCCTGGCCTCTACCCACTAGAATCCTACAATCTACCAGATCCTAGGATCTAGTTGATCCTAGAATGCTACCAAGGAGACTTGAATTTTG GTCCCATCATCAAAAATGCCTTCTGCATCAATCCTATGCCAGTTTCCCCTAAAAGAGGGCTAACTGGAATGTTCTAGGATGTACCAATCC TCCAGGACCCTCTTAGAGCTCATGCCATCAGAGACAGGCCTCTATCTCAGGGATCACCCCGGCTGACATCAAATTCCTCTTCTCTTTTCC CAACATTTCAAATTGTTCTTCGGACTCATTGAGTTCCCTAAGGTGACACGCCCCCCCCCCCCCCCACACCCACCTTGGGGGGTCACGGAT TGCTCCCTGTGGCCCTGGCTTCAGCCCACCTGCTCCATGACCCTATGCTCTTCTCCTCTGGTTCTCGAAGCTGTCTGTAACCAGGGCAGG GGAGATAACTGTATGGGACCATTCCCTGGCCTCCACCTGCCCCAGCACTCTCCAGGCCAGCACAGTCTGGCACTACAGACTACAGGGCCG GAGACACCAGGCTTGGAAGTGACCACTGGCCAAGAGCTGACATAAAAGCCAACTTCCCCAAACAGGAAGGGCCTGTTCTTCACAAAGATA AAGAAAACTGCCAGCATTTTGCTGGCCTTAAGAAGCCATGCAAAATTAACCAGTACATCATGAATTAACTTCTTTTACTCTGCTGTGCAC TGAATACCATTATGAAGCAATATACTTTTGGATTAAAAATAAGCATTTTTGTGGAAAATCTGCTACATGTTTTCATAAAATAAAGTGCTA >41625_41625_1_KCTD18-SNED1_KCTD18_chr2_201362476_ENST00000409157_SNED1_chr2_242007179_ENST00000310397_length(amino acids)=630AA_BP=220 MEGHKAEEEVLDVLRLNVGGCIYTARRESLCRFKDSMLASMFSGRFPLKTDESGACVIDRDGRLFKYLLDYLHGEVQIPTDEQTRIALQE EADYFGIPYPYSLSDHLANEMETYSLRSNIELKKALTDFCDSYGLVCNKPTVWVLHYLNTSGASCESRIIGVYATKTDGTDAIEKQLGGR IHSKGIFKREAGNNVQYIWSYYSVAELKKMMDAFDAWEGKGPRPVEGFEVTNVTASTISVQWALHRIRHATVSGVRVSIRHPEALRDQAT DVDRSVDRFTFRALLPGKRYTIQLTTLSGLRGEEHPTESLATAPTHVWTRPLPPANLTAARVTATSAHVVWDAPTPGSLLEAYVINVTTS QSTKSRYVPNGKLASYTVRDLLPGRRYQLSVIAVQSTELGPQHSEPAHLYIITSPRDGADRRWHQGGHHPRVLKNRPPPARLPELRLLND HSAPETPTQPPRFSELVDGRGRVSARFGGSPSKAATVRSQPTASAQLENMEEAPKRVSLALQLPEHGSKDIGNVPGNCSENPCQNGGTCV PGADAHSCDCGPGFKGRRCELACIKVSRPCTRLFSETKAFPVWEGGVCHHVYKRVYRVHQDICFKESCESTSLKKTPNRKQSKSQTLEKS -------------------------------------------------------------- >41625_41625_2_KCTD18-SNED1_KCTD18_chr2_201362476_ENST00000409157_SNED1_chr2_242007179_ENST00000342631_length(transcript)=3359nt_BP=794nt ACTTCCGCGTCCGCCCCGCGGCGTCCGGCGGCCGCGCCGAGGGTCTGGCGGGCCGCAGAGGGAGACCAAGACATCATTGTTTTGAAGTTT CTGAAAGTGTTGGGCGGCAAAAATTCAGGTGCCTGGGGGAGAAATGGAAGGCCACAAGGCAGAAGAAGAGGTGCTAGATGTTCTCCGACT GAACGTGGGTGGCTGTATTTACACAGCCCGGCGGGAGTCCTTGTGCCGCTTCAAGGACTCCATGTTGGCATCTATGTTCAGTGGTCGCTT TCCTCTAAAAACAGATGAGTCAGGGGCTTGTGTTATTGACCGTGATGGACGTCTATTTAAATACCTTTTGGATTACCTTCATGGAGAAGT TCAGATTCCCACAGATGAGCAAACCCGCATCGCCCTACAGGAAGAGGCTGATTACTTTGGCATCCCTTATCCATACAGCCTGTCTGACCA TTTGGCCAATGAAATGGAGACATATTCTTTAAGGTCAAATATAGAACTTAAAAAGGCTTTAACAGACTTCTGTGATTCATATGGCTTAGT CTGCAATAAACCAACAGTTTGGGTTCTCCACTATCTTAACACATCTGGTGCAAGCTGTGAGAGTAGAATTATTGGTGTATATGCCACAAA AACTGATGGAACAGACGCTATTGAAAAGCAGCTGGGAGGAAGAATTCACAGCAAAGGCATTTTTAAAAGAGAGGCGGGAAATAATGTTCA GTACATTTGGAGCTATTATTCAGTAGCAGAGTTGAAGAAAATGATGGATGCCTTTGATGCTTGGGAAGGAAAAGGACCCCGCCCTGTGGA AGGCTTCGAGGTCACCAATGTGACGGCTAGCACCATCTCAGTGCAGTGGGCCCTGCACAGGATCCGCCATGCCACCGTCAGTGGGGTCCG TGTGTCCATCCGCCACCCTGAGGCCCTCAGGGACCAGGCCACCGATGTGGACAGGAGTGTGGACAGGTTCACCTTTAGGGCCCTGCTGCC TGGGAAGAGGTACACCATCCAGCTGACCACCCTCAGTGGGCTCAGGGGAGAGGAGCACCCCACAGAGAGCCTGGCCACCGCGCCGACGCA CGTGTGGACCCGGCCCCTGCCTCCAGCAAACCTGACCGCCGCCCGAGTCACTGCCACCTCTGCCCACGTGGTCTGGGATGCCCCGACTCC AGGCAGCTTGCTGGAGGCTTATGTCATCAATGTGACCACCAGCCAGAGCACCAAGAGCCGCTATGTCCCCAACGGGAAGCTGGCGTCCTA CACGGTGCGCGACCTGCTGCCGGGACGGCGGTACCAGCTCTCTGTGATAGCAGTGCAGAGCACGGAGCTCGGGCCGCAGCACAGCGAGCC CGCCCACCTCTACATCATCACCTCCCCCAGGGATGGCGCTGACAGACGCTGGCACCAGGGAGGACACCACCCTCGGGTGCTCAAGAACAG ACCGCCCCCGGCGCGCCTGCCGGAGCTGCGCCTGCTCAATGACCACAGCGCCCCCGAGACCCCCACCCAGCCCCCCAGGTTCTCGGAGCT TGTGGACGGCAGAGGAAGAGTGAGCGCCAGGTTCGGTGGCTCACCCAGCAAAGCAGCCACCGTGAGATCACACGTCCCTGGCAACTGTTC AGAAAACCCCTGTCAGAACGGAGGCACTTGTGTGCCGGGCGCAGACGCCCACAGCTGTGACTGCGGGCCAGGGTTCAAAGGCAGACGCTG CGAGCTCGCCTGTATAAAGGTGTCCCGCCCCTGCACAAGGCTGTTCTCCGAGACAAAGGCCTTTCCAGTCTGGGAGGGAGGCGTCTGTCA CCACGTGTATAAAAGAGTCTACCGAGTTCACCAAGACATCTGCTTCAAAGAGAGCTGTGAAAGCACAAGCCTCAAGAAGACCCCAAACAG GAAACAAAGTAAGAGTCAGACACTGGAGAAATCTTAAGGATTTAAGACGTTCTTGTTACACTCCACCAACCTCACGAGTTTCTAACACCC AGGAAGATGAGGTCTAAAAACTGGATGAAAAAGGACACCCTGAGAAAAGGTCCTAGCTGGAGTCAGTCCCCTCTGTGACCTCTCTCCTCA GGCCTCTAGAGGACAGATGGCCAGGCCTGTGCACACACCAGCCCACCCTGAGAGACCCCTCTGGGACCAACCACCTGTGAGTCCTGCGAT GCGTTTAAGCAGCCTGTGCCCTCACCCAAGCTGCAGTTCCTGAAGGTGTAGTCTGTGTCTCTGCGGATGAGATGACAGCTCGCCATTCCC CGGAATCAGTGAGGCTGTCAGTCAGCCACGCTTCTGCAGTATGCAGAAACCTGTTCTTAGACTCCAAAGCCAGAGAAAGAATTCTCCCTT CGAGGCCCAACAAATTGAGAAGGAACTGTGATGGACCACTTCCAAAACAGAGACGGGGGCAGGGGCTGAAGGGCAGAGACCAGGTGATGT CAGAAGGAAAGCCGGGTTGCAGACACAGCCGCCCCTGCTCTGGTCCTCCAGCGTGTTTATGACGCTCGTGCAGGTCGACGAGCCATCCTA TGGACTAGTTAACACTAAGGTGGAGTTCAGACTTTTTTAGACAACGGCGCGACTGGCAGCCTTTCTCTATCAAGGGTCAGACGGTAAACG TTTTCAGCTTTGCAGACCAGAGGTCCCTGTGGCTACAGTAGCGCAGACACAGCCACAGGCATGTCATTGAATGGCTGCGGCTATGTTCCA ATAAAAACTTATTTACAATAACAGGTGGTGGCCAAATTGGCCCATGGGCCTTATTTGGTGAACCCTGTTCTATGAGATCACCTAGGCTTC AGCCTTAAACAGTGGAAGCCATCCCCTGAATGACAAGTCACAAGGGTATCAAAGAAAGACCCCTGAATTTTCATGGAAAAAGCTATTCAG ACCCCTGCTTGGAAAGCTAAGGCACACTGCCACGAAGCAGCAAGGACGCCTTACAAGTCTCAGTGCAACAGAGATGGACACCTGGGCTGG GCTGGACAATGTTTAAGGTTCCTTTTAGTCCATGACTCAAGTGATACTGTTTTAGGCTATCAGGTAGTAAACACGATCTTAGACATCCCC ATCTTTGTAAGCAGAACAGTACGGCACTTCACCACATCTGCTTCCCACCATGCTTCTAAGCAGCTGTCTTCCCCCTGCTAATGTTACAAC CAAAGCAGCCACCCCACCTCCTCTCGTGTTGAGCCTCACGACCGCTGACCCAGCTGGAAAGCCAGCGCCCTGCCGCGTCACCCTGACTCT GCTCAGAGCCAGCATTCCAGCCACAAAGAGGGCCTCCTTCCTTTCCTCTTTCATAAAAATGTTTTTTGAAGAGTTAGAGTATATTTTAGG >41625_41625_2_KCTD18-SNED1_KCTD18_chr2_201362476_ENST00000409157_SNED1_chr2_242007179_ENST00000342631_length(amino acids)=597AA_BP=220 MEGHKAEEEVLDVLRLNVGGCIYTARRESLCRFKDSMLASMFSGRFPLKTDESGACVIDRDGRLFKYLLDYLHGEVQIPTDEQTRIALQE EADYFGIPYPYSLSDHLANEMETYSLRSNIELKKALTDFCDSYGLVCNKPTVWVLHYLNTSGASCESRIIGVYATKTDGTDAIEKQLGGR IHSKGIFKREAGNNVQYIWSYYSVAELKKMMDAFDAWEGKGPRPVEGFEVTNVTASTISVQWALHRIRHATVSGVRVSIRHPEALRDQAT DVDRSVDRFTFRALLPGKRYTIQLTTLSGLRGEEHPTESLATAPTHVWTRPLPPANLTAARVTATSAHVVWDAPTPGSLLEAYVINVTTS QSTKSRYVPNGKLASYTVRDLLPGRRYQLSVIAVQSTELGPQHSEPAHLYIITSPRDGADRRWHQGGHHPRVLKNRPPPARLPELRLLND HSAPETPTQPPRFSELVDGRGRVSARFGGSPSKAATVRSHVPGNCSENPCQNGGTCVPGADAHSCDCGPGFKGRRCELACIKVSRPCTRL -------------------------------------------------------------- >41625_41625_3_KCTD18-SNED1_KCTD18_chr2_201362476_ENST00000409157_SNED1_chr2_242007179_ENST00000401884_length(transcript)=2322nt_BP=794nt ACTTCCGCGTCCGCCCCGCGGCGTCCGGCGGCCGCGCCGAGGGTCTGGCGGGCCGCAGAGGGAGACCAAGACATCATTGTTTTGAAGTTT CTGAAAGTGTTGGGCGGCAAAAATTCAGGTGCCTGGGGGAGAAATGGAAGGCCACAAGGCAGAAGAAGAGGTGCTAGATGTTCTCCGACT GAACGTGGGTGGCTGTATTTACACAGCCCGGCGGGAGTCCTTGTGCCGCTTCAAGGACTCCATGTTGGCATCTATGTTCAGTGGTCGCTT TCCTCTAAAAACAGATGAGTCAGGGGCTTGTGTTATTGACCGTGATGGACGTCTATTTAAATACCTTTTGGATTACCTTCATGGAGAAGT TCAGATTCCCACAGATGAGCAAACCCGCATCGCCCTACAGGAAGAGGCTGATTACTTTGGCATCCCTTATCCATACAGCCTGTCTGACCA TTTGGCCAATGAAATGGAGACATATTCTTTAAGGTCAAATATAGAACTTAAAAAGGCTTTAACAGACTTCTGTGATTCATATGGCTTAGT CTGCAATAAACCAACAGTTTGGGTTCTCCACTATCTTAACACATCTGGTGCAAGCTGTGAGAGTAGAATTATTGGTGTATATGCCACAAA AACTGATGGAACAGACGCTATTGAAAAGCAGCTGGGAGGAAGAATTCACAGCAAAGGCATTTTTAAAAGAGAGGCGGGAAATAATGTTCA GTACATTTGGAGCTATTATTCAGTAGCAGAGTTGAAGAAAATGATGGATGCCTTTGATGCTTGGGAAGGAAAAGGACCCCGCCCTGTGGA AGGCTTCGAGGTCACCAATGTGACGGCTAGCACCATCTCAGTGCAGTGGGCCCTGCACAGGATCCGCCATGCCACCGTCAGTGGGGTCCG TGTGTCCATCCGCCACCCTGAGGCCCTCAGGGACCAGGCCACCGATGTGGACAGGAGTGTGGACAGGTTCACCTTTAGGGCCCTGCTGCC TGGGAAGAGGTACACCATCCAGCTGACCACCCTCAGTGGGCTCAGGGGAGAGGAGCACCCCACAGAGAGCCTGGCCACCGCGCCGACGCA CGTGTGGACCCGGCCCCTGCCTCCAGCAAACCTGACCGCCGCCCGAGTCACTGCCACCTCTGCCCACGTGGTCTGGGATGCCCCGACTCC AGGCAGCTTGCTGGAGGCTTATGTCATCAATGTGACCACCAGCCAGAGCACCAAGAGCCGCTATGTCCCCAACGGGAAGCTGGCGTCCTA CACGGTGCGCGACCTGCTGCCGGGACGGCGGTACCAGCTCTCTGTGATAGCAGTGCAGAGCACGGAGCTCGGGCCGCAGCACAGCGAGCC CGCCCACCTCTACATCATCACCTCCCCCAGGGATGGCGCTGACAGACGCTGGCACCAGGGAGGACACCACCCTCGGGTGCTCAAGAACAG ACCGCCCCCGGCGCGCCTGCCGGAGCTGCGCCTGCTCAATGACCACAGCGCCCCCGAGACCCCCACCCAGCCCCCCAGGTTCTCGGAGCT TGTGGACGGCAGAGGAAGAGTGAGCGCCAGGTTCGGTGGCTCACCCAGCAAAGCAGCCACCGTGAGATCACAACCCACAGCCTCGGCGCA GCTCGAGAACATGGAGGAAGCCCCCAAGCGGGTCAGCCTGGCCCTCCAGCTCCCTGAACACGGCAGCAAGGACATCGGAAGTGAGTCAGC AGCGCTGGTGGGGACTTTGGGACTGACTGACTGCTCTCAGGGGCCTTAGAGGCTGCAGGCAGGAGGGACCACCCACGGTGAGGAATCAGG AGGCACAGAGCCTACCTGAGGGGAGGCTGAGCACCAGGCACCCCGGTGTGGGAAGATGGGGTGAAGCTACACCACCCAAGCAGTGGGACC CCACAGACGGGAACAGGCCAGGGGGCAGGACCCACCCAAACCACCCAGAGTCTGAGCTAGAGAGACTGGCTTTGATGCTGCCTCCCCTCC CCTCTCCTCCTTCGCCTCCACATGCAGCAGAGCCCACCCCAGCCCCTGCCTCTGGGCCCCTCACCCCTCACTTCTCCAAAGAGGAGCAGG CGGAGTCAGGATGGGAGAAGCAGAGGGAGCAGCCACTGGGCGAGCCCCAGCTTGAGGACTAGCTGGGCCCTGTGGACACTCAGGTTATGC AGGACCTGAACTGTCTCCTAGTCCGGGGCTCTGCCTCGTGAGGATCGAGGCCAGCACGTCCCTGCAGGGCACCAAGCATCTGCTGAGCAC >41625_41625_3_KCTD18-SNED1_KCTD18_chr2_201362476_ENST00000409157_SNED1_chr2_242007179_ENST00000401884_length(amino acids)=541AA_BP=220 MEGHKAEEEVLDVLRLNVGGCIYTARRESLCRFKDSMLASMFSGRFPLKTDESGACVIDRDGRLFKYLLDYLHGEVQIPTDEQTRIALQE EADYFGIPYPYSLSDHLANEMETYSLRSNIELKKALTDFCDSYGLVCNKPTVWVLHYLNTSGASCESRIIGVYATKTDGTDAIEKQLGGR IHSKGIFKREAGNNVQYIWSYYSVAELKKMMDAFDAWEGKGPRPVEGFEVTNVTASTISVQWALHRIRHATVSGVRVSIRHPEALRDQAT DVDRSVDRFTFRALLPGKRYTIQLTTLSGLRGEEHPTESLATAPTHVWTRPLPPANLTAARVTATSAHVVWDAPTPGSLLEAYVINVTTS QSTKSRYVPNGKLASYTVRDLLPGRRYQLSVIAVQSTELGPQHSEPAHLYIITSPRDGADRRWHQGGHHPRVLKNRPPPARLPELRLLND HSAPETPTQPPRFSELVDGRGRVSARFGGSPSKAATVRSQPTASAQLENMEEAPKRVSLALQLPEHGSKDIGSESAALVGTLGLTDCSQG -------------------------------------------------------------- >41625_41625_4_KCTD18-SNED1_KCTD18_chr2_201362476_ENST00000409157_SNED1_chr2_242007179_ENST00000405547_length(transcript)=5770nt_BP=794nt ACTTCCGCGTCCGCCCCGCGGCGTCCGGCGGCCGCGCCGAGGGTCTGGCGGGCCGCAGAGGGAGACCAAGACATCATTGTTTTGAAGTTT CTGAAAGTGTTGGGCGGCAAAAATTCAGGTGCCTGGGGGAGAAATGGAAGGCCACAAGGCAGAAGAAGAGGTGCTAGATGTTCTCCGACT GAACGTGGGTGGCTGTATTTACACAGCCCGGCGGGAGTCCTTGTGCCGCTTCAAGGACTCCATGTTGGCATCTATGTTCAGTGGTCGCTT TCCTCTAAAAACAGATGAGTCAGGGGCTTGTGTTATTGACCGTGATGGACGTCTATTTAAATACCTTTTGGATTACCTTCATGGAGAAGT TCAGATTCCCACAGATGAGCAAACCCGCATCGCCCTACAGGAAGAGGCTGATTACTTTGGCATCCCTTATCCATACAGCCTGTCTGACCA TTTGGCCAATGAAATGGAGACATATTCTTTAAGGTCAAATATAGAACTTAAAAAGGCTTTAACAGACTTCTGTGATTCATATGGCTTAGT CTGCAATAAACCAACAGTTTGGGTTCTCCACTATCTTAACACATCTGGTGCAAGCTGTGAGAGTAGAATTATTGGTGTATATGCCACAAA AACTGATGGAACAGACGCTATTGAAAAGCAGCTGGGAGGAAGAATTCACAGCAAAGGCATTTTTAAAAGAGAGGCGGGAAATAATGTTCA GTACATTTGGAGCTATTATTCAGTAGCAGAGTTGAAGAAAATGATGGATGCCTTTGATGCTTGGGAAGGAAAAGGACCCCGCCCTGTGGA AGGCTTCGAGGTCACCAATGTGACGGCTAGCACCATCTCAGTGCAGTGGGCCCTGCACAGGATCCGCCATGCCACCGTCAGTGGGGTCCG TGTGTCCATCCGCCACCCTGAGGCCCTCAGGGACCAGGCCACCGATGTGGACAGGAGTGTGGACAGGTTCACCTTTAGGGCCCTGCTGCC TGGGAAGAGGTACACCATCCAGCTGACCACCCTCAGTGGGCTCAGGGGAGAGGAGCACCCCACAGAGAGCCTGGCCACCGCGCCGACGCA CGTGTGGACCCGGCCCCTGCCTCCAGCAAACCTGACCGCCGCCCGAGTCACTGCCACCTCTGCCCACGTGGTCTGGGATGCCCCGACTCC AGGCAGCTTGCTGGAGGCTTATGTCATCAATGTGACCACCAGCCAGAGCACCAAGAGCCGCTATGTCCCCAACGGGAAGCTGGCGTCCTA CACGGTGCGCGACCTGCTGCCGGGACGGCGGTACCAGCTCTCTGTGATAGCAGTGCAGAGCACGGAGCTCGGGCCGCAGCACAGCGAGCC CGCCCACCTCTACATCATCACCTCCCCCAGGGATGGCGCTGACAGACGCTGGCACCAGGGAGGACACCACCCTCGGGTGCTCAAGAACAG ACCGCCCCCGGCGCGCCTGCCGGAGCTGCGCCTGCTCAATGACCACAGCGCCCCCGAGACCCCCACCCAGCCCCCCAGGTTCTCGGAGCT TGTGGACGGCAGAGGAAGAGTGAGCGCCAGGTTCGGTGGCTCACCCAGCAAAGCAGCCACCGTGAGATCACACGTCCCTGGCAACTGTTC AGAAAACCCCTGTCAGAACGGAGGCACTTGTGTGCCGGGCGCAGACGCCCACAGCTGTGACTGCGGGCCAGGGTTCAAAGGCAGACGCTG CGAGCTCGCCTGTATAAAGGTGTCCCGCCCCTGCACAAGGCTGTTCTCCGAGACAAAGGCCTTTCCAGTCTGGGAGGGAGGCGTCTGTCA CCACGTGAAACAAAGTAAGAGTCAGACACTGGAGAAATCTTAAGGATTTAAGACGTTCTTGTTACACTCCACCAACCTCACGAGTTTCTA ACACCCAGGAAGATGAGGTCTAAAAACTGGATGAAAAAGGACACCCTGAGAAAAGGTCCTAGCTGGAGTCAGTCCCCTCTGTGACCTCTC TCCTCAGGCCTCTAGAGGACAGATGGCCAGGCCTGTGCACACACCAGCCCACCCTGAGAGACCCCTCTGGGACCAACCACCTGTGAGTCC TGCGATGCGTTTAAGCAGCCTGTGCCCTCACCCAAGCTGCAGTTCCTGAAGGTGTAGTCTGTGTCTCTGCGGATGAGATGACAGCTCGCC ATTCCCCGGAATCAGTGAGGCTGTCAGTCAGCCACGCTTCTGCAGTATGCAGAAACCTGTTCTTAGACTCCAAAGCCAGAGAAAGAATTC TCCCTTCGAGGCCCAACAAATTGAGAAGGAACTGTGATGGACCACTTCCAAAACAGAGACGGGGGCAGGGGCTGAAGGGCAGAGACCAGG TGATGTCAGAAGGAAAGCCGGGTTGCAGACACAGCCGCCCCTGCTCTGGTCCTCCAGCGTGTTTATGACGCTCGTGCAGGTCGACGAGCC ATCCTATGGACTAGTTAACACTAAGGTGGAGTTCAGACTTTTTTAGACAACGGCGCGACTGGCAGCCTTTCTCTATCAAGGGTCAGACGG TAAACGTTTTCAGCTTTGCAGACCAGAGGTCCCTGTGGCTACAGTAGCGCAGACACAGCCACAGGCATGTCATTGAATGGCTGCGGCTAT GTTCCAATAAAAACTTATTTACAATAACAGGTGGTGGCCAAATTGGCCCATGGGCCTTATTTGGTGAACCCTGTTCTATGAGATCACCTA GGCTTCAGCCTTAAACAGTGGAAGCCATCCCCTGAATGACAAGTCACAAGGGTATCAAAGAAAGACCCCTGAATTTTCATGGAAAAAGCT ATTCAGACCCCTGCTTGGAAAGCTAAGGCACACTGCCACGAAGCAGCAAGGACGCCTTACAAGTCTCAGTGCAACAGAGATGGACACCTG GGCTGGGCTGGACAATGTTTAAGGTTCCTTTTAGTCCATGACTCAAGTGATACTGTTTTAGGCTATCAGGTAGTAAACACGATCTTAGAC ATCCCCATCTTTGTAAGCAGAACAGTACGGCACTTCACCACATCTGCTTCCCACCATGCTTCTAAGCAGCTGTCTTCCCCCTGCTAATGT TACAACCAAAGCAGCCACCCCACCTCCTCTCGTGTTGAGCCTCACGACCGCTGACCCAGCTGGAAAGCCAGCGCCCTGCCGCGTCACCCT GACTCTGCTCAGAGCCAGCATTCCAGCCACAAAGAGGGCCTCCTTCCTTTCCTCTTTCATAAAAATGTTTTTTGAAGAGTTAGAGTATAT TTTAGGCTTTTTATCTTTATTAAAATTTCATGTGCATGTGTCTGTGTATTCTGCAATTTGTCATTTTCAGAAAGAAGGAACAGGCAATTC GCGAAGTTTCACCTGTACTCCCGAGCTGTTCCCCAGGCTCCAGACCCACTTGAGAGCAGAAGGTGGAGCTCAATGAAGGGTCTCGAGCTC AGCGAAGGGTCACTGGGTGAACTGAGAGAAACCACGTTCACAAACGCGTACTGCGGACTTCCTGCCGCCCTGGGACCTGTCACTGTTTGT CCATGTAAGCTACAGCATTACTAGCAGATGCTAAGATCGAGTGATATCACTGGAAAAGTAGGTGAATCCTACTAGGAAACTTTCTACTCC CTACTAGGACCTCAAGCCCCTCAGCCACACAGCAAATGCTAATATGCTCCAGTGTTAGCTTAGAAGCCTTGTGTCAACAAGAACTGGCTC CTGAGTCCCAAGCTTGGTGCCACACAGCAAATGCTAATATGCTCCAGTGTTAGCTTAGAAGCCTTGTGTCAACAAGAACTGGCTCCTGAG TCCCAAGCTTGGTGCCACACAGCAAATGCTAATATGCTCCAGTGTTAGCTTAGAAGCCTTGTGTCAACAAGAACTGGCTCCTGAGTCCCA AGTGCTGTCACAGGACTTGCCCATTGGGATGTTTTCCACATTAAATATCAAGTAAAAAGACTTCCTGGTGCTCAGGAATTACAGTTCGTT CTTGAAACATTCCAAAGAGGCCACCACAGCTTTTCCCATGTGGCTTCTTTTAAAAACTCAAATGGCTTCCTTGAAAATACTCAAAGTCCA CCCAAGGAAATTAGTAATAATAGAATCAGAAAACTGTCAGGAGCATAAAGATTTCTGTATCAAAATGAAAGAAGCAATCCTGAGTTGCTG AATTACCCATCTGCTAATGAAACCGGGATGGACTGATCATCTAACCAAGTGCAGACTGAGGATTCTACTTAGTCCTCCGACTGGGTACAA CAACAGCCTAGGTTCTAGGGAGGGTGGCAGTGACCGGGATGCCACAATGGAAGAGAAAATGAAAACACTGGCACAGTGAAATGTCTCATT TCCAAACAGTTTTGCCTATGGCCAAGCAAGGCAATAAAGACAAACTCTCCCTTTTCCCCATTGCGTGTGGGCTGCCAGGTACAAGTAAGG GAATCTTTGCTGTGCCCACTGTCCTCCAGTAGAGAACCCAACAGGCAAGGGCCCCACTCAGGTATCAGCTCACCTCCTGCACCTCCCCTT AGCAGGAACTCCTTCCACTGGCAAAGGACTGCCACTGCCATCTGACTCAACTGTGGCGATGTGGACGGAGTCACCGAGCTGCTTTTCTTT TGCAAAACAAAAGTCTTTTTCTTTGCAGTCACGCTGTAAGACGAGGCTGCTGGAGAAAACAAAAGCACCTAGATTTCAGTGCTGAATCCC CACAATTGCATGCAGCTCACACCTACCAGGGGTATTCCAGTGCATAGGGGAAAGGAACCCGGCTGAAAAACCAGCTCCTTATTTTTCTTT TAAATAAAATAATACGATCCTAAGTCCATTTACCATCTGAAGTTGTCACGAGTGAACAGTCACATTACTGTTGTGGACCAGGCCTTAGAT GAGTTTCTCAGGCTCAGCACTGACATTCTGGGCCGGATCATCCTCTCTTGTGGGACCATCCTGTGCACTGCAGGATGTTTCACAGCACCC CTGGCCTCTACCCACTAGAATCCTACAATCTACCAGATCCTAGGATCTAGTTGATCCTAGAATGCTACCAAGGAGACTTGAATTTTGGTC CCATCATCAAAAATGCCTTCTGCATCAATCCTATGCCAGTTTCCCCTAAAAGAGGGCTAACTGGAATGTTCTAGGATGTACCAATCCTCC AGGACCCTCTTAGAGCTCATGCCATCAGAGACAGGCCTCTATCTCAGGGATCACCCCGGCTGACATCAAATTCCTCTTCTCTTTTCCCAA CATTTCAAATTGTTCTTCGGACTCATTGAGTTCCCTAAGGTGACACGCCCCCCCCCCCCCCCACACCCACCTTGGGGGGTCACGGATTGC TCCCTGTGGCCCTGGCTTCAGCCCACCTGCTCCATGACCCTATGCTCTTCTCCTCTGGTTCTCGAAGCTGTCTGTAACCAGGGCAGGGGA GATAACTGTATGGGACCATTCCCTGGCCTCCACCTGCCCCAGCACTCTCCAGGCCAGCACAGTCTGGCACTACAGACTACAGGGCCGGAG ACACCAGGCTTGGAAGTGACCACTGGCCAAGAGCTGACATAAAAGCCAACTTCCCCAAACAGGAAGGGCCTGTTCTTCACAAAGATAAAG AAAACTGCCAGCATTTTGCTGGCCTTAAGAAGCCATGCAAAATTAACCAGTACATCATGAATTAACTTCTTTTACTCTGCTGTGCACTGA ATACCATTATGAAGCAATATACTTTTGGATTAAAAATAAGCATTTTTGTGGAAAATCTGCTACATGTTTTCATAAAATAAAGTGCTATGA >41625_41625_4_KCTD18-SNED1_KCTD18_chr2_201362476_ENST00000409157_SNED1_chr2_242007179_ENST00000405547_length(amino acids)=569AA_BP=220 MEGHKAEEEVLDVLRLNVGGCIYTARRESLCRFKDSMLASMFSGRFPLKTDESGACVIDRDGRLFKYLLDYLHGEVQIPTDEQTRIALQE EADYFGIPYPYSLSDHLANEMETYSLRSNIELKKALTDFCDSYGLVCNKPTVWVLHYLNTSGASCESRIIGVYATKTDGTDAIEKQLGGR IHSKGIFKREAGNNVQYIWSYYSVAELKKMMDAFDAWEGKGPRPVEGFEVTNVTASTISVQWALHRIRHATVSGVRVSIRHPEALRDQAT DVDRSVDRFTFRALLPGKRYTIQLTTLSGLRGEEHPTESLATAPTHVWTRPLPPANLTAARVTATSAHVVWDAPTPGSLLEAYVINVTTS QSTKSRYVPNGKLASYTVRDLLPGRRYQLSVIAVQSTELGPQHSEPAHLYIITSPRDGADRRWHQGGHHPRVLKNRPPPARLPELRLLND HSAPETPTQPPRFSELVDGRGRVSARFGGSPSKAATVRSHVPGNCSENPCQNGGTCVPGADAHSCDCGPGFKGRRCELACIKVSRPCTRL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for KCTD18-SNED1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for KCTD18-SNED1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for KCTD18-SNED1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
