
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:KDM3A-SDHAF2 (FusionGDB2 ID:41768) |
Fusion Gene Summary for KDM3A-SDHAF2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: KDM3A-SDHAF2 | Fusion gene ID: 41768 | Hgene | Tgene | Gene symbol | KDM3A | SDHAF2 | Gene ID | 55818 | 54949 |
| Gene name | lysine demethylase 3A | succinate dehydrogenase complex assembly factor 2 | |
| Synonyms | JHDM2A|JHMD2A|JMJD1|JMJD1A|TSGA | C11orf79|PGL2|SDH5 | |
| Cytomap | 2p11.2 | 11q12.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | lysine-specific demethylase 3AjmjC domain-containing histone demethylation protein 2Ajumonji C domain-containing histone demethylase 2Ajumonji domain-containing protein 1Alysine (K)-specific demethylase 3Atestis-specific protein A | succinate dehydrogenase assembly factor 2, mitochondrialSDH assembly factor 2hSDH5succinate dehydrogenase subunit 5, mitochondrial | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9Y4C1 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000312912, ENST00000409064, ENST00000409556, ENST00000542128, ENST00000485171, | ENST00000537782, ENST00000543265, ENST00000534878, ENST00000542074, ENST00000301761, | |
| Fusion gene scores | * DoF score | 13 X 12 X 7=1092 | 4 X 4 X 3=48 |
| # samples | 15 | 4 | |
| ** MAII score | log2(15/1092*10)=-2.86393845042397 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(4/48*10)=-0.263034405833794 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: KDM3A [Title/Abstract] AND SDHAF2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | KDM3A(86683689)-SDHAF2(61213412), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | KDM3A-SDHAF2 seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor due to the frame-shifted ORF. KDM3A-SDHAF2 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. KDM3A-SDHAF2 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. KDM3A-SDHAF2 seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. KDM3A-SDHAF2 seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. KDM3A-SDHAF2 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | KDM3A | GO:0009755 | hormone-mediated signaling pathway | 16603237 |
| Hgene | KDM3A | GO:0030521 | androgen receptor signaling pathway | 16603237 |
| Hgene | KDM3A | GO:0033169 | histone H3-K9 demethylation | 16603237 |
| Hgene | KDM3A | GO:0046293 | formaldehyde biosynthetic process | 16603237 |
 Fusion gene breakpoints across KDM3A (5'-gene) Fusion gene breakpoints across KDM3A (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
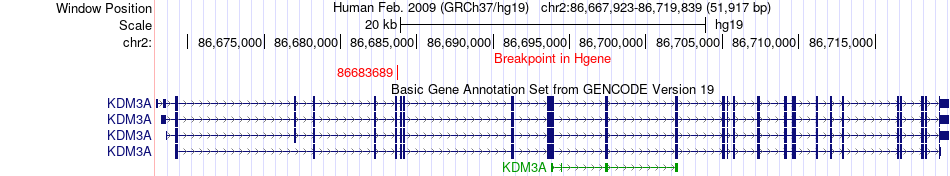 |
 Fusion gene breakpoints across SDHAF2 (3'-gene) Fusion gene breakpoints across SDHAF2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
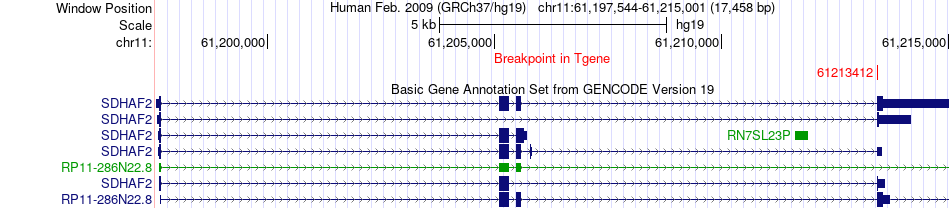 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-25-1877 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + |
Top |
Fusion Gene ORF analysis for KDM3A-SDHAF2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000312912 | ENST00000537782 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + |
| 5CDS-3UTR | ENST00000312912 | ENST00000543265 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + |
| 5CDS-3UTR | ENST00000409064 | ENST00000537782 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + |
| 5CDS-3UTR | ENST00000409064 | ENST00000543265 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + |
| 5CDS-3UTR | ENST00000409556 | ENST00000537782 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + |
| 5CDS-3UTR | ENST00000409556 | ENST00000543265 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + |
| 5CDS-3UTR | ENST00000542128 | ENST00000537782 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + |
| 5CDS-3UTR | ENST00000542128 | ENST00000543265 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + |
| 5CDS-intron | ENST00000312912 | ENST00000534878 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + |
| 5CDS-intron | ENST00000409064 | ENST00000534878 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + |
| 5CDS-intron | ENST00000409556 | ENST00000534878 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + |
| 5CDS-intron | ENST00000542128 | ENST00000534878 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + |
| Frame-shift | ENST00000312912 | ENST00000542074 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + |
| Frame-shift | ENST00000409064 | ENST00000542074 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + |
| Frame-shift | ENST00000409556 | ENST00000542074 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + |
| Frame-shift | ENST00000542128 | ENST00000542074 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + |
| In-frame | ENST00000312912 | ENST00000301761 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + |
| In-frame | ENST00000409064 | ENST00000301761 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + |
| In-frame | ENST00000409556 | ENST00000301761 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + |
| In-frame | ENST00000542128 | ENST00000301761 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + |
| intron-3CDS | ENST00000485171 | ENST00000301761 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + |
| intron-3CDS | ENST00000485171 | ENST00000542074 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + |
| intron-3UTR | ENST00000485171 | ENST00000537782 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + |
| intron-3UTR | ENST00000485171 | ENST00000543265 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + |
| intron-intron | ENST00000485171 | ENST00000534878 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000409556 | KDM3A | chr2 | 86683689 | + | ENST00000301761 | SDHAF2 | chr11 | 61213412 | + | 2635 | 1046 | 365 | 1096 | 243 |
| ENST00000312912 | KDM3A | chr2 | 86683689 | + | ENST00000301761 | SDHAF2 | chr11 | 61213412 | + | 2597 | 1008 | 327 | 1058 | 243 |
| ENST00000409064 | KDM3A | chr2 | 86683689 | + | ENST00000301761 | SDHAF2 | chr11 | 61213412 | + | 2328 | 739 | 58 | 789 | 243 |
| ENST00000542128 | KDM3A | chr2 | 86683689 | + | ENST00000301761 | SDHAF2 | chr11 | 61213412 | + | 2114 | 525 | 0 | 575 | 191 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000409556 | ENST00000301761 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + | 0.008817212 | 0.9911828 |
| ENST00000312912 | ENST00000301761 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + | 0.006809179 | 0.9931908 |
| ENST00000409064 | ENST00000301761 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + | 0.006415072 | 0.99358493 |
| ENST00000542128 | ENST00000301761 | KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + | 0.003445465 | 0.9965545 |
Top |
Fusion Genomic Features for KDM3A-SDHAF2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + | 0.013380871 | 0.9866192 |
| KDM3A | chr2 | 86683689 | + | SDHAF2 | chr11 | 61213412 | + | 0.013380871 | 0.9866192 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
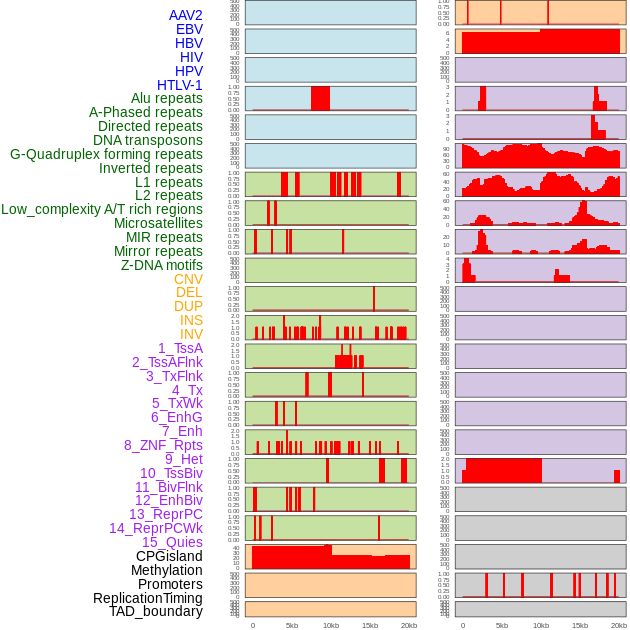 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
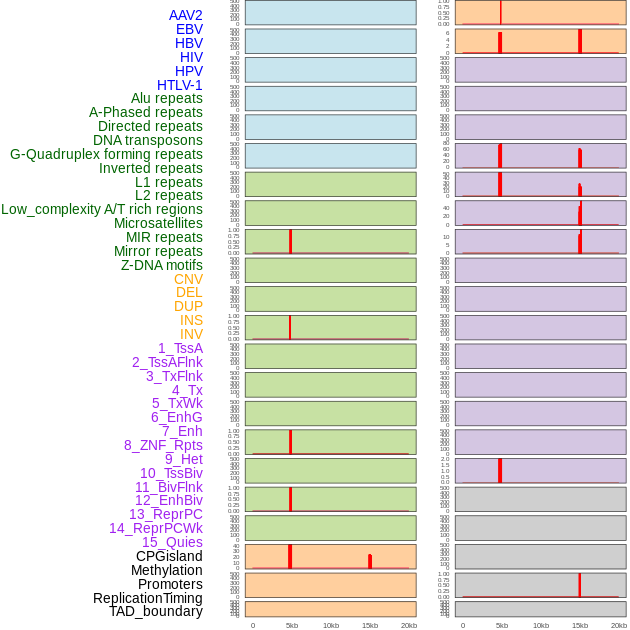 |
Top |
Fusion Protein Features for KDM3A-SDHAF2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:86683689/chr11:61213412) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| KDM3A | . |
| FUNCTION: Histone demethylase that specifically demethylates 'Lys-9' of histone H3, thereby playing a central role in histone code. Preferentially demethylates mono- and dimethylated H3 'Lys-9' residue, with a preference for dimethylated residue, while it has weak or no activity on trimethylated H3 'Lys-9'. Demethylation of Lys residue generates formaldehyde and succinate. Involved in hormone-dependent transcriptional activation, by participating in recruitment to androgen-receptor target genes, resulting in H3 'Lys-9' demethylation and transcriptional activation. Involved in spermatogenesis by regulating expression of target genes such as PRM1 and TNP1 which are required for packaging and condensation of sperm chromatin. Involved in obesity resistance through regulation of metabolic genes such as PPARA and UCP1. {ECO:0000269|PubMed:16603237, ECO:0000269|PubMed:28262558}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KDM3A | chr2:86683689 | chr11:61213412 | ENST00000312912 | + | 6 | 26 | 1058_1281 | 227 | 1322.0 | Domain | JmjC |
| Hgene | KDM3A | chr2:86683689 | chr11:61213412 | ENST00000409064 | + | 6 | 26 | 1058_1281 | 227 | 1322.0 | Domain | JmjC |
| Hgene | KDM3A | chr2:86683689 | chr11:61213412 | ENST00000409556 | + | 7 | 27 | 1058_1281 | 227 | 1322.0 | Domain | JmjC |
| Hgene | KDM3A | chr2:86683689 | chr11:61213412 | ENST00000312912 | + | 6 | 26 | 885_889 | 227 | 1322.0 | Motif | Note=LXXLL motif |
| Hgene | KDM3A | chr2:86683689 | chr11:61213412 | ENST00000409064 | + | 6 | 26 | 885_889 | 227 | 1322.0 | Motif | Note=LXXLL motif |
| Hgene | KDM3A | chr2:86683689 | chr11:61213412 | ENST00000409556 | + | 7 | 27 | 885_889 | 227 | 1322.0 | Motif | Note=LXXLL motif |
| Hgene | KDM3A | chr2:86683689 | chr11:61213412 | ENST00000312912 | + | 6 | 26 | 662_687 | 227 | 1322.0 | Zinc finger | C6-type |
| Hgene | KDM3A | chr2:86683689 | chr11:61213412 | ENST00000409064 | + | 6 | 26 | 662_687 | 227 | 1322.0 | Zinc finger | C6-type |
| Hgene | KDM3A | chr2:86683689 | chr11:61213412 | ENST00000409556 | + | 7 | 27 | 662_687 | 227 | 1322.0 | Zinc finger | C6-type |
Top |
Fusion Gene Sequence for KDM3A-SDHAF2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >41768_41768_1_KDM3A-SDHAF2_KDM3A_chr2_86683689_ENST00000312912_SDHAF2_chr11_61213412_ENST00000301761_length(transcript)=2597nt_BP=1008nt TAATGGGGGTCGCCCGGGAGTCGGAAGGGGGAGGGGAAAGGGAGGAGGCAGCCAAGGAATTGTTTTTTTCTCTGGCCCCGCCCTCGCCCG GGGGGCCAATGGTGATGATCTGTTTCCCCCGGAGCCTCGCCCAGCTCCTGTGTTTCAGCCAATGAGCGGCGGAAGCGGCTCCGAGGGGGG CGGGTCCGGGAGGCTGTGCGTGTCTTGTGAGAGCTCTTGAACCAAGTCAGCGCTGGAGTCGGCTAGGCGGCTGGAAACGGCGGCTGCCGC CGGTGACTCAGGGAGGCGGGAGGCGGGGGAGGAGCTCTTCCTGCAGGCGTGGAAACCATGGTGCTCACGCTCGGAGAAAGTTGGCCGGTA TTGGTGGGGAGGAGGTTTCTCAGTCTGTCCGCAGCCGACGGCAGCGATGGCAGCCACGACAGCTGGGACGTGGAGCGCGTCGCCGAGTGG CCCTGGCTCTCCGGGACCATTCGAGCTGTTTCCCACACCGACGTTACCAAGAAGGATCTGAAGGTGTGTGTGGAATTTGATGGGGAATCT TGGAGGAAAAGAAGATGGATAGAAGTCTACAGCCTTCTAAGGAGAGCATTTTTAGTAGAACATAATTTGGTTTTAGCTGAACGAAAGTCA CCTGAAATTTCTGAACGAATTGTACAGTGGCCTGCAATAACGTACAAACCTCTGTTGGACAAAGCTGGTTTGGGATCCATAACTTCTGTT CGCTTTCTGGGAGATCAACAAAGAGTATTTCTTTCTAAAGACCTTTTGAAGCCTATACAGGATGTAAACAGTCTTCGACTTTCTCTTACG GATAATCAGATTGTCAGTAAAGAATTTCAAGCTTTGATTGTGAAGCATTTAGATGAAAGCCATCTTTTAAAAGGTGACAAAAACTTAGTT GGTTCAGAAGTAAAAATTTATAGCTTGGACCCATCTACTCAGTGGTTTTCAGCAACCGTTATAAATGGAAACCCAGCATCAAAAACTCTT CAAGTCAACTGTGAGGAGAAGCTAAACCAGCCCCAGAAATATTTGAAAATGAAGTCATGGCCCTGCTGAGAGACTTTGCTAAAAACAAAA ACAAAGAGCAGAGACTGCGTGCCCCAGATCTTGAGTACCTCTTTGAAAAGCCACGTTGAGCTGTGCTCCACGGCCTGGCATGGGGGTTCA GTCTGTGGATGGTAACTACTTATGATGGACGTTAGCCTTGCTTCCGGCTTCTTAGATGCCCAGCTGCCCTACCCCAGACCACTGGTCCTG CCTCAAGTGATGGACATAACCCTCTCCTAGGACATACATGTAAATGCACAATGTGACTCATTCTCATACTTTTTTGTTCAGCTCTGAGCC TCAAAAGTGATTTGTCAGCAGCGCTGGCATGCAGGCTTTGCCTGGCTGCTATCTCTAGAGGCAAGTCTGCCACGAGAGGGCACTGCAGGC GAAGCAGTTGTGCTTGCTCATTGCCTCAGCCCAAGTGTACTCAAAGAAAAGAGGCAGCCAGCTGTGCGTGCTGTATGGAAAGCCTCCCGC CCTCCCTGCAGCTCCCCGCCCTCAGTGGCCCAGGGCTTTGTTGAAGTGGAACTCCCCACTTCCAACCTGGTATGGCTCCTTCTGCGAAGG GAAGCTGATCCCAGCCTCCTGAGCTGAATCCTTCAAAGGCACAGCAGGCAGAATGAGGGCCACAGGCAGGAGTGGTGTGGGCACACTGCT TAGGAGTCAACATCTTCATCATTGGGCTTTCTTTAAAACGTGCACTTTGTAATTTGAAAGAGAATTATTAAAGCATACTGAAAAAAGGAA ATTTACAATTTCCCAGCCCTCACAATTAATTTTAATTTTGTCTATTTTCATATAACCCATAGTCATGTTTATAGAGTTTTACAAGTTCAC AATTACAGGTTTTATTTTCATGTAGCACTATAGAAAACTTTCCCCCTCATTCCTACAGTCTTCACAATTAGTTGACTAAATAATAATACA TAAGAAATAATATATTAGAATGAATTAGTTAGAAGTGTTTTTAAAAGCTCTAGGGCTGTGTGACTTCTGTGTTGCTAGTGGTGCCCTCCC TGGAGGGGTTTTGCCCTCAGAGCTGTAATCTAGGCAGTAGAATCAGTCACCCAGACTGGACCAGGTGCATCTAAAGGTGTTGACTAGAAG AAGGTCGGCCAGATTTGATGAGCAGGTGGTTGGTGATTATAGGAGCCGCACACAGGGACTACACACGCACATGTTTCCAGCATTGTTGAG CTCCAGGGTTCCTACAGAGCCACCTCATGGTTCCTGCCTGCTCTTCAGTATCCCTGGTGGCTCAGGAGGGAAGGGAAGAGATCTAGCCTT TACGTAATCACAGTTAAGCCTCACAGCACTCTAGACTAGTGGTGTTACCTGTATTTTATACATAAGAAAGTGGGCTCAGAGAGAGGAAGT AATTTGTCCAAGGTCAGGTAGCTATTGAGTCGAAAAAGCAGAATTTAGTATAGCTCAGTCTGCCTCCAAAGCTCTTGAACTTTCTACTAG >41768_41768_1_KDM3A-SDHAF2_KDM3A_chr2_86683689_ENST00000312912_SDHAF2_chr11_61213412_ENST00000301761_length(amino acids)=243AA_BP=227 MVLTLGESWPVLVGRRFLSLSAADGSDGSHDSWDVERVAEWPWLSGTIRAVSHTDVTKKDLKVCVEFDGESWRKRRWIEVYSLLRRAFLV EHNLVLAERKSPEISERIVQWPAITYKPLLDKAGLGSITSVRFLGDQQRVFLSKDLLKPIQDVNSLRLSLTDNQIVSKEFQALIVKHLDE -------------------------------------------------------------- >41768_41768_2_KDM3A-SDHAF2_KDM3A_chr2_86683689_ENST00000409064_SDHAF2_chr11_61213412_ENST00000301761_length(transcript)=2328nt_BP=739nt CTCGGCTGCGGGGAGAGGGGAGAGAAAGGGAGGAGCTCTTCCTGCAGGCGTGGAAACCATGGTGCTCACGCTCGGAGAAAGTTGGCCGGT ATTGGTGGGGAGGAGGTTTCTCAGTCTGTCCGCAGCCGACGGCAGCGATGGCAGCCACGACAGCTGGGACGTGGAGCGCGTCGCCGAGTG GCCCTGGCTCTCCGGGACCATTCGAGCTGTTTCCCACACCGACGTTACCAAGAAGGATCTGAAGGTGTGTGTGGAATTTGATGGGGAATC TTGGAGGAAAAGAAGATGGATAGAAGTCTACAGCCTTCTAAGGAGAGCATTTTTAGTAGAACATAATTTGGTTTTAGCTGAACGAAAGTC ACCTGAAATTTCTGAACGAATTGTACAGTGGCCTGCAATAACGTACAAACCTCTGTTGGACAAAGCTGGTTTGGGATCCATAACTTCTGT TCGCTTTCTGGGAGATCAACAAAGAGTATTTCTTTCTAAAGACCTTTTGAAGCCTATACAGGATGTAAACAGTCTTCGACTTTCTCTTAC GGATAATCAGATTGTCAGTAAAGAATTTCAAGCTTTGATTGTGAAGCATTTAGATGAAAGCCATCTTTTAAAAGGTGACAAAAACTTAGT TGGTTCAGAAGTAAAAATTTATAGCTTGGACCCATCTACTCAGTGGTTTTCAGCAACCGTTATAAATGGAAACCCAGCATCAAAAACTCT TCAAGTCAACTGTGAGGAGAAGCTAAACCAGCCCCAGAAATATTTGAAAATGAAGTCATGGCCCTGCTGAGAGACTTTGCTAAAAACAAA AACAAAGAGCAGAGACTGCGTGCCCCAGATCTTGAGTACCTCTTTGAAAAGCCACGTTGAGCTGTGCTCCACGGCCTGGCATGGGGGTTC AGTCTGTGGATGGTAACTACTTATGATGGACGTTAGCCTTGCTTCCGGCTTCTTAGATGCCCAGCTGCCCTACCCCAGACCACTGGTCCT GCCTCAAGTGATGGACATAACCCTCTCCTAGGACATACATGTAAATGCACAATGTGACTCATTCTCATACTTTTTTGTTCAGCTCTGAGC CTCAAAAGTGATTTGTCAGCAGCGCTGGCATGCAGGCTTTGCCTGGCTGCTATCTCTAGAGGCAAGTCTGCCACGAGAGGGCACTGCAGG CGAAGCAGTTGTGCTTGCTCATTGCCTCAGCCCAAGTGTACTCAAAGAAAAGAGGCAGCCAGCTGTGCGTGCTGTATGGAAAGCCTCCCG CCCTCCCTGCAGCTCCCCGCCCTCAGTGGCCCAGGGCTTTGTTGAAGTGGAACTCCCCACTTCCAACCTGGTATGGCTCCTTCTGCGAAG GGAAGCTGATCCCAGCCTCCTGAGCTGAATCCTTCAAAGGCACAGCAGGCAGAATGAGGGCCACAGGCAGGAGTGGTGTGGGCACACTGC TTAGGAGTCAACATCTTCATCATTGGGCTTTCTTTAAAACGTGCACTTTGTAATTTGAAAGAGAATTATTAAAGCATACTGAAAAAAGGA AATTTACAATTTCCCAGCCCTCACAATTAATTTTAATTTTGTCTATTTTCATATAACCCATAGTCATGTTTATAGAGTTTTACAAGTTCA CAATTACAGGTTTTATTTTCATGTAGCACTATAGAAAACTTTCCCCCTCATTCCTACAGTCTTCACAATTAGTTGACTAAATAATAATAC ATAAGAAATAATATATTAGAATGAATTAGTTAGAAGTGTTTTTAAAAGCTCTAGGGCTGTGTGACTTCTGTGTTGCTAGTGGTGCCCTCC CTGGAGGGGTTTTGCCCTCAGAGCTGTAATCTAGGCAGTAGAATCAGTCACCCAGACTGGACCAGGTGCATCTAAAGGTGTTGACTAGAA GAAGGTCGGCCAGATTTGATGAGCAGGTGGTTGGTGATTATAGGAGCCGCACACAGGGACTACACACGCACATGTTTCCAGCATTGTTGA GCTCCAGGGTTCCTACAGAGCCACCTCATGGTTCCTGCCTGCTCTTCAGTATCCCTGGTGGCTCAGGAGGGAAGGGAAGAGATCTAGCCT TTACGTAATCACAGTTAAGCCTCACAGCACTCTAGACTAGTGGTGTTACCTGTATTTTATACATAAGAAAGTGGGCTCAGAGAGAGGAAG TAATTTGTCCAAGGTCAGGTAGCTATTGAGTCGAAAAAGCAGAATTTAGTATAGCTCAGTCTGCCTCCAAAGCTCTTGAACTTTCTACTA >41768_41768_2_KDM3A-SDHAF2_KDM3A_chr2_86683689_ENST00000409064_SDHAF2_chr11_61213412_ENST00000301761_length(amino acids)=243AA_BP=227 MVLTLGESWPVLVGRRFLSLSAADGSDGSHDSWDVERVAEWPWLSGTIRAVSHTDVTKKDLKVCVEFDGESWRKRRWIEVYSLLRRAFLV EHNLVLAERKSPEISERIVQWPAITYKPLLDKAGLGSITSVRFLGDQQRVFLSKDLLKPIQDVNSLRLSLTDNQIVSKEFQALIVKHLDE -------------------------------------------------------------- >41768_41768_3_KDM3A-SDHAF2_KDM3A_chr2_86683689_ENST00000409556_SDHAF2_chr11_61213412_ENST00000301761_length(transcript)=2635nt_BP=1046nt ACTATCGCTCCTCCCTCTAGCTGAGTACGCCTGAAACCGCACGTTCTCTTCCCTGCCCTCCCCCGCTAACCCTGCACCGCCCTCACCCTT TCCTGTGAGATTCTTCCGCCAAGTGGAAGGCTCATCTTCGGTCGACAGCCTACGCGGTTGAAGAACAATCCACCTCGCCCAGCTCCTGTG TTTCAGCCAATGAGCGGCGGAAGCGGCTCCGAGGGGGGCGGGTCCGGGAGGCTGTGCGTGTCTTGTGAGAGCTCTTGAACCAAGTCAGCG CTGGAGTCGGCTAGGCGGCTGGAAACGGCGGCTGCCGCCGGTGACTCAGGGAGGCGGGAGGCGGGGGAGGAGCTCTTCCTGCAGGCGTGG AAACCATGGTGCTCACGCTCGGAGAAAGTTGGCCGGTATTGGTGGGGAGGAGGTTTCTCAGTCTGTCCGCAGCCGACGGCAGCGATGGCA GCCACGACAGCTGGGACGTGGAGCGCGTCGCCGAGTGGCCCTGGCTCTCCGGGACCATTCGAGCTGTTTCCCACACCGACGTTACCAAGA AGGATCTGAAGGTGTGTGTGGAATTTGATGGGGAATCTTGGAGGAAAAGAAGATGGATAGAAGTCTACAGCCTTCTAAGGAGAGCATTTT TAGTAGAACATAATTTGGTTTTAGCTGAACGAAAGTCACCTGAAATTTCTGAACGAATTGTACAGTGGCCTGCAATAACGTACAAACCTC TGTTGGACAAAGCTGGTTTGGGATCCATAACTTCTGTTCGCTTTCTGGGAGATCAACAAAGAGTATTTCTTTCTAAAGACCTTTTGAAGC CTATACAGGATGTAAACAGTCTTCGACTTTCTCTTACGGATAATCAGATTGTCAGTAAAGAATTTCAAGCTTTGATTGTGAAGCATTTAG ATGAAAGCCATCTTTTAAAAGGTGACAAAAACTTAGTTGGTTCAGAAGTAAAAATTTATAGCTTGGACCCATCTACTCAGTGGTTTTCAG CAACCGTTATAAATGGAAACCCAGCATCAAAAACTCTTCAAGTCAACTGTGAGGAGAAGCTAAACCAGCCCCAGAAATATTTGAAAATGA AGTCATGGCCCTGCTGAGAGACTTTGCTAAAAACAAAAACAAAGAGCAGAGACTGCGTGCCCCAGATCTTGAGTACCTCTTTGAAAAGCC ACGTTGAGCTGTGCTCCACGGCCTGGCATGGGGGTTCAGTCTGTGGATGGTAACTACTTATGATGGACGTTAGCCTTGCTTCCGGCTTCT TAGATGCCCAGCTGCCCTACCCCAGACCACTGGTCCTGCCTCAAGTGATGGACATAACCCTCTCCTAGGACATACATGTAAATGCACAAT GTGACTCATTCTCATACTTTTTTGTTCAGCTCTGAGCCTCAAAAGTGATTTGTCAGCAGCGCTGGCATGCAGGCTTTGCCTGGCTGCTAT CTCTAGAGGCAAGTCTGCCACGAGAGGGCACTGCAGGCGAAGCAGTTGTGCTTGCTCATTGCCTCAGCCCAAGTGTACTCAAAGAAAAGA GGCAGCCAGCTGTGCGTGCTGTATGGAAAGCCTCCCGCCCTCCCTGCAGCTCCCCGCCCTCAGTGGCCCAGGGCTTTGTTGAAGTGGAAC TCCCCACTTCCAACCTGGTATGGCTCCTTCTGCGAAGGGAAGCTGATCCCAGCCTCCTGAGCTGAATCCTTCAAAGGCACAGCAGGCAGA ATGAGGGCCACAGGCAGGAGTGGTGTGGGCACACTGCTTAGGAGTCAACATCTTCATCATTGGGCTTTCTTTAAAACGTGCACTTTGTAA TTTGAAAGAGAATTATTAAAGCATACTGAAAAAAGGAAATTTACAATTTCCCAGCCCTCACAATTAATTTTAATTTTGTCTATTTTCATA TAACCCATAGTCATGTTTATAGAGTTTTACAAGTTCACAATTACAGGTTTTATTTTCATGTAGCACTATAGAAAACTTTCCCCCTCATTC CTACAGTCTTCACAATTAGTTGACTAAATAATAATACATAAGAAATAATATATTAGAATGAATTAGTTAGAAGTGTTTTTAAAAGCTCTA GGGCTGTGTGACTTCTGTGTTGCTAGTGGTGCCCTCCCTGGAGGGGTTTTGCCCTCAGAGCTGTAATCTAGGCAGTAGAATCAGTCACCC AGACTGGACCAGGTGCATCTAAAGGTGTTGACTAGAAGAAGGTCGGCCAGATTTGATGAGCAGGTGGTTGGTGATTATAGGAGCCGCACA CAGGGACTACACACGCACATGTTTCCAGCATTGTTGAGCTCCAGGGTTCCTACAGAGCCACCTCATGGTTCCTGCCTGCTCTTCAGTATC CCTGGTGGCTCAGGAGGGAAGGGAAGAGATCTAGCCTTTACGTAATCACAGTTAAGCCTCACAGCACTCTAGACTAGTGGTGTTACCTGT ATTTTATACATAAGAAAGTGGGCTCAGAGAGAGGAAGTAATTTGTCCAAGGTCAGGTAGCTATTGAGTCGAAAAAGCAGAATTTAGTATA GCTCAGTCTGCCTCCAAAGCTCTTGAACTTTCTACTAGAACACACTGCCTTACTGCTGGAAATTTAATAGAAGTCTCTGTTTGGGAAAGC >41768_41768_3_KDM3A-SDHAF2_KDM3A_chr2_86683689_ENST00000409556_SDHAF2_chr11_61213412_ENST00000301761_length(amino acids)=243AA_BP=227 MVLTLGESWPVLVGRRFLSLSAADGSDGSHDSWDVERVAEWPWLSGTIRAVSHTDVTKKDLKVCVEFDGESWRKRRWIEVYSLLRRAFLV EHNLVLAERKSPEISERIVQWPAITYKPLLDKAGLGSITSVRFLGDQQRVFLSKDLLKPIQDVNSLRLSLTDNQIVSKEFQALIVKHLDE -------------------------------------------------------------- >41768_41768_4_KDM3A-SDHAF2_KDM3A_chr2_86683689_ENST00000542128_SDHAF2_chr11_61213412_ENST00000301761_length(transcript)=2114nt_BP=525nt ATGGTGCTCACGCTCGGAGAAAGTTGGCCGGTATTGGTGGGGAGGAGGTTTCTCAGTCTGTCCGCAGCCGACGGCAGCGATGGCAGCCAC GACAGCTGGGACGTGGAGCGCGTCGCCGAGTGGCCCTGGCTCTCCGGGACCATTCGAGCTGTTTCCCACACCGACGTTACCAAGAAGGAT CTGAAGACGTACAAACCTCTGTTGGACAAAGCTGGTTTGGGATCCATAACTTCTGTTCGCTTTCTGGGAGATCAACAAAGAGTATTTCTT TCTAAAGACCTTTTGAAGCCTATACAGGATGTAAACAGTCTTCGACTTTCTCTTACGGATAATCAGATTGTCAGTAAAGAATTTCAAGCT TTGATTGTGAAGCATTTAGATGAAAGCCATCTTTTAAAAGGTGACAAAAACTTAGTTGGTTCAGAAGTAAAAATTTATAGCTTGGACCCA TCTACTCAGTGGTTTTCAGCAACCGTTATAAATGGAAACCCAGCATCAAAAACTCTTCAAGTCAACTGTGAGGAGAAGCTAAACCAGCCC CAGAAATATTTGAAAATGAAGTCATGGCCCTGCTGAGAGACTTTGCTAAAAACAAAAACAAAGAGCAGAGACTGCGTGCCCCAGATCTTG AGTACCTCTTTGAAAAGCCACGTTGAGCTGTGCTCCACGGCCTGGCATGGGGGTTCAGTCTGTGGATGGTAACTACTTATGATGGACGTT AGCCTTGCTTCCGGCTTCTTAGATGCCCAGCTGCCCTACCCCAGACCACTGGTCCTGCCTCAAGTGATGGACATAACCCTCTCCTAGGAC ATACATGTAAATGCACAATGTGACTCATTCTCATACTTTTTTGTTCAGCTCTGAGCCTCAAAAGTGATTTGTCAGCAGCGCTGGCATGCA GGCTTTGCCTGGCTGCTATCTCTAGAGGCAAGTCTGCCACGAGAGGGCACTGCAGGCGAAGCAGTTGTGCTTGCTCATTGCCTCAGCCCA AGTGTACTCAAAGAAAAGAGGCAGCCAGCTGTGCGTGCTGTATGGAAAGCCTCCCGCCCTCCCTGCAGCTCCCCGCCCTCAGTGGCCCAG GGCTTTGTTGAAGTGGAACTCCCCACTTCCAACCTGGTATGGCTCCTTCTGCGAAGGGAAGCTGATCCCAGCCTCCTGAGCTGAATCCTT CAAAGGCACAGCAGGCAGAATGAGGGCCACAGGCAGGAGTGGTGTGGGCACACTGCTTAGGAGTCAACATCTTCATCATTGGGCTTTCTT TAAAACGTGCACTTTGTAATTTGAAAGAGAATTATTAAAGCATACTGAAAAAAGGAAATTTACAATTTCCCAGCCCTCACAATTAATTTT AATTTTGTCTATTTTCATATAACCCATAGTCATGTTTATAGAGTTTTACAAGTTCACAATTACAGGTTTTATTTTCATGTAGCACTATAG AAAACTTTCCCCCTCATTCCTACAGTCTTCACAATTAGTTGACTAAATAATAATACATAAGAAATAATATATTAGAATGAATTAGTTAGA AGTGTTTTTAAAAGCTCTAGGGCTGTGTGACTTCTGTGTTGCTAGTGGTGCCCTCCCTGGAGGGGTTTTGCCCTCAGAGCTGTAATCTAG GCAGTAGAATCAGTCACCCAGACTGGACCAGGTGCATCTAAAGGTGTTGACTAGAAGAAGGTCGGCCAGATTTGATGAGCAGGTGGTTGG TGATTATAGGAGCCGCACACAGGGACTACACACGCACATGTTTCCAGCATTGTTGAGCTCCAGGGTTCCTACAGAGCCACCTCATGGTTC CTGCCTGCTCTTCAGTATCCCTGGTGGCTCAGGAGGGAAGGGAAGAGATCTAGCCTTTACGTAATCACAGTTAAGCCTCACAGCACTCTA GACTAGTGGTGTTACCTGTATTTTATACATAAGAAAGTGGGCTCAGAGAGAGGAAGTAATTTGTCCAAGGTCAGGTAGCTATTGAGTCGA AAAAGCAGAATTTAGTATAGCTCAGTCTGCCTCCAAAGCTCTTGAACTTTCTACTAGAACACACTGCCTTACTGCTGGAAATTTAATAGA >41768_41768_4_KDM3A-SDHAF2_KDM3A_chr2_86683689_ENST00000542128_SDHAF2_chr11_61213412_ENST00000301761_length(amino acids)=191AA_BP=175 MVLTLGESWPVLVGRRFLSLSAADGSDGSHDSWDVERVAEWPWLSGTIRAVSHTDVTKKDLKTYKPLLDKAGLGSITSVRFLGDQQRVFL SKDLLKPIQDVNSLRLSLTDNQIVSKEFQALIVKHLDESHLLKGDKNLVGSEVKIYSLDPSTQWFSATVINGNPASKTLQVNCEEKLNQP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for KDM3A-SDHAF2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for KDM3A-SDHAF2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for KDM3A-SDHAF2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
