
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:KDM4B-CHAF1A (FusionGDB2 ID:41799) |
Fusion Gene Summary for KDM4B-CHAF1A |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: KDM4B-CHAF1A | Fusion gene ID: 41799 | Hgene | Tgene | Gene symbol | KDM4B | CHAF1A | Gene ID | 23030 | 10036 |
| Gene name | lysine demethylase 4B | chromatin assembly factor 1 subunit A | |
| Synonyms | JMJD2B|TDRD14B | CAF-1|CAF1|CAF1B|CAF1P150|P150 | |
| Cytomap | 19p13.3 | 19p13.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | lysine-specific demethylase 4BjmjC domain-containing histone demethylation protein 3Bjumonji domain containing 2Bjumonji domain-containing protein 2Blysine (K)-specific demethylase 4Btudor domain containing 14B | chromatin assembly factor 1 subunit ACAF-1 subunit ACAF-I 150 kDa subunitCAF-I p150CTB-50L17.7chromatin assembly factor I (150 kDa)chromatin assembly factor I p150 subunithp150 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | O94953 | Q13111 | |
| Ensembl transtripts involved in fusion gene | ENST00000159111, ENST00000381759, ENST00000536461, ENST00000592175, | ENST00000587368, ENST00000301280, | |
| Fusion gene scores | * DoF score | 30 X 16 X 14=6720 | 5 X 6 X 6=180 |
| # samples | 38 | 7 | |
| ** MAII score | log2(38/6720*10)=-4.14438990933518 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/180*10)=-1.36257007938471 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: KDM4B [Title/Abstract] AND CHAF1A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | KDM4B(5082515)-CHAF1A(4418017), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | KDM4B | GO:0033169 | histone H3-K9 demethylation | 21914792 |
| Hgene | KDM4B | GO:0070544 | histone H3-K36 demethylation | 21914792 |
| Tgene | CHAF1A | GO:0006335 | DNA replication-dependent nucleosome assembly | 14718166 |
| Tgene | CHAF1A | GO:0031497 | chromatin assembly | 8858152 |
 Fusion gene breakpoints across KDM4B (5'-gene) Fusion gene breakpoints across KDM4B (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
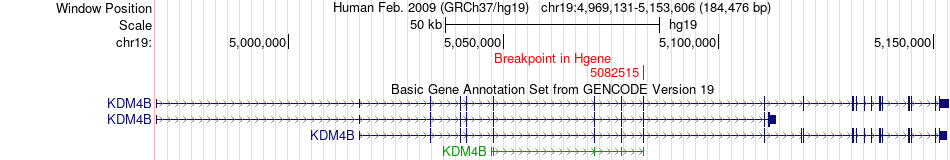 |
 Fusion gene breakpoints across CHAF1A (3'-gene) Fusion gene breakpoints across CHAF1A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
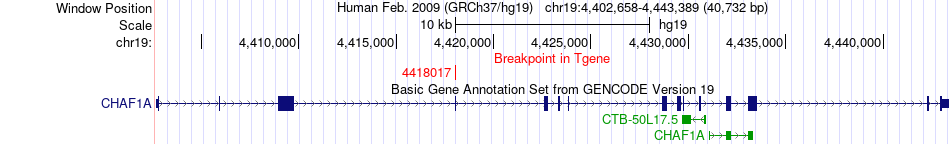 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LGG | TCGA-DU-8163-01A | KDM4B | chr19 | 5082515 | - | CHAF1A | chr19 | 4418017 | + |
| ChimerDB4 | LGG | TCGA-DU-8163-01A | KDM4B | chr19 | 5082515 | + | CHAF1A | chr19 | 4418017 | + |
Top |
Fusion Gene ORF analysis for KDM4B-CHAF1A |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000159111 | ENST00000587368 | KDM4B | chr19 | 5082515 | + | CHAF1A | chr19 | 4418017 | + |
| 5CDS-intron | ENST00000381759 | ENST00000587368 | KDM4B | chr19 | 5082515 | + | CHAF1A | chr19 | 4418017 | + |
| 5CDS-intron | ENST00000536461 | ENST00000587368 | KDM4B | chr19 | 5082515 | + | CHAF1A | chr19 | 4418017 | + |
| In-frame | ENST00000159111 | ENST00000301280 | KDM4B | chr19 | 5082515 | + | CHAF1A | chr19 | 4418017 | + |
| In-frame | ENST00000381759 | ENST00000301280 | KDM4B | chr19 | 5082515 | + | CHAF1A | chr19 | 4418017 | + |
| In-frame | ENST00000536461 | ENST00000301280 | KDM4B | chr19 | 5082515 | + | CHAF1A | chr19 | 4418017 | + |
| intron-3CDS | ENST00000592175 | ENST00000301280 | KDM4B | chr19 | 5082515 | + | CHAF1A | chr19 | 4418017 | + |
| intron-intron | ENST00000592175 | ENST00000587368 | KDM4B | chr19 | 5082515 | + | CHAF1A | chr19 | 4418017 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000159111 | KDM4B | chr19 | 5082515 | + | ENST00000301280 | CHAF1A | chr19 | 4418017 | + | 3414 | 1136 | 218 | 3046 | 942 |
| ENST00000381759 | KDM4B | chr19 | 5082515 | + | ENST00000301280 | CHAF1A | chr19 | 4418017 | + | 3403 | 1125 | 207 | 3035 | 942 |
| ENST00000536461 | KDM4B | chr19 | 5082515 | + | ENST00000301280 | CHAF1A | chr19 | 4418017 | + | 3236 | 958 | 40 | 2868 | 942 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000159111 | ENST00000301280 | KDM4B | chr19 | 5082515 | + | CHAF1A | chr19 | 4418017 | + | 0.001380299 | 0.9986197 |
| ENST00000381759 | ENST00000301280 | KDM4B | chr19 | 5082515 | + | CHAF1A | chr19 | 4418017 | + | 0.001337451 | 0.9986626 |
| ENST00000536461 | ENST00000301280 | KDM4B | chr19 | 5082515 | + | CHAF1A | chr19 | 4418017 | + | 0.00129263 | 0.9987074 |
Top |
Fusion Genomic Features for KDM4B-CHAF1A |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| KDM4B | chr19 | 5082515 | + | CHAF1A | chr19 | 4418016 | + | 4.92E-05 | 0.99995077 |
| KDM4B | chr19 | 5082515 | + | CHAF1A | chr19 | 4418016 | + | 4.92E-05 | 0.99995077 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
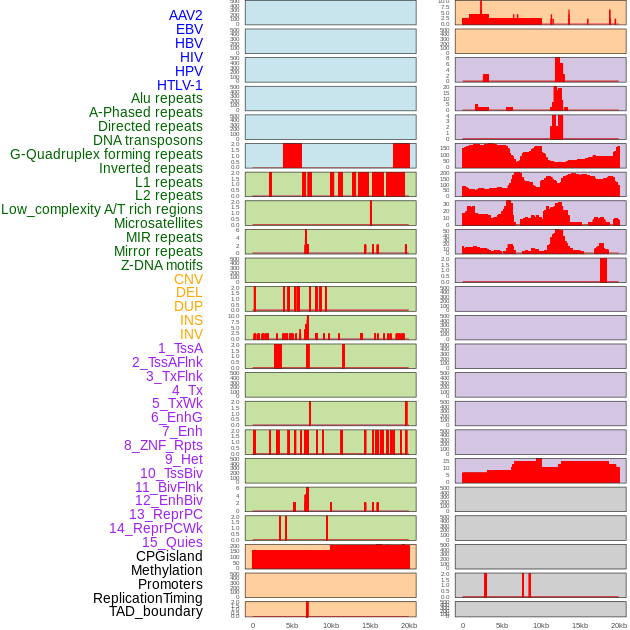 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
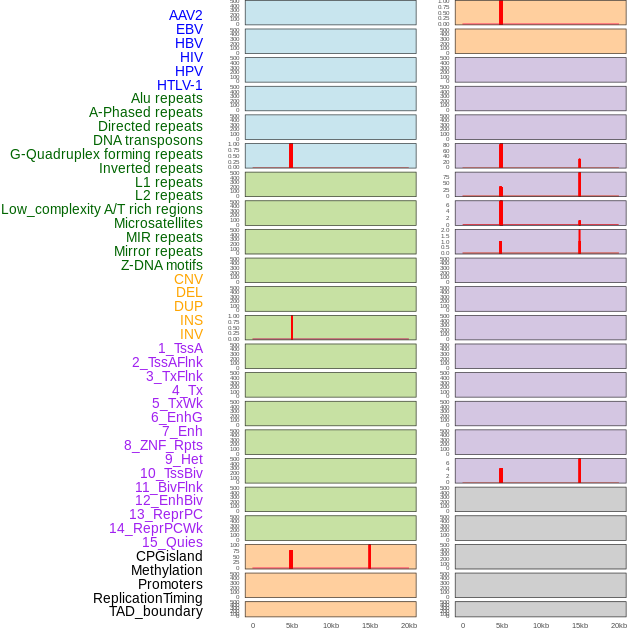 |
Top |
Fusion Protein Features for KDM4B-CHAF1A |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:5082515/chr19:4418017) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| KDM4B | CHAF1A |
| FUNCTION: Histone demethylase that specifically demethylates 'Lys-9' of histone H3, thereby playing a role in histone code. Does not demethylate histone H3 'Lys-4', H3 'Lys-27', H3 'Lys-36' nor H4 'Lys-20'. Only able to demethylate trimethylated H3 'Lys-9', with a weaker activity than KDM4A, KDM4C and KDM4D. Demethylation of Lys residue generates formaldehyde and succinate. {ECO:0000269|PubMed:16603238, ECO:0000269|PubMed:28262558}. | FUNCTION: Core component of the CAF-1 complex, a complex that is thought to mediate chromatin assembly in DNA replication and DNA repair. Assembles histone octamers onto replicating DNA in vitro. CAF-1 performs the first step of the nucleosome assembly process, bringing newly synthesized histones H3 and H4 to replicating DNA; histones H2A/H2B can bind to this chromatin precursor subsequent to DNA replication to complete the histone octamer. It may play a role in heterochromatin maintenance in proliferating cells by bringing newly synthesized cbx proteins to heterochromatic DNA replication foci. {ECO:0000250|UniProtKB:Q5R1T0}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KDM4B | chr19:5082515 | chr19:4418017 | ENST00000159111 | + | 9 | 23 | 146_309 | 306 | 1097.0 | Domain | JmjC |
| Hgene | KDM4B | chr19:5082515 | chr19:4418017 | ENST00000159111 | + | 9 | 23 | 15_57 | 306 | 1097.0 | Domain | JmjN |
| Hgene | KDM4B | chr19:5082515 | chr19:4418017 | ENST00000381759 | + | 9 | 12 | 146_309 | 306 | 449.0 | Domain | JmjC |
| Hgene | KDM4B | chr19:5082515 | chr19:4418017 | ENST00000381759 | + | 9 | 12 | 15_57 | 306 | 449.0 | Domain | JmjN |
| Tgene | CHAF1A | chr19:5082515 | chr19:4418017 | ENST00000301280 | 2 | 15 | 323_453 | 320 | 957.0 | Compositional bias | Note=Arg/Glu/Lys-rich | |
| Tgene | CHAF1A | chr19:5082515 | chr19:4418017 | ENST00000301280 | 2 | 15 | 602_608 | 320 | 957.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | CHAF1A | chr19:5082515 | chr19:4418017 | ENST00000301280 | 2 | 15 | 619_623 | 320 | 957.0 | Compositional bias | Note=Poly-Asp | |
| Tgene | CHAF1A | chr19:5082515 | chr19:4418017 | ENST00000301280 | 2 | 15 | 905_911 | 320 | 957.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | CHAF1A | chr19:5082515 | chr19:4418017 | ENST00000301280 | 2 | 15 | 642_678 | 320 | 957.0 | Region | Note=Necessary for homodimerization and competence for chromatin assembly | |
| Tgene | CHAF1A | chr19:5082515 | chr19:4418017 | ENST00000301280 | 2 | 15 | 660_956 | 320 | 957.0 | Region | Note=Binds to p60 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KDM4B | chr19:5082515 | chr19:4418017 | ENST00000159111 | + | 9 | 23 | 463_535 | 306 | 1097.0 | Compositional bias | Note=Pro-rich |
| Hgene | KDM4B | chr19:5082515 | chr19:4418017 | ENST00000381759 | + | 9 | 12 | 463_535 | 306 | 449.0 | Compositional bias | Note=Pro-rich |
| Hgene | KDM4B | chr19:5082515 | chr19:4418017 | ENST00000159111 | + | 9 | 23 | 917_974 | 306 | 1097.0 | Domain | Note=Tudor 1 |
| Hgene | KDM4B | chr19:5082515 | chr19:4418017 | ENST00000159111 | + | 9 | 23 | 975_1031 | 306 | 1097.0 | Domain | Note=Tudor 2 |
| Hgene | KDM4B | chr19:5082515 | chr19:4418017 | ENST00000381759 | + | 9 | 12 | 917_974 | 306 | 449.0 | Domain | Note=Tudor 1 |
| Hgene | KDM4B | chr19:5082515 | chr19:4418017 | ENST00000381759 | + | 9 | 12 | 975_1031 | 306 | 449.0 | Domain | Note=Tudor 2 |
| Hgene | KDM4B | chr19:5082515 | chr19:4418017 | ENST00000159111 | + | 9 | 23 | 731_789 | 306 | 1097.0 | Zinc finger | Note=PHD-type 1 |
| Hgene | KDM4B | chr19:5082515 | chr19:4418017 | ENST00000159111 | + | 9 | 23 | 794_827 | 306 | 1097.0 | Zinc finger | C2HC pre-PHD-type |
| Hgene | KDM4B | chr19:5082515 | chr19:4418017 | ENST00000159111 | + | 9 | 23 | 850_907 | 306 | 1097.0 | Zinc finger | PHD-type 2 |
| Hgene | KDM4B | chr19:5082515 | chr19:4418017 | ENST00000381759 | + | 9 | 12 | 731_789 | 306 | 449.0 | Zinc finger | Note=PHD-type 1 |
| Hgene | KDM4B | chr19:5082515 | chr19:4418017 | ENST00000381759 | + | 9 | 12 | 794_827 | 306 | 449.0 | Zinc finger | C2HC pre-PHD-type |
| Hgene | KDM4B | chr19:5082515 | chr19:4418017 | ENST00000381759 | + | 9 | 12 | 850_907 | 306 | 449.0 | Zinc finger | PHD-type 2 |
| Tgene | CHAF1A | chr19:5082515 | chr19:4418017 | ENST00000301280 | 2 | 15 | 233_246 | 320 | 957.0 | Motif | Note=PxVxL motif | |
| Tgene | CHAF1A | chr19:5082515 | chr19:4418017 | ENST00000301280 | 2 | 15 | 1_314 | 320 | 957.0 | Region | Note=Binds to CBX1 chromo shadow domain | |
| Tgene | CHAF1A | chr19:5082515 | chr19:4418017 | ENST00000301280 | 2 | 15 | 1_49 | 320 | 957.0 | Region | Note=Binds to PCNA |
Top |
Fusion Gene Sequence for KDM4B-CHAF1A |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >41799_41799_1_KDM4B-CHAF1A_KDM4B_chr19_5082515_ENST00000159111_CHAF1A_chr19_4418017_ENST00000301280_length(transcript)=3414nt_BP=1136nt GTCGCCAGCAACCGAGCGGGGCCCGGCCCGAGCGGGGCCTGGGGGTGCGACGCCGAGGGCGGGGGAGAGCGCGCCGCTGCTCCCGGACCG GGCCGCGCACGCCGCCTCAGGAACCATCACTGTTGCTGGAGGCACCTGACAAATCCTAGCGAATTTTTGGAGCATCTCCACCCAGGAACC TCGCCATCCAGAAGTGTGCTTCCCGCACAGCTGCAGCCATGGGGTCTGAGGACCACGGCGCCCAGAACCCCAGCTGTAAAATCATGACGT TTCGCCCAACCATGGAAGAATTTAAAGACTTCAACAAATACGTGGCCTACATAGAGTCGCAGGGAGCCCACCGGGCGGGCCTGGCCAAGA TCATCCCCCCGAAGGAGTGGAAGCCGCGGCAGACGTATGATGACATCGACGACGTGGTGATCCCGGCGCCCATCCAGCAGGTGGTGACGG GCCAGTCGGGCCTCTTCACGCAGTACAATATCCAGAAGAAGGCCATGACAGTGGGCGAGTACCGCCGCCTGGCCAACAGCGAGAAGTACT GTACCCCGCGGCACCAGGACTTTGATGACCTTGAACGCAAATACTGGAAGAACCTCACCTTTGTCTCCCCGATCTACGGGGCTGACATCA GCGGCTCTTTGTATGATGACGACGTGGCCCAGTGGAACATCGGGAGCCTCCGGACCATCCTGGACATGGTGGAGCGCGAGTGCGGCACCA TCATCGAGGGCGTGAACACGCCCTACCTGTACTTCGGCATGTGGAAGACCACCTTCGCCTGGCACACCGAGGACATGGACCTGTACAGCA TCAACTACCTGCACTTTGGGGAGCCTAAGTCCTGGTACGCCATCCCACCAGAGCACGGCAAGCGCCTGGAGCGGCTGGCCATCGGCTTCT TCCCCGGGAGCTCGCAGGGCTGCGACGCCTTCCTGCGGCATAAGATGACCCTCATCTCGCCCATCATCCTGAAGAAGTACGGGATCCCCT TCAGCCGGATCACGCAGGAGGCCGGGGAATTCATGATCACATTTCCCTACGGCTACCACGCCGGCTTCAATCACGGGTTCAACTGCGCAG AATCTACCAACTTCGCCACCCTGCGGTGGATTGACTACGGCAAAGTGGCCACTCAGATAACTAAGAAATTCGTCAAAGGCTCTACAGAGA AGAACAAGCTCAGACTGCAAAGAGATCAGGAGCGTCTGGGCAAGCAGCTCAAGTTACGTGCAGAAAGGGAAGAAAAGGAGAAGCTGAAAG AGGAGGCCAAGCGGGCCAAGGAGGAGGCCAAGAAGAAGAAGGAGGAAGAGAAGGAGCTTAAGGAAAAGGAGAGGCGGGAGAAGCGGGAGA AGGATGAGAAGGAGAAGGCGGAGAAGCAGCGGCTCAAGGAGGAGCGGCGCAAGGAGAGACAGGAAGCCCTGGAGGCTAAACTTGAGGAAA AAAGGAAAAAGGAAGAAGAGAAACGGTTAAGAGAAGAAGAGAAGCGCATTAAAGCAGAGAAGGCCGAAATCACGAGGTTCTTCCAGAAAC CAAAGACTCCACAGGCCCCCAAGACCCTGGCCGGCTCCTGTGGGAAGTTTGCCCCCTTTGAAATTAAAGAGCACATGGTCCTGGCCCCTC GGCGTCGGACCGCTTTCCATCCAGACCTCTGCAGTCAGCTGGACCAGCTCCTCCAGCAGCAGAGCGGCGAGTTCTCCTTCTTGAAAGACC TCAAAGGCCGGCAGCCCCTGAGGTCCGGACCCACGCACGTTTCCACCCGGAATGCAGATATTTTTAACAGTGATGTCGTCATCGTGGAGC GTGGGAAGGGCGACGGTGTTCCCGAGAGGAGGAAGTTTGGCAGGATGAAGCTCCTGCAGTTCTGTGAGAACCACCGGCCTGCCTACTGGG GTACCTGGAATAAGAAGACGGCACTCATCCGCGCGCGAGACCCCTGGGCCCAGGACACGAAGCTCCTGGACTATGAGGTGGACAGTGATG AGGAGTGGGAAGAAGAGGAGCCTGGGGAGTCCCTGTCCCACAGTGAGGGGGATGATGATGACGACATGGGAGAGGATGAAGATGAGGACG ATGGTTTCTTTGTGCCCCATGGGTACCTGTCTGAGGACGAAGGTGTGACAGAGGAGTGTGCCGACCCTGAGAACCATAAGGTCCGCCAGA AACTGAAGGCCAAGGAGTGGGACGAGTTCCTGGCTAAGGGGAAGCGCTTTCGCGTCCTGCAACCTGTGAAGATCGGCTGCGTGTGGGCGG CTGACAGAGACTGCGCAGGCGATGACCTGAAGGTACTGCAGCAGTTCGCAGCCTGCTTCCTGGAGACCCTGCCGGCCCAGGAGGAGCAGA CGCCCAAGGCCTCCAAGCGGGAGAGGAGAGACGAGCAGATCCTGGCCCAGCTGCTGCCGCTCCTGCACGGCAATGTGAACGGGAGCAAGG TCATCATCCGGGAGTTCCAGGAGCACTGCCGCCGGGGACTGCTCAGCAACCACACCGGCAGCCCGCGGAGCCCCTCCACCACCTACCTGC ACACCCCCACCCCCAGCGAGGATGCCGCCATCCCCTCTAAGTCCCGGCTCAAGCGGCTCATTTCCGAGAACTCAGTGTATGAGAAGCGGC CTGACTTCAGGATGTGCTGGTACGTGCACCCGCAGGTGCTACAGAGCTTCCAGCAGGAGCACCTGCCCGTGCCGTGCCAGTGGAGCTATG TGACATCGGTGCCCTCGGCCCCCAAAGAGGACAGTGGCAGCGTCCCCTCCACGGGGCCCAGCCAGGGCACTCCCATCTCGCTGAAGAGGA AGTCAGCGGGCAGCATGTGCATCACCCAATTCATGAAGAAGCGCAGGCACGACGGCCAGATTGGTGCTGAAGACATGGACGGCTTCCAGG CAGACACGGAGGAGGAGGAAGAGGAGGAGGGCGACTGTATGATCGTGGATGTCCCGGATGCTGCGGAGGTCCAAGCCCCGTGTGGAGCCG CTTCCGGAGCTGGGGGTGGTGTGGGGGTGGACACCGGCAAGGCCACCCTGACCTCGAGCCCACTGGGTGCATCCTGAGAGCAGGGGTGAC GTATGTAGAATGCTTAGGGTGTCCTCCCCACAGAGCAGATACTTGAACCGACTCAATTCCTGTGTAAAGAGCACTTTGTCCTGCTTCACG GACCTCCCCAAAGTGTGCAGAGTTCTATATAGGATGCTGGATTAGTTCCTTTGATATTTGTAAAAATTCCCCCAAGAGCCGCATATGAAT CTGCCCTTTAATAAAGCATTATTGAGATTGCTGGCCTATTGGGGAAGCCTGCGGGCACAGGAGCAGGCGTGGAATCCAATACTTGTAAAT >41799_41799_1_KDM4B-CHAF1A_KDM4B_chr19_5082515_ENST00000159111_CHAF1A_chr19_4418017_ENST00000301280_length(amino acids)=942AA_BP=305 MGSEDHGAQNPSCKIMTFRPTMEEFKDFNKYVAYIESQGAHRAGLAKIIPPKEWKPRQTYDDIDDVVIPAPIQQVVTGQSGLFTQYNIQK KAMTVGEYRRLANSEKYCTPRHQDFDDLERKYWKNLTFVSPIYGADISGSLYDDDVAQWNIGSLRTILDMVERECGTIIEGVNTPYLYFG MWKTTFAWHTEDMDLYSINYLHFGEPKSWYAIPPEHGKRLERLAIGFFPGSSQGCDAFLRHKMTLISPIILKKYGIPFSRITQEAGEFMI TFPYGYHAGFNHGFNCAESTNFATLRWIDYGKVATQITKKFVKGSTEKNKLRLQRDQERLGKQLKLRAEREEKEKLKEEAKRAKEEAKKK KEEEKELKEKERREKREKDEKEKAEKQRLKEERRKERQEALEAKLEEKRKKEEEKRLREEEKRIKAEKAEITRFFQKPKTPQAPKTLAGS CGKFAPFEIKEHMVLAPRRRTAFHPDLCSQLDQLLQQQSGEFSFLKDLKGRQPLRSGPTHVSTRNADIFNSDVVIVERGKGDGVPERRKF GRMKLLQFCENHRPAYWGTWNKKTALIRARDPWAQDTKLLDYEVDSDEEWEEEEPGESLSHSEGDDDDDMGEDEDEDDGFFVPHGYLSED EGVTEECADPENHKVRQKLKAKEWDEFLAKGKRFRVLQPVKIGCVWAADRDCAGDDLKVLQQFAACFLETLPAQEEQTPKASKRERRDEQ ILAQLLPLLHGNVNGSKVIIREFQEHCRRGLLSNHTGSPRSPSTTYLHTPTPSEDAAIPSKSRLKRLISENSVYEKRPDFRMCWYVHPQV LQSFQQEHLPVPCQWSYVTSVPSAPKEDSGSVPSTGPSQGTPISLKRKSAGSMCITQFMKKRRHDGQIGAEDMDGFQADTEEEEEEEGDC -------------------------------------------------------------- >41799_41799_2_KDM4B-CHAF1A_KDM4B_chr19_5082515_ENST00000381759_CHAF1A_chr19_4418017_ENST00000301280_length(transcript)=3403nt_BP=1125nt CCGAGCGGGGCCCGGCCCGAGCGGGGCCTGGGGGTGCGACGCCGAGGGCGGGGGAGAGCGCGCCGCTGCTCCCGGACCGGGCCGCGCACG CCGCCTCAGGAACCATCACTGTTGCTGGAGGCACCTGACAAATCCTAGCGAATTTTTGGAGCATCTCCACCCAGGAACCTCGCCATCCAG AAGTGTGCTTCCCGCACAGCTGCAGCCATGGGGTCTGAGGACCACGGCGCCCAGAACCCCAGCTGTAAAATCATGACGTTTCGCCCAACC ATGGAAGAATTTAAAGACTTCAACAAATACGTGGCCTACATAGAGTCGCAGGGAGCCCACCGGGCGGGCCTGGCCAAGATCATCCCCCCG AAGGAGTGGAAGCCGCGGCAGACGTATGATGACATCGACGACGTGGTGATCCCGGCGCCCATCCAGCAGGTGGTGACGGGCCAGTCGGGC CTCTTCACGCAGTACAATATCCAGAAGAAGGCCATGACAGTGGGCGAGTACCGCCGCCTGGCCAACAGCGAGAAGTACTGTACCCCGCGG CACCAGGACTTTGATGACCTTGAACGCAAATACTGGAAGAACCTCACCTTTGTCTCCCCGATCTACGGGGCTGACATCAGCGGCTCTTTG TATGATGACGACGTGGCCCAGTGGAACATCGGGAGCCTCCGGACCATCCTGGACATGGTGGAGCGCGAGTGCGGCACCATCATCGAGGGC GTGAACACGCCCTACCTGTACTTCGGCATGTGGAAGACCACCTTCGCCTGGCACACCGAGGACATGGACCTGTACAGCATCAACTACCTG CACTTTGGGGAGCCTAAGTCCTGGTACGCCATCCCACCAGAGCACGGCAAGCGCCTGGAGCGGCTGGCCATCGGCTTCTTCCCCGGGAGC TCGCAGGGCTGCGACGCCTTCCTGCGGCATAAGATGACCCTCATCTCGCCCATCATCCTGAAGAAGTACGGGATCCCCTTCAGCCGGATC ACGCAGGAGGCCGGGGAATTCATGATCACATTTCCCTACGGCTACCACGCCGGCTTCAATCACGGGTTCAACTGCGCAGAATCTACCAAC TTCGCCACCCTGCGGTGGATTGACTACGGCAAAGTGGCCACTCAGATAACTAAGAAATTCGTCAAAGGCTCTACAGAGAAGAACAAGCTC AGACTGCAAAGAGATCAGGAGCGTCTGGGCAAGCAGCTCAAGTTACGTGCAGAAAGGGAAGAAAAGGAGAAGCTGAAAGAGGAGGCCAAG CGGGCCAAGGAGGAGGCCAAGAAGAAGAAGGAGGAAGAGAAGGAGCTTAAGGAAAAGGAGAGGCGGGAGAAGCGGGAGAAGGATGAGAAG GAGAAGGCGGAGAAGCAGCGGCTCAAGGAGGAGCGGCGCAAGGAGAGACAGGAAGCCCTGGAGGCTAAACTTGAGGAAAAAAGGAAAAAG GAAGAAGAGAAACGGTTAAGAGAAGAAGAGAAGCGCATTAAAGCAGAGAAGGCCGAAATCACGAGGTTCTTCCAGAAACCAAAGACTCCA CAGGCCCCCAAGACCCTGGCCGGCTCCTGTGGGAAGTTTGCCCCCTTTGAAATTAAAGAGCACATGGTCCTGGCCCCTCGGCGTCGGACC GCTTTCCATCCAGACCTCTGCAGTCAGCTGGACCAGCTCCTCCAGCAGCAGAGCGGCGAGTTCTCCTTCTTGAAAGACCTCAAAGGCCGG CAGCCCCTGAGGTCCGGACCCACGCACGTTTCCACCCGGAATGCAGATATTTTTAACAGTGATGTCGTCATCGTGGAGCGTGGGAAGGGC GACGGTGTTCCCGAGAGGAGGAAGTTTGGCAGGATGAAGCTCCTGCAGTTCTGTGAGAACCACCGGCCTGCCTACTGGGGTACCTGGAAT AAGAAGACGGCACTCATCCGCGCGCGAGACCCCTGGGCCCAGGACACGAAGCTCCTGGACTATGAGGTGGACAGTGATGAGGAGTGGGAA GAAGAGGAGCCTGGGGAGTCCCTGTCCCACAGTGAGGGGGATGATGATGACGACATGGGAGAGGATGAAGATGAGGACGATGGTTTCTTT GTGCCCCATGGGTACCTGTCTGAGGACGAAGGTGTGACAGAGGAGTGTGCCGACCCTGAGAACCATAAGGTCCGCCAGAAACTGAAGGCC AAGGAGTGGGACGAGTTCCTGGCTAAGGGGAAGCGCTTTCGCGTCCTGCAACCTGTGAAGATCGGCTGCGTGTGGGCGGCTGACAGAGAC TGCGCAGGCGATGACCTGAAGGTACTGCAGCAGTTCGCAGCCTGCTTCCTGGAGACCCTGCCGGCCCAGGAGGAGCAGACGCCCAAGGCC TCCAAGCGGGAGAGGAGAGACGAGCAGATCCTGGCCCAGCTGCTGCCGCTCCTGCACGGCAATGTGAACGGGAGCAAGGTCATCATCCGG GAGTTCCAGGAGCACTGCCGCCGGGGACTGCTCAGCAACCACACCGGCAGCCCGCGGAGCCCCTCCACCACCTACCTGCACACCCCCACC CCCAGCGAGGATGCCGCCATCCCCTCTAAGTCCCGGCTCAAGCGGCTCATTTCCGAGAACTCAGTGTATGAGAAGCGGCCTGACTTCAGG ATGTGCTGGTACGTGCACCCGCAGGTGCTACAGAGCTTCCAGCAGGAGCACCTGCCCGTGCCGTGCCAGTGGAGCTATGTGACATCGGTG CCCTCGGCCCCCAAAGAGGACAGTGGCAGCGTCCCCTCCACGGGGCCCAGCCAGGGCACTCCCATCTCGCTGAAGAGGAAGTCAGCGGGC AGCATGTGCATCACCCAATTCATGAAGAAGCGCAGGCACGACGGCCAGATTGGTGCTGAAGACATGGACGGCTTCCAGGCAGACACGGAG GAGGAGGAAGAGGAGGAGGGCGACTGTATGATCGTGGATGTCCCGGATGCTGCGGAGGTCCAAGCCCCGTGTGGAGCCGCTTCCGGAGCT GGGGGTGGTGTGGGGGTGGACACCGGCAAGGCCACCCTGACCTCGAGCCCACTGGGTGCATCCTGAGAGCAGGGGTGACGTATGTAGAAT GCTTAGGGTGTCCTCCCCACAGAGCAGATACTTGAACCGACTCAATTCCTGTGTAAAGAGCACTTTGTCCTGCTTCACGGACCTCCCCAA AGTGTGCAGAGTTCTATATAGGATGCTGGATTAGTTCCTTTGATATTTGTAAAAATTCCCCCAAGAGCCGCATATGAATCTGCCCTTTAA TAAAGCATTATTGAGATTGCTGGCCTATTGGGGAAGCCTGCGGGCACAGGAGCAGGCGTGGAATCCAATACTTGTAAATGAATTGAAGCG >41799_41799_2_KDM4B-CHAF1A_KDM4B_chr19_5082515_ENST00000381759_CHAF1A_chr19_4418017_ENST00000301280_length(amino acids)=942AA_BP=305 MGSEDHGAQNPSCKIMTFRPTMEEFKDFNKYVAYIESQGAHRAGLAKIIPPKEWKPRQTYDDIDDVVIPAPIQQVVTGQSGLFTQYNIQK KAMTVGEYRRLANSEKYCTPRHQDFDDLERKYWKNLTFVSPIYGADISGSLYDDDVAQWNIGSLRTILDMVERECGTIIEGVNTPYLYFG MWKTTFAWHTEDMDLYSINYLHFGEPKSWYAIPPEHGKRLERLAIGFFPGSSQGCDAFLRHKMTLISPIILKKYGIPFSRITQEAGEFMI TFPYGYHAGFNHGFNCAESTNFATLRWIDYGKVATQITKKFVKGSTEKNKLRLQRDQERLGKQLKLRAEREEKEKLKEEAKRAKEEAKKK KEEEKELKEKERREKREKDEKEKAEKQRLKEERRKERQEALEAKLEEKRKKEEEKRLREEEKRIKAEKAEITRFFQKPKTPQAPKTLAGS CGKFAPFEIKEHMVLAPRRRTAFHPDLCSQLDQLLQQQSGEFSFLKDLKGRQPLRSGPTHVSTRNADIFNSDVVIVERGKGDGVPERRKF GRMKLLQFCENHRPAYWGTWNKKTALIRARDPWAQDTKLLDYEVDSDEEWEEEEPGESLSHSEGDDDDDMGEDEDEDDGFFVPHGYLSED EGVTEECADPENHKVRQKLKAKEWDEFLAKGKRFRVLQPVKIGCVWAADRDCAGDDLKVLQQFAACFLETLPAQEEQTPKASKRERRDEQ ILAQLLPLLHGNVNGSKVIIREFQEHCRRGLLSNHTGSPRSPSTTYLHTPTPSEDAAIPSKSRLKRLISENSVYEKRPDFRMCWYVHPQV LQSFQQEHLPVPCQWSYVTSVPSAPKEDSGSVPSTGPSQGTPISLKRKSAGSMCITQFMKKRRHDGQIGAEDMDGFQADTEEEEEEEGDC -------------------------------------------------------------- >41799_41799_3_KDM4B-CHAF1A_KDM4B_chr19_5082515_ENST00000536461_CHAF1A_chr19_4418017_ENST00000301280_length(transcript)=3236nt_BP=958nt CCTCGCCATCCAGAAGTGTGCTTCCCGCACAGCTGCAGCCATGGGGTCTGAGGACCACGGCGCCCAGAACCCCAGCTGTAAAATCATGAC GTTTCGCCCAACCATGGAAGAATTTAAAGACTTCAACAAATACGTGGCCTACATAGAGTCGCAGGGAGCCCACCGGGCGGGCCTGGCCAA GATCATCCCCCCGAAGGAGTGGAAGCCGCGGCAGACGTATGATGACATCGACGACGTGGTGATCCCGGCGCCCATCCAGCAGGTGGTGAC GGGCCAGTCGGGCCTCTTCACGCAGTACAATATCCAGAAGAAGGCCATGACAGTGGGCGAGTACCGCCGCCTGGCCAACAGCGAGAAGTA CTGTACCCCGCGGCACCAGGACTTTGATGACCTTGAACGCAAATACTGGAAGAACCTCACCTTTGTCTCCCCGATCTACGGGGCTGACAT CAGCGGCTCTTTGTATGATGACGACGTGGCCCAGTGGAACATCGGGAGCCTCCGGACCATCCTGGACATGGTGGAGCGCGAGTGCGGCAC CATCATCGAGGGCGTGAACACGCCCTACCTGTACTTCGGCATGTGGAAGACCACCTTCGCCTGGCACACCGAGGACATGGACCTGTACAG CATCAACTACCTGCACTTTGGGGAGCCTAAGTCCTGGTACGCCATCCCACCAGAGCACGGCAAGCGCCTGGAGCGGCTGGCCATCGGCTT CTTCCCCGGGAGCTCGCAGGGCTGCGACGCCTTCCTGCGGCATAAGATGACCCTCATCTCGCCCATCATCCTGAAGAAGTACGGGATCCC CTTCAGCCGGATCACGCAGGAGGCCGGGGAATTCATGATCACATTTCCCTACGGCTACCACGCCGGCTTCAATCACGGGTTCAACTGCGC AGAATCTACCAACTTCGCCACCCTGCGGTGGATTGACTACGGCAAAGTGGCCACTCAGATAACTAAGAAATTCGTCAAAGGCTCTACAGA GAAGAACAAGCTCAGACTGCAAAGAGATCAGGAGCGTCTGGGCAAGCAGCTCAAGTTACGTGCAGAAAGGGAAGAAAAGGAGAAGCTGAA AGAGGAGGCCAAGCGGGCCAAGGAGGAGGCCAAGAAGAAGAAGGAGGAAGAGAAGGAGCTTAAGGAAAAGGAGAGGCGGGAGAAGCGGGA GAAGGATGAGAAGGAGAAGGCGGAGAAGCAGCGGCTCAAGGAGGAGCGGCGCAAGGAGAGACAGGAAGCCCTGGAGGCTAAACTTGAGGA AAAAAGGAAAAAGGAAGAAGAGAAACGGTTAAGAGAAGAAGAGAAGCGCATTAAAGCAGAGAAGGCCGAAATCACGAGGTTCTTCCAGAA ACCAAAGACTCCACAGGCCCCCAAGACCCTGGCCGGCTCCTGTGGGAAGTTTGCCCCCTTTGAAATTAAAGAGCACATGGTCCTGGCCCC TCGGCGTCGGACCGCTTTCCATCCAGACCTCTGCAGTCAGCTGGACCAGCTCCTCCAGCAGCAGAGCGGCGAGTTCTCCTTCTTGAAAGA CCTCAAAGGCCGGCAGCCCCTGAGGTCCGGACCCACGCACGTTTCCACCCGGAATGCAGATATTTTTAACAGTGATGTCGTCATCGTGGA GCGTGGGAAGGGCGACGGTGTTCCCGAGAGGAGGAAGTTTGGCAGGATGAAGCTCCTGCAGTTCTGTGAGAACCACCGGCCTGCCTACTG GGGTACCTGGAATAAGAAGACGGCACTCATCCGCGCGCGAGACCCCTGGGCCCAGGACACGAAGCTCCTGGACTATGAGGTGGACAGTGA TGAGGAGTGGGAAGAAGAGGAGCCTGGGGAGTCCCTGTCCCACAGTGAGGGGGATGATGATGACGACATGGGAGAGGATGAAGATGAGGA CGATGGTTTCTTTGTGCCCCATGGGTACCTGTCTGAGGACGAAGGTGTGACAGAGGAGTGTGCCGACCCTGAGAACCATAAGGTCCGCCA GAAACTGAAGGCCAAGGAGTGGGACGAGTTCCTGGCTAAGGGGAAGCGCTTTCGCGTCCTGCAACCTGTGAAGATCGGCTGCGTGTGGGC GGCTGACAGAGACTGCGCAGGCGATGACCTGAAGGTACTGCAGCAGTTCGCAGCCTGCTTCCTGGAGACCCTGCCGGCCCAGGAGGAGCA GACGCCCAAGGCCTCCAAGCGGGAGAGGAGAGACGAGCAGATCCTGGCCCAGCTGCTGCCGCTCCTGCACGGCAATGTGAACGGGAGCAA GGTCATCATCCGGGAGTTCCAGGAGCACTGCCGCCGGGGACTGCTCAGCAACCACACCGGCAGCCCGCGGAGCCCCTCCACCACCTACCT GCACACCCCCACCCCCAGCGAGGATGCCGCCATCCCCTCTAAGTCCCGGCTCAAGCGGCTCATTTCCGAGAACTCAGTGTATGAGAAGCG GCCTGACTTCAGGATGTGCTGGTACGTGCACCCGCAGGTGCTACAGAGCTTCCAGCAGGAGCACCTGCCCGTGCCGTGCCAGTGGAGCTA TGTGACATCGGTGCCCTCGGCCCCCAAAGAGGACAGTGGCAGCGTCCCCTCCACGGGGCCCAGCCAGGGCACTCCCATCTCGCTGAAGAG GAAGTCAGCGGGCAGCATGTGCATCACCCAATTCATGAAGAAGCGCAGGCACGACGGCCAGATTGGTGCTGAAGACATGGACGGCTTCCA GGCAGACACGGAGGAGGAGGAAGAGGAGGAGGGCGACTGTATGATCGTGGATGTCCCGGATGCTGCGGAGGTCCAAGCCCCGTGTGGAGC CGCTTCCGGAGCTGGGGGTGGTGTGGGGGTGGACACCGGCAAGGCCACCCTGACCTCGAGCCCACTGGGTGCATCCTGAGAGCAGGGGTG ACGTATGTAGAATGCTTAGGGTGTCCTCCCCACAGAGCAGATACTTGAACCGACTCAATTCCTGTGTAAAGAGCACTTTGTCCTGCTTCA CGGACCTCCCCAAAGTGTGCAGAGTTCTATATAGGATGCTGGATTAGTTCCTTTGATATTTGTAAAAATTCCCCCAAGAGCCGCATATGA ATCTGCCCTTTAATAAAGCATTATTGAGATTGCTGGCCTATTGGGGAAGCCTGCGGGCACAGGAGCAGGCGTGGAATCCAATACTTGTAA >41799_41799_3_KDM4B-CHAF1A_KDM4B_chr19_5082515_ENST00000536461_CHAF1A_chr19_4418017_ENST00000301280_length(amino acids)=942AA_BP=305 MGSEDHGAQNPSCKIMTFRPTMEEFKDFNKYVAYIESQGAHRAGLAKIIPPKEWKPRQTYDDIDDVVIPAPIQQVVTGQSGLFTQYNIQK KAMTVGEYRRLANSEKYCTPRHQDFDDLERKYWKNLTFVSPIYGADISGSLYDDDVAQWNIGSLRTILDMVERECGTIIEGVNTPYLYFG MWKTTFAWHTEDMDLYSINYLHFGEPKSWYAIPPEHGKRLERLAIGFFPGSSQGCDAFLRHKMTLISPIILKKYGIPFSRITQEAGEFMI TFPYGYHAGFNHGFNCAESTNFATLRWIDYGKVATQITKKFVKGSTEKNKLRLQRDQERLGKQLKLRAEREEKEKLKEEAKRAKEEAKKK KEEEKELKEKERREKREKDEKEKAEKQRLKEERRKERQEALEAKLEEKRKKEEEKRLREEEKRIKAEKAEITRFFQKPKTPQAPKTLAGS CGKFAPFEIKEHMVLAPRRRTAFHPDLCSQLDQLLQQQSGEFSFLKDLKGRQPLRSGPTHVSTRNADIFNSDVVIVERGKGDGVPERRKF GRMKLLQFCENHRPAYWGTWNKKTALIRARDPWAQDTKLLDYEVDSDEEWEEEEPGESLSHSEGDDDDDMGEDEDEDDGFFVPHGYLSED EGVTEECADPENHKVRQKLKAKEWDEFLAKGKRFRVLQPVKIGCVWAADRDCAGDDLKVLQQFAACFLETLPAQEEQTPKASKRERRDEQ ILAQLLPLLHGNVNGSKVIIREFQEHCRRGLLSNHTGSPRSPSTTYLHTPTPSEDAAIPSKSRLKRLISENSVYEKRPDFRMCWYVHPQV LQSFQQEHLPVPCQWSYVTSVPSAPKEDSGSVPSTGPSQGTPISLKRKSAGSMCITQFMKKRRHDGQIGAEDMDGFQADTEEEEEEEGDC -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for KDM4B-CHAF1A |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for KDM4B-CHAF1A |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for KDM4B-CHAF1A |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
