
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:KDM4C-DMRT3 (FusionGDB2 ID:41828) |
Fusion Gene Summary for KDM4C-DMRT3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: KDM4C-DMRT3 | Fusion gene ID: 41828 | Hgene | Tgene | Gene symbol | KDM4C | DMRT3 | Gene ID | 23081 | 58524 |
| Gene name | lysine demethylase 4C | doublesex and mab-3 related transcription factor 3 | |
| Synonyms | GASC1|JHDM3C|JMJD2C|TDRD14C | DMRTA3 | |
| Cytomap | 9p24.1 | 9p24.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | lysine-specific demethylase 4CJmjC domain-containing histone demethylation protein 3Cgene amplified in squamous cell carcinoma 1 proteinjumonji domain-containing protein 2Clysine (K)-specific demethylase 4Ctudor domain containing 14C | doublesex- and mab-3-related transcription factor 3DMRT-like family A3testis-specific protein | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | Q9H3R0 | Q9NQL9 | |
| Ensembl transtripts involved in fusion gene | ENST00000489243, ENST00000442236, ENST00000401787, ENST00000381306, ENST00000381309, ENST00000535193, ENST00000543771, ENST00000428870, ENST00000536108, | ENST00000190165, | |
| Fusion gene scores | * DoF score | 22 X 15 X 12=3960 | 1 X 1 X 1=1 |
| # samples | 22 | 1 | |
| ** MAII score | log2(22/3960*10)=-4.16992500144231 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(1/1*10)=3.32192809488736 | |
| Context | PubMed: KDM4C [Title/Abstract] AND DMRT3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | KDM4C(6814745)-DMRT3(990041), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | KDM4C-DMRT3 seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor due to the frame-shifted ORF. KDM4C-DMRT3 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. KDM4C-DMRT3 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. KDM4C-DMRT3 seems lost the major protein functional domain in Tgene partner, which is a transcription factor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | KDM4C | GO:0006357 | regulation of transcription by RNA polymerase II | 17277772 |
| Hgene | KDM4C | GO:0033169 | histone H3-K9 demethylation | 18066052|21914792 |
| Hgene | KDM4C | GO:0070544 | histone H3-K36 demethylation | 21914792 |
 Fusion gene breakpoints across KDM4C (5'-gene) Fusion gene breakpoints across KDM4C (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
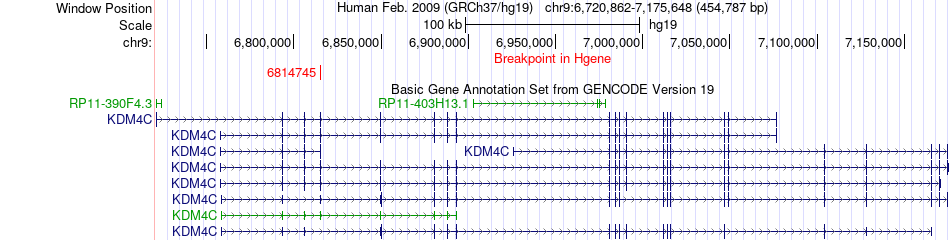 |
 Fusion gene breakpoints across DMRT3 (3'-gene) Fusion gene breakpoints across DMRT3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
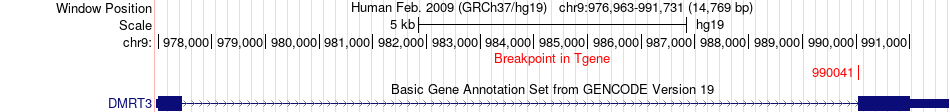 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | CESC | TCGA-EK-A2IR-01A | KDM4C | chr9 | 6814745 | - | DMRT3 | chr9 | 990041 | + |
| ChimerDB4 | CESC | TCGA-EK-A2IR-01A | KDM4C | chr9 | 6814745 | + | DMRT3 | chr9 | 990041 | + |
Top |
Fusion Gene ORF analysis for KDM4C-DMRT3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000489243 | ENST00000190165 | KDM4C | chr9 | 6814745 | + | DMRT3 | chr9 | 990041 | + |
| 5UTR-3CDS | ENST00000442236 | ENST00000190165 | KDM4C | chr9 | 6814745 | + | DMRT3 | chr9 | 990041 | + |
| Frame-shift | ENST00000401787 | ENST00000190165 | KDM4C | chr9 | 6814745 | + | DMRT3 | chr9 | 990041 | + |
| In-frame | ENST00000381306 | ENST00000190165 | KDM4C | chr9 | 6814745 | + | DMRT3 | chr9 | 990041 | + |
| In-frame | ENST00000381309 | ENST00000190165 | KDM4C | chr9 | 6814745 | + | DMRT3 | chr9 | 990041 | + |
| In-frame | ENST00000535193 | ENST00000190165 | KDM4C | chr9 | 6814745 | + | DMRT3 | chr9 | 990041 | + |
| In-frame | ENST00000543771 | ENST00000190165 | KDM4C | chr9 | 6814745 | + | DMRT3 | chr9 | 990041 | + |
| intron-3CDS | ENST00000428870 | ENST00000190165 | KDM4C | chr9 | 6814745 | + | DMRT3 | chr9 | 990041 | + |
| intron-3CDS | ENST00000536108 | ENST00000190165 | KDM4C | chr9 | 6814745 | + | DMRT3 | chr9 | 990041 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000535193 | KDM4C | chr9 | 6814745 | + | ENST00000190165 | DMRT3 | chr9 | 990041 | + | 2278 | 587 | 589 | 1551 | 320 |
| ENST00000543771 | KDM4C | chr9 | 6814745 | + | ENST00000190165 | DMRT3 | chr9 | 990041 | + | 2706 | 1015 | 1017 | 1979 | 320 |
| ENST00000381306 | KDM4C | chr9 | 6814745 | + | ENST00000190165 | DMRT3 | chr9 | 990041 | + | 2691 | 1000 | 1002 | 1964 | 320 |
| ENST00000381309 | KDM4C | chr9 | 6814745 | + | ENST00000190165 | DMRT3 | chr9 | 990041 | + | 2691 | 1000 | 1002 | 1964 | 320 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000535193 | ENST00000190165 | KDM4C | chr9 | 6814745 | + | DMRT3 | chr9 | 990041 | + | 0.008777275 | 0.99122274 |
| ENST00000543771 | ENST00000190165 | KDM4C | chr9 | 6814745 | + | DMRT3 | chr9 | 990041 | + | 0.029290805 | 0.9707092 |
| ENST00000381306 | ENST00000190165 | KDM4C | chr9 | 6814745 | + | DMRT3 | chr9 | 990041 | + | 0.014075289 | 0.98592466 |
| ENST00000381309 | ENST00000190165 | KDM4C | chr9 | 6814745 | + | DMRT3 | chr9 | 990041 | + | 0.014075289 | 0.98592466 |
Top |
Fusion Genomic Features for KDM4C-DMRT3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| KDM4C | chr9 | 6814745 | + | DMRT3 | chr9 | 990040 | + | 0.000302733 | 0.9996973 |
| KDM4C | chr9 | 6814745 | + | DMRT3 | chr9 | 990040 | + | 0.000302733 | 0.9996973 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
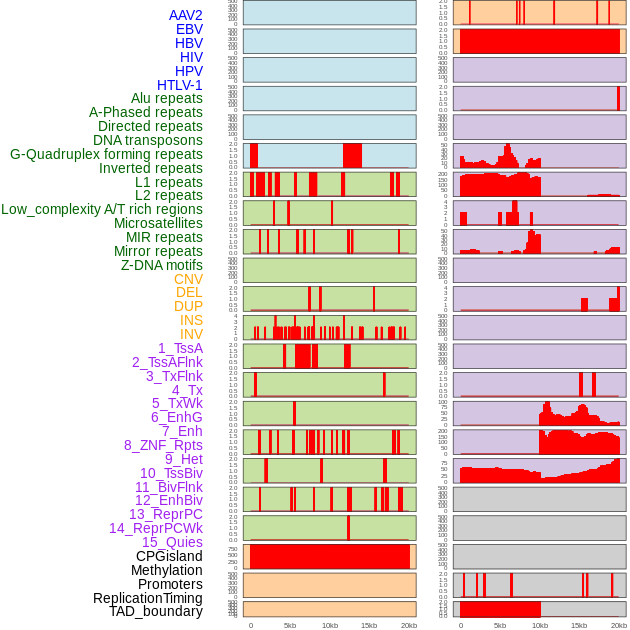 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for KDM4C-DMRT3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr9:6814745/chr9:990041) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| KDM4C | DMRT3 |
| FUNCTION: Histone demethylase that specifically demethylates 'Lys-9' and 'Lys-36' residues of histone H3, thereby playing a central role in histone code. Does not demethylate histone H3 'Lys-4', H3 'Lys-27' nor H4 'Lys-20'. Demethylates trimethylated H3 'Lys-9' and H3 'Lys-36' residue, while it has no activity on mono- and dimethylated residues. Demethylation of Lys residue generates formaldehyde and succinate. {ECO:0000269|PubMed:16603238, ECO:0000269|PubMed:28262558}. | FUNCTION: Probable transcription factor that plays a role in configuring the spinal circuits controlling stride in vertebrates. Involved in neuronal specification within specific subdivision of spinal cord neurons and in the development of a coordinated locomotor network controlling limb movements. May regulate transcription during sexual development (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000381306 | + | 4 | 21 | 16_58 | 145 | 1048.0 | Domain | JmjN |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000381309 | + | 4 | 22 | 16_58 | 145 | 1057.0 | Domain | JmjN |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000535193 | + | 4 | 18 | 16_58 | 167 | 836.0 | Domain | JmjN |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000543771 | + | 4 | 18 | 16_58 | 145 | 814.0 | Domain | JmjN |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000381306 | + | 4 | 21 | 144_310 | 145 | 1048.0 | Domain | JmjC |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000381306 | + | 4 | 21 | 877_934 | 145 | 1048.0 | Domain | Note=Tudor 1 |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000381306 | + | 4 | 21 | 935_991 | 145 | 1048.0 | Domain | Note=Tudor 2 |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000381309 | + | 4 | 22 | 144_310 | 145 | 1057.0 | Domain | JmjC |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000381309 | + | 4 | 22 | 877_934 | 145 | 1057.0 | Domain | Note=Tudor 1 |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000381309 | + | 4 | 22 | 935_991 | 145 | 1057.0 | Domain | Note=Tudor 2 |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000535193 | + | 4 | 18 | 144_310 | 167 | 836.0 | Domain | JmjC |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000535193 | + | 4 | 18 | 877_934 | 167 | 836.0 | Domain | Note=Tudor 1 |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000535193 | + | 4 | 18 | 935_991 | 167 | 836.0 | Domain | Note=Tudor 2 |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000543771 | + | 4 | 18 | 144_310 | 145 | 814.0 | Domain | JmjC |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000543771 | + | 4 | 18 | 877_934 | 145 | 814.0 | Domain | Note=Tudor 1 |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000543771 | + | 4 | 18 | 935_991 | 145 | 814.0 | Domain | Note=Tudor 2 |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000381306 | + | 4 | 21 | 689_747 | 145 | 1048.0 | Zinc finger | Note=PHD-type 1 |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000381306 | + | 4 | 21 | 752_785 | 145 | 1048.0 | Zinc finger | C2HC pre-PHD-type |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000381306 | + | 4 | 21 | 808_865 | 145 | 1048.0 | Zinc finger | PHD-type 2 |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000381309 | + | 4 | 22 | 689_747 | 145 | 1057.0 | Zinc finger | Note=PHD-type 1 |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000381309 | + | 4 | 22 | 752_785 | 145 | 1057.0 | Zinc finger | C2HC pre-PHD-type |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000381309 | + | 4 | 22 | 808_865 | 145 | 1057.0 | Zinc finger | PHD-type 2 |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000535193 | + | 4 | 18 | 689_747 | 167 | 836.0 | Zinc finger | Note=PHD-type 1 |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000535193 | + | 4 | 18 | 752_785 | 167 | 836.0 | Zinc finger | C2HC pre-PHD-type |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000535193 | + | 4 | 18 | 808_865 | 167 | 836.0 | Zinc finger | PHD-type 2 |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000543771 | + | 4 | 18 | 689_747 | 145 | 814.0 | Zinc finger | Note=PHD-type 1 |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000543771 | + | 4 | 18 | 752_785 | 145 | 814.0 | Zinc finger | C2HC pre-PHD-type |
| Hgene | KDM4C | chr9:6814745 | chr9:990041 | ENST00000543771 | + | 4 | 18 | 808_865 | 145 | 814.0 | Zinc finger | PHD-type 2 |
| Tgene | DMRT3 | chr9:6814745 | chr9:990041 | ENST00000190165 | 0 | 2 | 29_76 | 151 | 473.0 | DNA binding | DM |
Top |
Fusion Gene Sequence for KDM4C-DMRT3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >41828_41828_1_KDM4C-DMRT3_KDM4C_chr9_6814745_ENST00000381306_DMRT3_chr9_990041_ENST00000190165_length(transcript)=2691nt_BP=1000nt CGGCGCCCAGGAGGACGTGTGGCGCGTGGACTACATCAGGTCCAGCCCTGCGGGACCCCAGCCAGCGCTTCCGGGCAAGGTTCTGTGCAC CTGTTTTCTCCTTCTACGCGAGTATCTTTCCCCTCCGGAAAGAATGGGATATGCCTGTGTCCAAAGGACAAGAAGATGCGCGCCAGCAAG CCTAAGTTAACCACAGCGCGGAAGTTGAGCCCAAAGCAAGAGCGTGCCGGGCACCTTTAAGCTGTTTGTAAGCCCACGTGACTCACCAAG TGCGGGCCCCAGCGGTCACGTGACGGCGCGCGCGCCCTCGCGCAGGGAGAGCCGGCGGTGCGCGCGCCTTCGCCGCTGCCTCCCACCCAC CCCCTCGACGGGAGGGTGAGGCGCGGCGCAGTGATCGGGCGGCCGGGGTCCTGTGCGCGTGCGCAGCGAACAGCTGTCACCTAGTGCGGA ACAAGTCTCCCAAATTTCCCAAATCTCCCTGGGCCGGAGGCCACTGTCTTCTCTTCCTCCTCCACCGAGTCGTGCTCTCGCCCCAACCCG CGCGCCAGACACTGCCCTAACCATCATGGAGGTGGCCGAGGTGGAAAGTCCTCTGAACCCCAGCTGTAAGATAATGACCTTCAGACCCTC CATGGAGGAGTTCCGGGAGTTCAACAAATACCTTGCATACATGGAGTCTAAAGGAGCCCATCGTGCGGGTCTTGCAAAGGTGATTCCTCC TAAGGAGTGGAAGCCAAGACAGTGCTATGATGACATTGATAATTTGCTCATTCCAGCACCAATTCAGCAGATGGTCACAGGGCAGTCAGG ACTGTTCACTCAGTACAACATCCAGAAAAAAGCGATGACTGTGAAGGAGTTCAGGCAGCTGGCCAACAGTGGCAAATATTGTACTCCAAG ATACTTGGATTACGAAGATTTGGAGCGCAAGTACTGGAAGAACTTAACTTTTGTGGCACCTATCTATGGTGCAGATATTAATGGGAGCAT ATATGATGAGATTTGACTGAAGAACGACTTGGAGACGGCAAGTCGGCAGACAATACAGAGGTCTTCAGTGACAAAGACACTGACCAGAGG AGTTCCCCAGATGTGGCAAAGAGTAAGGGCTGCTTCACCCCTGAGAGCCCTGAGATAGTGTCCGTGGAGGAAGGGGGATACGCTGTCCAG AAAAACGGAGGCAACCCCGAGAGCCGCCCTGACAGCCCCAAGTGTCACGCGGAGCAGAATCACCTCCTGATTGAGGGCCCCTCGGGGACT GTTTCTCTGCCCTTCAGCTTGAAAGCCAACAGACCGCCGCTTGAAGTGTTAAAAAAGATATTCCCCAACCAGAAGCCAACGGTGCTTGAG CTCATCCTCAAGGGCTGTGGCGGGGACCTGGTGAGCGCCGTGGAAGTCCTTCTGTCCAGCCGATCCTCAGTCACGGGAGCAGAGCGAACT TCCGCAGAACCTGAGAGTCTAGCGTTGCCCTCCAATGGGCACATCTTTGAACACACCTTGAGCTCCTACCCCATCTCGTCTTCCAAATGG TCTGTGGGATCAGCCTTTCGAGTCCCAGACACGTTGAGGTTTTCTGCCGACTCTAGCAACGTTGTCCCCAGTCCCTTGGCTGGGCCTCTG CAGCCCCCTTTCCCCCAGCCACCCCGGTACCCGCTGATGCTGAGGAATACTTTGGCGAGAAGCCAGTCGAGCCCCTTTTTGCCCAATGAT GTCACCCTGTGGAACACCATGACGCTGCAGCAGCAGTATCAGCTGAGGTCCCAGTATGTCAGTCCTTTCCCCAGTAACTCTACCAGCGTC TTCAGAAGCTCGCCCGTCCTTCCTGCCCGCGCCACGGAAGACCCTCGGATTTCCATCCCTGATGATGGGTGTCCATTTGTGTCAAAGCAG TCCATTTACACCGAGGACGACTATGACGAGAGGTCTGACTCCTCAGACTCTAGAACACTCAACACATCATCTTAAAGTGGTGCTGGATGG GTGGTGGCCAGGTGACATTTTCTGTGCGTTTTGACCCTGAGGCATCTGAGGAGAGGCCACATCTTGTGTATGCCCTTTCCTTCTGTTTGA CAAAGTGACTGTGCTTGATTCTATACATTAGCAATAAAAACATAACTTATTTAACTTCTTGCACTTCACTGGAAAATGCCAAATAGCTCT GTTCTGTGGCTTTAGTGCTGAATGTTTATTGTAAAAGAGAGTCTAATGTTAAGAATAGTCTTGGGAAGGCTGGGTCCGTGGAAGATTTAT TTGGGGATGTAAAGCTGAAGGTCAGCCTTGCACCTAAACCCAACCTGGAATGTTAAATGAAATAATATACTTGAATGCAATTTTGTAAAA GTGGATTCCTCAGGATATGTGAAACCTAAAGGAAGTGGTTCTGTTGCAAATGGACTATAAACAGGGACATTATATTCTTACACTAAAAAT CCTTGCATTTTAAAGAGAGATGCACTTAAGAATAGAGTGAACTGCTCATATGCTTATTTAAGCTTGGACAGTTTTCAGAGACAAACCCCA TTAAGAATTACTCTTTTCACATGGCTGAATCGAAACATGTGTAATGTCAATGTAAAACCAATCACAGCTGTGAACTGCATGAAATGTATT >41828_41828_1_KDM4C-DMRT3_KDM4C_chr9_6814745_ENST00000381306_DMRT3_chr9_990041_ENST00000190165_length(amino acids)=320AA_BP=1 MTEERLGDGKSADNTEVFSDKDTDQRSSPDVAKSKGCFTPESPEIVSVEEGGYAVQKNGGNPESRPDSPKCHAEQNHLLIEGPSGTVSLP FSLKANRPPLEVLKKIFPNQKPTVLELILKGCGGDLVSAVEVLLSSRSSVTGAERTSAEPESLALPSNGHIFEHTLSSYPISSSKWSVGS AFRVPDTLRFSADSSNVVPSPLAGPLQPPFPQPPRYPLMLRNTLARSQSSPFLPNDVTLWNTMTLQQQYQLRSQYVSPFPSNSTSVFRSS -------------------------------------------------------------- >41828_41828_2_KDM4C-DMRT3_KDM4C_chr9_6814745_ENST00000381309_DMRT3_chr9_990041_ENST00000190165_length(transcript)=2691nt_BP=1000nt CGGCGCCCAGGAGGACGTGTGGCGCGTGGACTACATCAGGTCCAGCCCTGCGGGACCCCAGCCAGCGCTTCCGGGCAAGGTTCTGTGCAC CTGTTTTCTCCTTCTACGCGAGTATCTTTCCCCTCCGGAAAGAATGGGATATGCCTGTGTCCAAAGGACAAGAAGATGCGCGCCAGCAAG CCTAAGTTAACCACAGCGCGGAAGTTGAGCCCAAAGCAAGAGCGTGCCGGGCACCTTTAAGCTGTTTGTAAGCCCACGTGACTCACCAAG TGCGGGCCCCAGCGGTCACGTGACGGCGCGCGCGCCCTCGCGCAGGGAGAGCCGGCGGTGCGCGCGCCTTCGCCGCTGCCTCCCACCCAC CCCCTCGACGGGAGGGTGAGGCGCGGCGCAGTGATCGGGCGGCCGGGGTCCTGTGCGCGTGCGCAGCGAACAGCTGTCACCTAGTGCGGA ACAAGTCTCCCAAATTTCCCAAATCTCCCTGGGCCGGAGGCCACTGTCTTCTCTTCCTCCTCCACCGAGTCGTGCTCTCGCCCCAACCCG CGCGCCAGACACTGCCCTAACCATCATGGAGGTGGCCGAGGTGGAAAGTCCTCTGAACCCCAGCTGTAAGATAATGACCTTCAGACCCTC CATGGAGGAGTTCCGGGAGTTCAACAAATACCTTGCATACATGGAGTCTAAAGGAGCCCATCGTGCGGGTCTTGCAAAGGTGATTCCTCC TAAGGAGTGGAAGCCAAGACAGTGCTATGATGACATTGATAATTTGCTCATTCCAGCACCAATTCAGCAGATGGTCACAGGGCAGTCAGG ACTGTTCACTCAGTACAACATCCAGAAAAAAGCGATGACTGTGAAGGAGTTCAGGCAGCTGGCCAACAGTGGCAAATATTGTACTCCAAG ATACTTGGATTACGAAGATTTGGAGCGCAAGTACTGGAAGAACTTAACTTTTGTGGCACCTATCTATGGTGCAGATATTAATGGGAGCAT ATATGATGAGATTTGACTGAAGAACGACTTGGAGACGGCAAGTCGGCAGACAATACAGAGGTCTTCAGTGACAAAGACACTGACCAGAGG AGTTCCCCAGATGTGGCAAAGAGTAAGGGCTGCTTCACCCCTGAGAGCCCTGAGATAGTGTCCGTGGAGGAAGGGGGATACGCTGTCCAG AAAAACGGAGGCAACCCCGAGAGCCGCCCTGACAGCCCCAAGTGTCACGCGGAGCAGAATCACCTCCTGATTGAGGGCCCCTCGGGGACT GTTTCTCTGCCCTTCAGCTTGAAAGCCAACAGACCGCCGCTTGAAGTGTTAAAAAAGATATTCCCCAACCAGAAGCCAACGGTGCTTGAG CTCATCCTCAAGGGCTGTGGCGGGGACCTGGTGAGCGCCGTGGAAGTCCTTCTGTCCAGCCGATCCTCAGTCACGGGAGCAGAGCGAACT TCCGCAGAACCTGAGAGTCTAGCGTTGCCCTCCAATGGGCACATCTTTGAACACACCTTGAGCTCCTACCCCATCTCGTCTTCCAAATGG TCTGTGGGATCAGCCTTTCGAGTCCCAGACACGTTGAGGTTTTCTGCCGACTCTAGCAACGTTGTCCCCAGTCCCTTGGCTGGGCCTCTG CAGCCCCCTTTCCCCCAGCCACCCCGGTACCCGCTGATGCTGAGGAATACTTTGGCGAGAAGCCAGTCGAGCCCCTTTTTGCCCAATGAT GTCACCCTGTGGAACACCATGACGCTGCAGCAGCAGTATCAGCTGAGGTCCCAGTATGTCAGTCCTTTCCCCAGTAACTCTACCAGCGTC TTCAGAAGCTCGCCCGTCCTTCCTGCCCGCGCCACGGAAGACCCTCGGATTTCCATCCCTGATGATGGGTGTCCATTTGTGTCAAAGCAG TCCATTTACACCGAGGACGACTATGACGAGAGGTCTGACTCCTCAGACTCTAGAACACTCAACACATCATCTTAAAGTGGTGCTGGATGG GTGGTGGCCAGGTGACATTTTCTGTGCGTTTTGACCCTGAGGCATCTGAGGAGAGGCCACATCTTGTGTATGCCCTTTCCTTCTGTTTGA CAAAGTGACTGTGCTTGATTCTATACATTAGCAATAAAAACATAACTTATTTAACTTCTTGCACTTCACTGGAAAATGCCAAATAGCTCT GTTCTGTGGCTTTAGTGCTGAATGTTTATTGTAAAAGAGAGTCTAATGTTAAGAATAGTCTTGGGAAGGCTGGGTCCGTGGAAGATTTAT TTGGGGATGTAAAGCTGAAGGTCAGCCTTGCACCTAAACCCAACCTGGAATGTTAAATGAAATAATATACTTGAATGCAATTTTGTAAAA GTGGATTCCTCAGGATATGTGAAACCTAAAGGAAGTGGTTCTGTTGCAAATGGACTATAAACAGGGACATTATATTCTTACACTAAAAAT CCTTGCATTTTAAAGAGAGATGCACTTAAGAATAGAGTGAACTGCTCATATGCTTATTTAAGCTTGGACAGTTTTCAGAGACAAACCCCA TTAAGAATTACTCTTTTCACATGGCTGAATCGAAACATGTGTAATGTCAATGTAAAACCAATCACAGCTGTGAACTGCATGAAATGTATT >41828_41828_2_KDM4C-DMRT3_KDM4C_chr9_6814745_ENST00000381309_DMRT3_chr9_990041_ENST00000190165_length(amino acids)=320AA_BP=1 MTEERLGDGKSADNTEVFSDKDTDQRSSPDVAKSKGCFTPESPEIVSVEEGGYAVQKNGGNPESRPDSPKCHAEQNHLLIEGPSGTVSLP FSLKANRPPLEVLKKIFPNQKPTVLELILKGCGGDLVSAVEVLLSSRSSVTGAERTSAEPESLALPSNGHIFEHTLSSYPISSSKWSVGS AFRVPDTLRFSADSSNVVPSPLAGPLQPPFPQPPRYPLMLRNTLARSQSSPFLPNDVTLWNTMTLQQQYQLRSQYVSPFPSNSTSVFRSS -------------------------------------------------------------- >41828_41828_3_KDM4C-DMRT3_KDM4C_chr9_6814745_ENST00000535193_DMRT3_chr9_990041_ENST00000190165_length(transcript)=2278nt_BP=587nt ATGGCTGACATGATGGTCTGTGACCTGAAAGTTGTTGAATAGTTGGAGTGTTCTGCATTCATGTGGAAGAGGCTAAAGGATGTTTGATGA AGCACTATGGGCTGCCCTGGAAGAGGACTGAAGAAGCAGCTGCAGACACTGCCCTAACCATCATGGAGGTGGCCGAGGTGGAAAGTCCTC TGAACCCCAGCTGTAAGATAATGACCTTCAGACCCTCCATGGAGGAGTTCCGGGAGTTCAACAAATACCTTGCATACATGGAGTCTAAAG GAGCCCATCGTGCGGGTCTTGCAAAGGTGATTCCTCCTAAGGAGTGGAAGCCAAGACAGTGCTATGATGACATTGATAATTTGCTCATTC CAGCACCAATTCAGCAGATGGTCACAGGGCAGTCAGGACTGTTCACTCAGTACAACATCCAGAAAAAAGCGATGACTGTGAAGGAGTTCA GGCAGCTGGCCAACAGTGGCAAATATTGTACTCCAAGATACTTGGATTACGAAGATTTGGAGCGCAAGTACTGGAAGAACTTAACTTTTG TGGCACCTATCTATGGTGCAGATATTAATGGGAGCATATATGATGAGATTTGACTGAAGAACGACTTGGAGACGGCAAGTCGGCAGACAA TACAGAGGTCTTCAGTGACAAAGACACTGACCAGAGGAGTTCCCCAGATGTGGCAAAGAGTAAGGGCTGCTTCACCCCTGAGAGCCCTGA GATAGTGTCCGTGGAGGAAGGGGGATACGCTGTCCAGAAAAACGGAGGCAACCCCGAGAGCCGCCCTGACAGCCCCAAGTGTCACGCGGA GCAGAATCACCTCCTGATTGAGGGCCCCTCGGGGACTGTTTCTCTGCCCTTCAGCTTGAAAGCCAACAGACCGCCGCTTGAAGTGTTAAA AAAGATATTCCCCAACCAGAAGCCAACGGTGCTTGAGCTCATCCTCAAGGGCTGTGGCGGGGACCTGGTGAGCGCCGTGGAAGTCCTTCT GTCCAGCCGATCCTCAGTCACGGGAGCAGAGCGAACTTCCGCAGAACCTGAGAGTCTAGCGTTGCCCTCCAATGGGCACATCTTTGAACA CACCTTGAGCTCCTACCCCATCTCGTCTTCCAAATGGTCTGTGGGATCAGCCTTTCGAGTCCCAGACACGTTGAGGTTTTCTGCCGACTC TAGCAACGTTGTCCCCAGTCCCTTGGCTGGGCCTCTGCAGCCCCCTTTCCCCCAGCCACCCCGGTACCCGCTGATGCTGAGGAATACTTT GGCGAGAAGCCAGTCGAGCCCCTTTTTGCCCAATGATGTCACCCTGTGGAACACCATGACGCTGCAGCAGCAGTATCAGCTGAGGTCCCA GTATGTCAGTCCTTTCCCCAGTAACTCTACCAGCGTCTTCAGAAGCTCGCCCGTCCTTCCTGCCCGCGCCACGGAAGACCCTCGGATTTC CATCCCTGATGATGGGTGTCCATTTGTGTCAAAGCAGTCCATTTACACCGAGGACGACTATGACGAGAGGTCTGACTCCTCAGACTCTAG AACACTCAACACATCATCTTAAAGTGGTGCTGGATGGGTGGTGGCCAGGTGACATTTTCTGTGCGTTTTGACCCTGAGGCATCTGAGGAG AGGCCACATCTTGTGTATGCCCTTTCCTTCTGTTTGACAAAGTGACTGTGCTTGATTCTATACATTAGCAATAAAAACATAACTTATTTA ACTTCTTGCACTTCACTGGAAAATGCCAAATAGCTCTGTTCTGTGGCTTTAGTGCTGAATGTTTATTGTAAAAGAGAGTCTAATGTTAAG AATAGTCTTGGGAAGGCTGGGTCCGTGGAAGATTTATTTGGGGATGTAAAGCTGAAGGTCAGCCTTGCACCTAAACCCAACCTGGAATGT TAAATGAAATAATATACTTGAATGCAATTTTGTAAAAGTGGATTCCTCAGGATATGTGAAACCTAAAGGAAGTGGTTCTGTTGCAAATGG ACTATAAACAGGGACATTATATTCTTACACTAAAAATCCTTGCATTTTAAAGAGAGATGCACTTAAGAATAGAGTGAACTGCTCATATGC TTATTTAAGCTTGGACAGTTTTCAGAGACAAACCCCATTAAGAATTACTCTTTTCACATGGCTGAATCGAAACATGTGTAATGTCAATGT AAAACCAATCACAGCTGTGAACTGCATGAAATGTATTGTGAAACGAACACAAGATTAAGCTTTGTCAGGTTAATGTAGCATGTTAAGGAC >41828_41828_3_KDM4C-DMRT3_KDM4C_chr9_6814745_ENST00000535193_DMRT3_chr9_990041_ENST00000190165_length(amino acids)=320AA_BP=1 MTEERLGDGKSADNTEVFSDKDTDQRSSPDVAKSKGCFTPESPEIVSVEEGGYAVQKNGGNPESRPDSPKCHAEQNHLLIEGPSGTVSLP FSLKANRPPLEVLKKIFPNQKPTVLELILKGCGGDLVSAVEVLLSSRSSVTGAERTSAEPESLALPSNGHIFEHTLSSYPISSSKWSVGS AFRVPDTLRFSADSSNVVPSPLAGPLQPPFPQPPRYPLMLRNTLARSQSSPFLPNDVTLWNTMTLQQQYQLRSQYVSPFPSNSTSVFRSS -------------------------------------------------------------- >41828_41828_4_KDM4C-DMRT3_KDM4C_chr9_6814745_ENST00000543771_DMRT3_chr9_990041_ENST00000190165_length(transcript)=2706nt_BP=1015nt GCCATAGGTGCGCGTCGGCGCCCAGGAGGACGTGTGGCGCGTGGACTACATCAGGTCCAGCCCTGCGGGACCCCAGCCAGCGCTTCCGGG CAAGGTTCTGTGCACCTGTTTTCTCCTTCTACGCGAGTATCTTTCCCCTCCGGAAAGAATGGGATATGCCTGTGTCCAAAGGACAAGAAG ATGCGCGCCAGCAAGCCTAAGTTAACCACAGCGCGGAAGTTGAGCCCAAAGCAAGAGCGTGCCGGGCACCTTTAAGCTGTTTGTAAGCCC ACGTGACTCACCAAGTGCGGGCCCCAGCGGTCACGTGACGGCGCGCGCGCCCTCGCGCAGGGAGAGCCGGCGGTGCGCGCGCCTTCGCCG CTGCCTCCCACCCACCCCCTCGACGGGAGGGTGAGGCGCGGCGCAGTGATCGGGCGGCCGGGGTCCTGTGCGCGTGCGCAGCGAACAGCT GTCACCTAGTGCGGAACAAGTCTCCCAAATTTCCCAAATCTCCCTGGGCCGGAGGCCACTGTCTTCTCTTCCTCCTCCACCGAGTCGTGC TCTCGCCCCAACCCGCGCGCCAGACACTGCCCTAACCATCATGGAGGTGGCCGAGGTGGAAAGTCCTCTGAACCCCAGCTGTAAGATAAT GACCTTCAGACCCTCCATGGAGGAGTTCCGGGAGTTCAACAAATACCTTGCATACATGGAGTCTAAAGGAGCCCATCGTGCGGGTCTTGC AAAGGTGATTCCTCCTAAGGAGTGGAAGCCAAGACAGTGCTATGATGACATTGATAATTTGCTCATTCCAGCACCAATTCAGCAGATGGT CACAGGGCAGTCAGGACTGTTCACTCAGTACAACATCCAGAAAAAAGCGATGACTGTGAAGGAGTTCAGGCAGCTGGCCAACAGTGGCAA ATATTGTACTCCAAGATACTTGGATTACGAAGATTTGGAGCGCAAGTACTGGAAGAACTTAACTTTTGTGGCACCTATCTATGGTGCAGA TATTAATGGGAGCATATATGATGAGATTTGACTGAAGAACGACTTGGAGACGGCAAGTCGGCAGACAATACAGAGGTCTTCAGTGACAAA GACACTGACCAGAGGAGTTCCCCAGATGTGGCAAAGAGTAAGGGCTGCTTCACCCCTGAGAGCCCTGAGATAGTGTCCGTGGAGGAAGGG GGATACGCTGTCCAGAAAAACGGAGGCAACCCCGAGAGCCGCCCTGACAGCCCCAAGTGTCACGCGGAGCAGAATCACCTCCTGATTGAG GGCCCCTCGGGGACTGTTTCTCTGCCCTTCAGCTTGAAAGCCAACAGACCGCCGCTTGAAGTGTTAAAAAAGATATTCCCCAACCAGAAG CCAACGGTGCTTGAGCTCATCCTCAAGGGCTGTGGCGGGGACCTGGTGAGCGCCGTGGAAGTCCTTCTGTCCAGCCGATCCTCAGTCACG GGAGCAGAGCGAACTTCCGCAGAACCTGAGAGTCTAGCGTTGCCCTCCAATGGGCACATCTTTGAACACACCTTGAGCTCCTACCCCATC TCGTCTTCCAAATGGTCTGTGGGATCAGCCTTTCGAGTCCCAGACACGTTGAGGTTTTCTGCCGACTCTAGCAACGTTGTCCCCAGTCCC TTGGCTGGGCCTCTGCAGCCCCCTTTCCCCCAGCCACCCCGGTACCCGCTGATGCTGAGGAATACTTTGGCGAGAAGCCAGTCGAGCCCC TTTTTGCCCAATGATGTCACCCTGTGGAACACCATGACGCTGCAGCAGCAGTATCAGCTGAGGTCCCAGTATGTCAGTCCTTTCCCCAGT AACTCTACCAGCGTCTTCAGAAGCTCGCCCGTCCTTCCTGCCCGCGCCACGGAAGACCCTCGGATTTCCATCCCTGATGATGGGTGTCCA TTTGTGTCAAAGCAGTCCATTTACACCGAGGACGACTATGACGAGAGGTCTGACTCCTCAGACTCTAGAACACTCAACACATCATCTTAA AGTGGTGCTGGATGGGTGGTGGCCAGGTGACATTTTCTGTGCGTTTTGACCCTGAGGCATCTGAGGAGAGGCCACATCTTGTGTATGCCC TTTCCTTCTGTTTGACAAAGTGACTGTGCTTGATTCTATACATTAGCAATAAAAACATAACTTATTTAACTTCTTGCACTTCACTGGAAA ATGCCAAATAGCTCTGTTCTGTGGCTTTAGTGCTGAATGTTTATTGTAAAAGAGAGTCTAATGTTAAGAATAGTCTTGGGAAGGCTGGGT CCGTGGAAGATTTATTTGGGGATGTAAAGCTGAAGGTCAGCCTTGCACCTAAACCCAACCTGGAATGTTAAATGAAATAATATACTTGAA TGCAATTTTGTAAAAGTGGATTCCTCAGGATATGTGAAACCTAAAGGAAGTGGTTCTGTTGCAAATGGACTATAAACAGGGACATTATAT TCTTACACTAAAAATCCTTGCATTTTAAAGAGAGATGCACTTAAGAATAGAGTGAACTGCTCATATGCTTATTTAAGCTTGGACAGTTTT CAGAGACAAACCCCATTAAGAATTACTCTTTTCACATGGCTGAATCGAAACATGTGTAATGTCAATGTAAAACCAATCACAGCTGTGAAC TGCATGAAATGTATTGTGAAACGAACACAAGATTAAGCTTTGTCAGGTTAATGTAGCATGTTAAGGACTCTAGAAAAAAATAAACTAAGG >41828_41828_4_KDM4C-DMRT3_KDM4C_chr9_6814745_ENST00000543771_DMRT3_chr9_990041_ENST00000190165_length(amino acids)=320AA_BP=1 MTEERLGDGKSADNTEVFSDKDTDQRSSPDVAKSKGCFTPESPEIVSVEEGGYAVQKNGGNPESRPDSPKCHAEQNHLLIEGPSGTVSLP FSLKANRPPLEVLKKIFPNQKPTVLELILKGCGGDLVSAVEVLLSSRSSVTGAERTSAEPESLALPSNGHIFEHTLSSYPISSSKWSVGS AFRVPDTLRFSADSSNVVPSPLAGPLQPPFPQPPRYPLMLRNTLARSQSSPFLPNDVTLWNTMTLQQQYQLRSQYVSPFPSNSTSVFRSS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for KDM4C-DMRT3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for KDM4C-DMRT3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for KDM4C-DMRT3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
