
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:KHDRBS1-LRRC37A (FusionGDB2 ID:41955) |
Fusion Gene Summary for KHDRBS1-LRRC37A |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: KHDRBS1-LRRC37A | Fusion gene ID: 41955 | Hgene | Tgene | Gene symbol | KHDRBS1 | LRRC37A | Gene ID | 10657 | 105376839 |
| Gene name | KH RNA binding domain containing, signal transduction associated 1 | uncharacterized LOC105376839 | |
| Synonyms | Sam68|p62|p68 | LRRC37A | |
| Cytomap | 1p35.2 | 17q21.31 | |
| Type of gene | protein-coding | ncRNA | |
| Description | KH domain-containing, RNA-binding, signal transduction-associated protein 1GAP-associated tyrosine phosphoprotein p62 (Sam68)KH domain containing, RNA binding, signal transduction associated 1p21 Ras GTPase-activating protein-associated p62src-associa | leucine rich repeat containing 37A | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | Q07666 | O60309 | |
| Ensembl transtripts involved in fusion gene | ENST00000307714, ENST00000327300, ENST00000492989, | ENST00000320254, ENST00000393465, ENST00000496930, | |
| Fusion gene scores | * DoF score | 10 X 7 X 7=490 | 7 X 7 X 6=294 |
| # samples | 11 | 7 | |
| ** MAII score | log2(11/490*10)=-2.15527822547791 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/294*10)=-2.0703893278914 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: KHDRBS1 [Title/Abstract] AND LRRC37A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | KHDRBS1(32479978)-LRRC37A(44407816), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | KHDRBS1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | 24514149 |
| Hgene | KHDRBS1 | GO:0007166 | cell surface receptor signaling pathway | 9045636 |
| Hgene | KHDRBS1 | GO:0031647 | regulation of protein stability | 21613532 |
| Hgene | KHDRBS1 | GO:0045948 | positive regulation of translational initiation | 21613532 |
| Hgene | KHDRBS1 | GO:0046833 | positive regulation of RNA export from nucleus | 21613532 |
 Fusion gene breakpoints across KHDRBS1 (5'-gene) Fusion gene breakpoints across KHDRBS1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
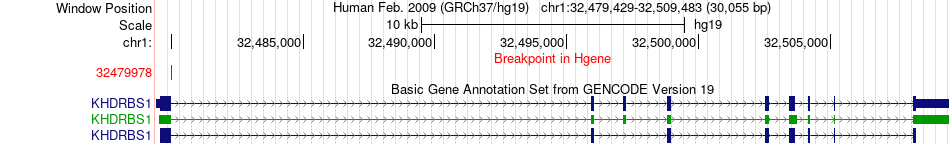 |
 Fusion gene breakpoints across LRRC37A (3'-gene) Fusion gene breakpoints across LRRC37A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
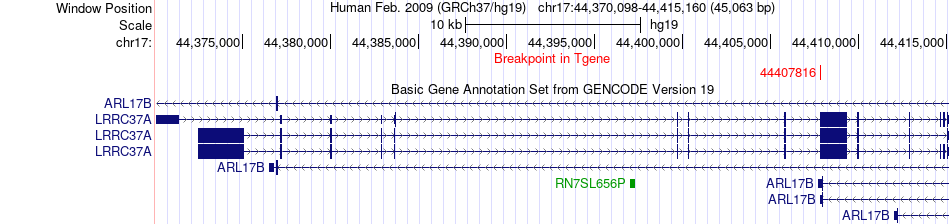 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | Non-Cancer | TCGA-IN-8462-11A | KHDRBS1 | chr1 | 32479978 | + | LRRC37A | chr17 | 44407816 | + |
Top |
Fusion Gene ORF analysis for KHDRBS1-LRRC37A |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000307714 | ENST00000320254 | KHDRBS1 | chr1 | 32479978 | + | LRRC37A | chr17 | 44407816 | + |
| 3UTR-3CDS | ENST00000307714 | ENST00000393465 | KHDRBS1 | chr1 | 32479978 | + | LRRC37A | chr17 | 44407816 | + |
| 3UTR-3CDS | ENST00000307714 | ENST00000496930 | KHDRBS1 | chr1 | 32479978 | + | LRRC37A | chr17 | 44407816 | + |
| In-frame | ENST00000327300 | ENST00000320254 | KHDRBS1 | chr1 | 32479978 | + | LRRC37A | chr17 | 44407816 | + |
| In-frame | ENST00000327300 | ENST00000393465 | KHDRBS1 | chr1 | 32479978 | + | LRRC37A | chr17 | 44407816 | + |
| In-frame | ENST00000327300 | ENST00000496930 | KHDRBS1 | chr1 | 32479978 | + | LRRC37A | chr17 | 44407816 | + |
| In-frame | ENST00000492989 | ENST00000320254 | KHDRBS1 | chr1 | 32479978 | + | LRRC37A | chr17 | 44407816 | + |
| In-frame | ENST00000492989 | ENST00000393465 | KHDRBS1 | chr1 | 32479978 | + | LRRC37A | chr17 | 44407816 | + |
| In-frame | ENST00000492989 | ENST00000496930 | KHDRBS1 | chr1 | 32479978 | + | LRRC37A | chr17 | 44407816 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000327300 | KHDRBS1 | chr1 | 32479978 | + | ENST00000496930 | LRRC37A | chr17 | 44407816 | + | 2549 | 549 | 5 | 2479 | 824 |
| ENST00000327300 | KHDRBS1 | chr1 | 32479978 | + | ENST00000393465 | LRRC37A | chr17 | 44407816 | + | 2339 | 549 | 5 | 2284 | 759 |
| ENST00000327300 | KHDRBS1 | chr1 | 32479978 | + | ENST00000320254 | LRRC37A | chr17 | 44407816 | + | 2551 | 549 | 5 | 2479 | 824 |
| ENST00000492989 | KHDRBS1 | chr1 | 32479978 | + | ENST00000496930 | LRRC37A | chr17 | 44407816 | + | 2382 | 382 | 0 | 2312 | 770 |
| ENST00000492989 | KHDRBS1 | chr1 | 32479978 | + | ENST00000393465 | LRRC37A | chr17 | 44407816 | + | 2172 | 382 | 0 | 2117 | 705 |
| ENST00000492989 | KHDRBS1 | chr1 | 32479978 | + | ENST00000320254 | LRRC37A | chr17 | 44407816 | + | 2384 | 382 | 0 | 2312 | 770 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000327300 | ENST00000496930 | KHDRBS1 | chr1 | 32479978 | + | LRRC37A | chr17 | 44407816 | + | 0.007929902 | 0.99207 |
| ENST00000327300 | ENST00000393465 | KHDRBS1 | chr1 | 32479978 | + | LRRC37A | chr17 | 44407816 | + | 0.009107705 | 0.9908923 |
| ENST00000327300 | ENST00000320254 | KHDRBS1 | chr1 | 32479978 | + | LRRC37A | chr17 | 44407816 | + | 0.008083405 | 0.9919166 |
| ENST00000492989 | ENST00000496930 | KHDRBS1 | chr1 | 32479978 | + | LRRC37A | chr17 | 44407816 | + | 0.008011001 | 0.99198896 |
| ENST00000492989 | ENST00000393465 | KHDRBS1 | chr1 | 32479978 | + | LRRC37A | chr17 | 44407816 | + | 0.009087944 | 0.9909121 |
| ENST00000492989 | ENST00000320254 | KHDRBS1 | chr1 | 32479978 | + | LRRC37A | chr17 | 44407816 | + | 0.008158351 | 0.9918417 |
Top |
Fusion Genomic Features for KHDRBS1-LRRC37A |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| KHDRBS1 | chr1 | 32479978 | + | LRRC37A | chr17 | 44407815 | + | 3.86E-07 | 0.99999964 |
| KHDRBS1 | chr1 | 32479978 | + | LRRC37A | chr17 | 44407815 | + | 3.86E-07 | 0.99999964 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
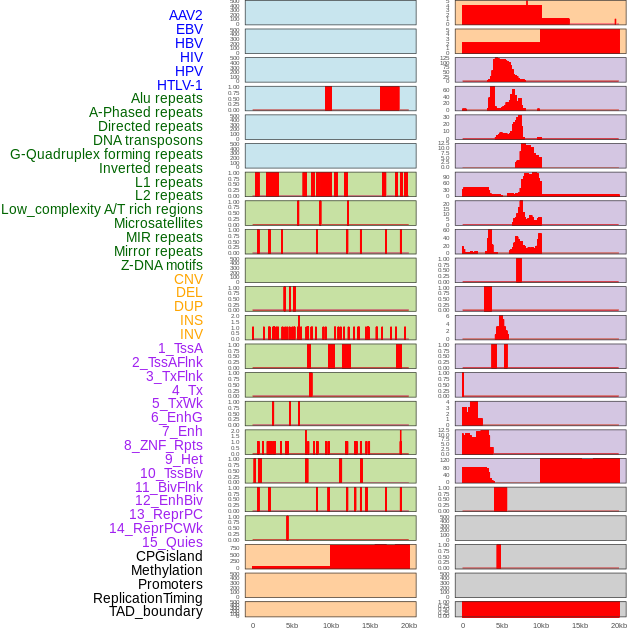 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
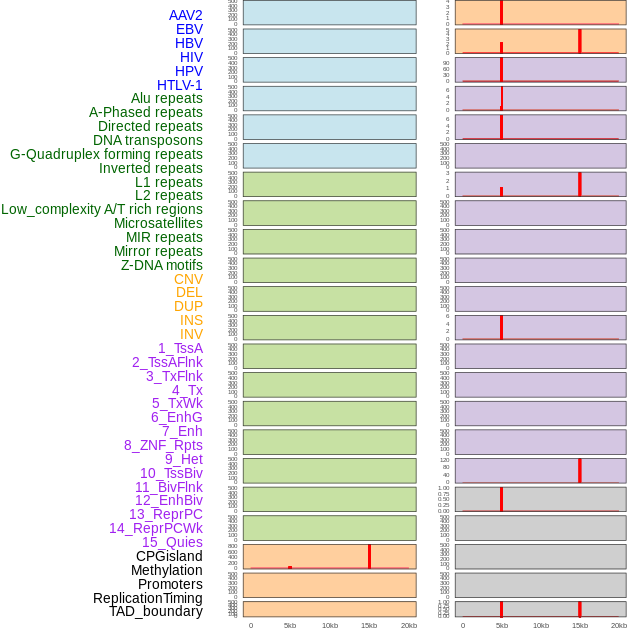 |
Top |
Fusion Protein Features for KHDRBS1-LRRC37A |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:32479978/chr17:44407816) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| KHDRBS1 | LRRC37A |
| FUNCTION: Recruited and tyrosine phosphorylated by several receptor systems, for example the T-cell, leptin and insulin receptors. Once phosphorylated, functions as an adapter protein in signal transduction cascades by binding to SH2 and SH3 domain-containing proteins. Role in G2-M progression in the cell cycle. Represses CBP-dependent transcriptional activation apparently by competing with other nuclear factors for binding to CBP. Also acts as a putative regulator of mRNA stability and/or translation rates and mediates mRNA nuclear export. Positively regulates the association of constitutive transport element (CTE)-containing mRNA with large polyribosomes and translation initiation. According to some authors, is not involved in the nucleocytoplasmic export of unspliced (CTE)-containing RNA species according to (PubMed:22253824). RNA-binding protein that plays a role in the regulation of alternative splicing and influences mRNA splice site selection and exon inclusion. Binds to RNA containing 5'-[AU]UAA-3' as a bipartite motif spaced by more than 15 nucleotides. Binds poly(A). Can regulate CD44 alternative splicing in a Ras pathway-dependent manner (By similarity). In cooperation with HNRNPA1 modulates alternative splicing of BCL2L1 by promoting splicing toward isoform Bcl-X(S), and of SMN1 (PubMed:17371836, PubMed:20186123). Can regulate alternative splicing of NRXN1 and NRXN3 in the laminin G-like domain 6 containing the evolutionary conserved neurexin alternative spliced segment 4 (AS4) involved in neurexin selective targeting to postsynaptic partners. In a neuronal activity-dependent manner cooperates synergistically with KHDRBS2/SLIM-1 in regulation of NRXN1 exon skipping at AS4. The cooperation with KHDRBS2/SLIM-1 is antagonistic for regulation of NXRN3 alternative splicing at AS4 (By similarity). {ECO:0000250|UniProtKB:Q60749, ECO:0000269|PubMed:15021911, ECO:0000269|PubMed:17371836, ECO:0000269|PubMed:20186123, ECO:0000269|PubMed:20610388, ECO:0000269|PubMed:22253824, ECO:0000269|PubMed:26758068}.; FUNCTION: Isoform 3, which is expressed in growth-arrested cells only, inhibits S phase. {ECO:0000269|PubMed:9013542}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KHDRBS1 | chr1:32479978 | chr17:44407816 | ENST00000327300 | + | 1 | 9 | 34_41 | 127 | 444.0 | Compositional bias | Pro-rich |
| Hgene | KHDRBS1 | chr1:32479978 | chr17:44407816 | ENST00000327300 | + | 1 | 9 | 44_55 | 127 | 444.0 | Compositional bias | DMA/Gly-rich |
| Hgene | KHDRBS1 | chr1:32479978 | chr17:44407816 | ENST00000327300 | + | 1 | 9 | 59_89 | 127 | 444.0 | Compositional bias | Pro-rich |
| Hgene | KHDRBS1 | chr1:32479978 | chr17:44407816 | ENST00000492989 | + | 1 | 8 | 34_41 | 127 | 405.0 | Compositional bias | Pro-rich |
| Hgene | KHDRBS1 | chr1:32479978 | chr17:44407816 | ENST00000492989 | + | 1 | 8 | 44_55 | 127 | 405.0 | Compositional bias | DMA/Gly-rich |
| Hgene | KHDRBS1 | chr1:32479978 | chr17:44407816 | ENST00000492989 | + | 1 | 8 | 59_89 | 127 | 405.0 | Compositional bias | Pro-rich |
| Tgene | LRRC37A | chr1:32479978 | chr17:44407816 | ENST00000320254 | 7 | 14 | 1604_1700 | 1057 | 1701.0 | Topological domain | Cytoplasmic | |
| Tgene | LRRC37A | chr1:32479978 | chr17:44407816 | ENST00000320254 | 7 | 14 | 1583_1603 | 1057 | 1701.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KHDRBS1 | chr1:32479978 | chr17:44407816 | ENST00000327300 | + | 1 | 9 | 282_292 | 127 | 444.0 | Compositional bias | DMA/Gly-rich |
| Hgene | KHDRBS1 | chr1:32479978 | chr17:44407816 | ENST00000327300 | + | 1 | 9 | 295_301 | 127 | 444.0 | Compositional bias | Pro-rich |
| Hgene | KHDRBS1 | chr1:32479978 | chr17:44407816 | ENST00000327300 | + | 1 | 9 | 302_332 | 127 | 444.0 | Compositional bias | Arg/Gly-rich |
| Hgene | KHDRBS1 | chr1:32479978 | chr17:44407816 | ENST00000327300 | + | 1 | 9 | 334_363 | 127 | 444.0 | Compositional bias | Pro-rich |
| Hgene | KHDRBS1 | chr1:32479978 | chr17:44407816 | ENST00000492989 | + | 1 | 8 | 282_292 | 127 | 405.0 | Compositional bias | DMA/Gly-rich |
| Hgene | KHDRBS1 | chr1:32479978 | chr17:44407816 | ENST00000492989 | + | 1 | 8 | 295_301 | 127 | 405.0 | Compositional bias | Pro-rich |
| Hgene | KHDRBS1 | chr1:32479978 | chr17:44407816 | ENST00000492989 | + | 1 | 8 | 302_332 | 127 | 405.0 | Compositional bias | Arg/Gly-rich |
| Hgene | KHDRBS1 | chr1:32479978 | chr17:44407816 | ENST00000492989 | + | 1 | 8 | 334_363 | 127 | 405.0 | Compositional bias | Pro-rich |
| Hgene | KHDRBS1 | chr1:32479978 | chr17:44407816 | ENST00000327300 | + | 1 | 9 | 171_197 | 127 | 444.0 | Domain | KH |
| Hgene | KHDRBS1 | chr1:32479978 | chr17:44407816 | ENST00000492989 | + | 1 | 8 | 171_197 | 127 | 405.0 | Domain | KH |
| Hgene | KHDRBS1 | chr1:32479978 | chr17:44407816 | ENST00000327300 | + | 1 | 9 | 100_260 | 127 | 444.0 | Region | Involved in homodimerization |
| Hgene | KHDRBS1 | chr1:32479978 | chr17:44407816 | ENST00000492989 | + | 1 | 8 | 100_260 | 127 | 405.0 | Region | Involved in homodimerization |
| Tgene | LRRC37A | chr1:32479978 | chr17:44407816 | ENST00000320254 | 7 | 14 | 866_887 | 1057 | 1701.0 | Repeat | Note=LRR 1 | |
| Tgene | LRRC37A | chr1:32479978 | chr17:44407816 | ENST00000320254 | 7 | 14 | 890_911 | 1057 | 1701.0 | Repeat | Note=LRR 2 | |
| Tgene | LRRC37A | chr1:32479978 | chr17:44407816 | ENST00000320254 | 7 | 14 | 914_935 | 1057 | 1701.0 | Repeat | Note=LRR 3 | |
| Tgene | LRRC37A | chr1:32479978 | chr17:44407816 | ENST00000320254 | 7 | 14 | 938_959 | 1057 | 1701.0 | Repeat | Note=LRR 4 | |
| Tgene | LRRC37A | chr1:32479978 | chr17:44407816 | ENST00000320254 | 7 | 14 | 965_986 | 1057 | 1701.0 | Repeat | Note=LRR 5 | |
| Tgene | LRRC37A | chr1:32479978 | chr17:44407816 | ENST00000320254 | 7 | 14 | 36_1582 | 1057 | 1701.0 | Topological domain | Extracellular |
Top |
Fusion Gene Sequence for KHDRBS1-LRRC37A |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >41955_41955_1_KHDRBS1-LRRC37A_KHDRBS1_chr1_32479978_ENST00000327300_LRRC37A_chr17_44407816_ENST00000320254_length(transcript)=2551nt_BP=549nt GCTGGCTGCGCACGCGCGCCGCCTCATTTCCGGTGCTCTCTCTCGCTGGGTCGCTCGGGTCGGCTTCGGTCGCTACCGCTCCCGCTCTGC CACCCCCGCCAACCGCCGCTCGGGCCTCCGTCGCTGCCGCGTCGCTTTCTCGCTCCTTGGATCGCACATCCTCCCAGATGCAGCGCCGGG ACGACCCCGCCGCGCGCATGAGCCGGTCTTCGGGCCGTAGCGGCTCCATGGACCCCTCCGGTGCCCACCCCTCGGTGCGTCAGACGCCGT CTCGGCAGCCGCCGCTGCCTCACCGGTCCCGGGGAGGCGGAGGGGGATCCCGCGGGGGCGCCCGGGCCTCGCCCGCCACGCAGCCGCCAC CGCTGCTGCCGCCCTCGGCCACGGGTCCCGACGCGACAGTGGGCGGGCCAGCGCCGACCCCGCTGCTGCCCCCCTCGGCCACAGCCTCGG TCAAGATGGAGCCAGAGAACAAGTACCTGCCCGAACTCATGGCCGAGAAGGACTCGCTCGACCCGTCCTTCACTCACGCCATGCAGCTGC TGACGGCAGCTGAAGAAGCATCGGTAGGGAATCCAGAAGGAGCGTTCATGAAGGTGTTACAAGCCCGGAAGAACTACACAAGCACTGAGC TGATTGTTGAGCCAGAGGAGCCCTCAGACAGCAGTGGCATCAACTTGTCAGGCTTTGGGAGTGAGCAGCTAGACACCAATGACGAGAGTG ATTTTATCAGTACACTAAGTTACATCTTGCCTTATTTCTCAGCGGTAAACCTAGATGTGAAATCACTGTTACTACCGTTCATTAAACTGC CAACCACAGGAAACAGCCTGGCAAAGATTCAAACTGTAGGCCAAAACCGGCAGAGAGTGAAGAGAGTCCTCATGGGCCCAAGGAGCATCC AGAAAAGGCACTTCAAAGAGGTAGGAAGGCAGAGCATCAGGAGGGAACAGGGTGCCCAGGCATCTGTGGAGAACGCTGCCGAAGAAAAAA GGCTCACGAGTCCAGCCCCAAGGGAGGTGGAACAGCCCCACACACAGCAGGGGCCTGAGAAGTTAGCGGGAAACGCCGTCTACACCAAGC CTTCGTTCACCCAAGAGCATAAGGCAGCAGTCTCTGTGCTGAAACCCTTCTCCAAGGGCACGCCTTCTACCTCCAGCCCTGCAAAAGCCC TACCACAGGTGAGAGACAGATCGAAAGACTTAACCCACGCTATTTCCATTTTAGAAAGTGCAAAGGCTAGAGTTACAAATACGAAGACGT CTAAACCAATCGTACATGCCAGAAAAAAATACCGCTTTCACAAAACTCGCTCCCACGTGACCCACAGAACAACCAAAGTCAAAAAGAGTC CAAAGGTCAGAAAGAAAAGTTATCTGAGTAGACTGATGCTCGCAAACAGGCTTCCATTCTCTGCAGCGAAGAGCCTCATAAATTCCCCTT CACAAGGGGCTTTTTCATCCTTAGGAGACCTGAGTCCTCAAGAAAACCCTTTTCTGGAAGTATCTGCTCTTTCAGAACATTTTATAGAGA AGAATAATACAAAACACACAACTGCAAGAAATGCCTTTGAAGAAAATGATTTTATGGAAAACACTAACATGCCAGAAGGAACCATCTCTG AAAACACAAACTACAATCATCCTCCTGAGGCAGATTCCGCTGGGACTGCATTCAACTTAGGGCCAACTGTTAAACAAACTGAGACAAAAT GGGAATACAACAACGTGGGCACTGACCTGTCCCCCGAGCCCAAAAGCTTCAATTACCCATTGCTCTCGTCCCCAGGTGATCAGTTTGAAA TTCAGCTAACCCAGCAGCTACAGTCCCTTATCCCCAACAACAATGTGAGAAGGCTCATTGCTCATGTTATCCGGACCTTGAAGATGGACT GCTCTGGGGCCCATGTGCAAGTGACCTGTGCCAAGCTCATCTCCAGGACAGGCCACCTGATGAAGCTTCTCAGTGGGCAGCAGGAAGTAA AGGCATCCAAGATAGAATGGGATACGGACCAATGGAAGATTGAGAACTACATTAATGAGAGCACAGAAGCCCAGAGTGAACAGAAAGAGA AGTCGCTTGAGCTCAAAAAAGAAGTTCCAGGATATGGCTATACTGACAAACTCATCTTGGCATTAATTGTTACTGGAATACTAACGATTT TGATTATACTTTTCTGCCTCATTGTGATATGTTGTCACCGAAGGTCATTACAAGAAGATGAAGAAGGATTCTCAAGGGGCATTTTCAGAT TTCTGCCATGGAGGGGATGCTCTTCGCGAAGGGAGAGTCAGGATGGACTTTCCTCATTTGGACAGCCGCTCTGGTTTAAAGATCTGTACA AACCTCTCAGTGCCACAAGAATAAATAATCATGCATGGAAGCTGCACAAGAAGTCATCTAATGAGGACAAGATCCTCAACAGGGACCCTG GGGACAGCGAAGCCCCAACGGAGGAGGAGGAGAGTGAAGCCCTGCCATAGGAGGAGAACACAGCCCACCTCAGGCCTCCTGCAAAAATAC >41955_41955_1_KHDRBS1-LRRC37A_KHDRBS1_chr1_32479978_ENST00000327300_LRRC37A_chr17_44407816_ENST00000320254_length(amino acids)=824AA_BP=181 MRTRAASFPVLSLAGSLGSASVATAPALPPPPTAARASVAAASLSRSLDRTSSQMQRRDDPAARMSRSSGRSGSMDPSGAHPSVRQTPSR QPPLPHRSRGGGGGSRGGARASPATQPPPLLPPSATGPDATVGGPAPTPLLPPSATASVKMEPENKYLPELMAEKDSLDPSFTHAMQLLT AAEEASVGNPEGAFMKVLQARKNYTSTELIVEPEEPSDSSGINLSGFGSEQLDTNDESDFISTLSYILPYFSAVNLDVKSLLLPFIKLPT TGNSLAKIQTVGQNRQRVKRVLMGPRSIQKRHFKEVGRQSIRREQGAQASVENAAEEKRLTSPAPREVEQPHTQQGPEKLAGNAVYTKPS FTQEHKAAVSVLKPFSKGTPSTSSPAKALPQVRDRSKDLTHAISILESAKARVTNTKTSKPIVHARKKYRFHKTRSHVTHRTTKVKKSPK VRKKSYLSRLMLANRLPFSAAKSLINSPSQGAFSSLGDLSPQENPFLEVSALSEHFIEKNNTKHTTARNAFEENDFMENTNMPEGTISEN TNYNHPPEADSAGTAFNLGPTVKQTETKWEYNNVGTDLSPEPKSFNYPLLSSPGDQFEIQLTQQLQSLIPNNNVRRLIAHVIRTLKMDCS GAHVQVTCAKLISRTGHLMKLLSGQQEVKASKIEWDTDQWKIENYINESTEAQSEQKEKSLELKKEVPGYGYTDKLILALIVTGILTILI ILFCLIVICCHRRSLQEDEEGFSRGIFRFLPWRGCSSRRESQDGLSSFGQPLWFKDLYKPLSATRINNHAWKLHKKSSNEDKILNRDPGD -------------------------------------------------------------- >41955_41955_2_KHDRBS1-LRRC37A_KHDRBS1_chr1_32479978_ENST00000327300_LRRC37A_chr17_44407816_ENST00000393465_length(transcript)=2339nt_BP=549nt GCTGGCTGCGCACGCGCGCCGCCTCATTTCCGGTGCTCTCTCTCGCTGGGTCGCTCGGGTCGGCTTCGGTCGCTACCGCTCCCGCTCTGC CACCCCCGCCAACCGCCGCTCGGGCCTCCGTCGCTGCCGCGTCGCTTTCTCGCTCCTTGGATCGCACATCCTCCCAGATGCAGCGCCGGG ACGACCCCGCCGCGCGCATGAGCCGGTCTTCGGGCCGTAGCGGCTCCATGGACCCCTCCGGTGCCCACCCCTCGGTGCGTCAGACGCCGT CTCGGCAGCCGCCGCTGCCTCACCGGTCCCGGGGAGGCGGAGGGGGATCCCGCGGGGGCGCCCGGGCCTCGCCCGCCACGCAGCCGCCAC CGCTGCTGCCGCCCTCGGCCACGGGTCCCGACGCGACAGTGGGCGGGCCAGCGCCGACCCCGCTGCTGCCCCCCTCGGCCACAGCCTCGG TCAAGATGGAGCCAGAGAACAAGTACCTGCCCGAACTCATGGCCGAGAAGGACTCGCTCGACCCGTCCTTCACTCACGCCATGCAGCTGC TGACGGCAGCTGAAGAAGCATCGGTAGGGAATCCAGAAGGAGCGTTCATGAAGGTGTTACAAGCCCGGAAGAACTACACAAGCACTGAGC TGATTGTTGAGCCAGAGGAGCCCTCAGACAGCAGTGGCATCAACTTGTCAGGCTTTGGGAGTGAGCAGCTAGACACCAATGACGAGAGTG ATTTTATCAGTACACTAAGTTACATCTTGCCTTATTTCTCAGCGGTAAACCTAGATGTGAAATCACTGTTACTACCGTTCATTAAACTGC CAACCACAGGAAACAGCCTGGCAAAGATTCAAACTGTAGGCCAAAACCGGCAGAGAGTGAAGAGAGTCCTCATGGGCCCAAGGAGCATCC AGAAAAGGCACTTCAAAGAGGTAGGAAGGCAGAGCATCAGGAGGGAACAGGGTGCCCAGGCATCTGTGGAGAACGCTGCCGAAGAAAAAA GGCTCACGAGTCCAGCCCCAAGGGAGGTGGAACAGCCCCACACACAGCAGGGGCCTGAGAAGTTAGCGGGAAACGCCGTCTACACCAAGC CTTCGTTCACCCAAGAGCATAAGGCAGCAGTCTCTGTGCTGAAACCCTTCTCCAAGGGCACGCCTTCTACCTCCAGCCCTGCAAAAGCCC TACCACAGGTGAGAGACAGATCGAAAGACTTAACCCACGCTATTTCCATTTTAGAAAGTGCAAAGGCTAGAGTTACAAATACGAAGACGT CTAAACCAATCGTACATGCCAGAAAAAAATACCGCTTTCACAAAACTCGCTCCCACGTGACCCACAGAACAACCAAAGTCAAAAAGAGTC CAAAGGTCAGAAAGAAAAGTTATCTGAGTAGACTGATGCTCGCAAACAGGCTTCCATTCTCTGCAGCGAAGAGCCTCATAAATTCCCCTT CACAAGGGGCTTTTTCATCCTTAGGAGACCTGAGTCCTCAAGAAAACCCTTTTCTGGAAGTATCTGCTCTTTCAGAACATTTTATAGAGA AGAATAATACAAAACACACAACTGCAAGAAATGCCTTTGAAGAAAATGATTTTATGGAAAACACTAACATGCCAGAAGGAACCATCTCTG AAAACACAAACTACAATCATCCTCCTGAGGCAGATTCCGCTGGGACTGCATTCAACTTAGGGCCAACTGTTAAACAAACTGAGACAAAAT GGGAATACAACAACGTGGGCACTGACCTGTCCCCCGAGCCCAAAAGCTTCAATTACCCATTGCTCTCGTCCCCAGGTGATCAGTTTGAAA TTCAGCTAACCCAGCAGCTACAGTCCCTTATCCCCAACAACAATGTGAGAAGGCTCATTGCTCATGTTATCCGGACCTTGAAGATGGACT GCTCTGGGGCCCATGTGCAAGTGACCTGTGCCAAGCTCATCTCCAGGACAGGCCACCTGATGAAGCTTCTCAGTGGGCAGCAGGAAGTAA AGGCATCCAAGATAGAATGGGATACGGACCAATGGAAGATTGAGAACTACATTAATGAGAGCACAGAAGCCCAGAGTGAACAGAAAGAGA AGTCGCTTGAGCTCAAAAAAGAAGTTCCAGGATATGGCTATACTGACAAACTCATCTTGGCATTAATTGTTACTGGAATACTAACGATTT TGATTATACTTTTCTGCCTCATTGTGATATGTTGTCACCGAAGGTCATTACAAGAAGATGAAGAAGGATTCTCAAGGGACAGCGAAGCCC >41955_41955_2_KHDRBS1-LRRC37A_KHDRBS1_chr1_32479978_ENST00000327300_LRRC37A_chr17_44407816_ENST00000393465_length(amino acids)=759AA_BP=181 MRTRAASFPVLSLAGSLGSASVATAPALPPPPTAARASVAAASLSRSLDRTSSQMQRRDDPAARMSRSSGRSGSMDPSGAHPSVRQTPSR QPPLPHRSRGGGGGSRGGARASPATQPPPLLPPSATGPDATVGGPAPTPLLPPSATASVKMEPENKYLPELMAEKDSLDPSFTHAMQLLT AAEEASVGNPEGAFMKVLQARKNYTSTELIVEPEEPSDSSGINLSGFGSEQLDTNDESDFISTLSYILPYFSAVNLDVKSLLLPFIKLPT TGNSLAKIQTVGQNRQRVKRVLMGPRSIQKRHFKEVGRQSIRREQGAQASVENAAEEKRLTSPAPREVEQPHTQQGPEKLAGNAVYTKPS FTQEHKAAVSVLKPFSKGTPSTSSPAKALPQVRDRSKDLTHAISILESAKARVTNTKTSKPIVHARKKYRFHKTRSHVTHRTTKVKKSPK VRKKSYLSRLMLANRLPFSAAKSLINSPSQGAFSSLGDLSPQENPFLEVSALSEHFIEKNNTKHTTARNAFEENDFMENTNMPEGTISEN TNYNHPPEADSAGTAFNLGPTVKQTETKWEYNNVGTDLSPEPKSFNYPLLSSPGDQFEIQLTQQLQSLIPNNNVRRLIAHVIRTLKMDCS GAHVQVTCAKLISRTGHLMKLLSGQQEVKASKIEWDTDQWKIENYINESTEAQSEQKEKSLELKKEVPGYGYTDKLILALIVTGILTILI -------------------------------------------------------------- >41955_41955_3_KHDRBS1-LRRC37A_KHDRBS1_chr1_32479978_ENST00000327300_LRRC37A_chr17_44407816_ENST00000496930_length(transcript)=2549nt_BP=549nt GCTGGCTGCGCACGCGCGCCGCCTCATTTCCGGTGCTCTCTCTCGCTGGGTCGCTCGGGTCGGCTTCGGTCGCTACCGCTCCCGCTCTGC CACCCCCGCCAACCGCCGCTCGGGCCTCCGTCGCTGCCGCGTCGCTTTCTCGCTCCTTGGATCGCACATCCTCCCAGATGCAGCGCCGGG ACGACCCCGCCGCGCGCATGAGCCGGTCTTCGGGCCGTAGCGGCTCCATGGACCCCTCCGGTGCCCACCCCTCGGTGCGTCAGACGCCGT CTCGGCAGCCGCCGCTGCCTCACCGGTCCCGGGGAGGCGGAGGGGGATCCCGCGGGGGCGCCCGGGCCTCGCCCGCCACGCAGCCGCCAC CGCTGCTGCCGCCCTCGGCCACGGGTCCCGACGCGACAGTGGGCGGGCCAGCGCCGACCCCGCTGCTGCCCCCCTCGGCCACAGCCTCGG TCAAGATGGAGCCAGAGAACAAGTACCTGCCCGAACTCATGGCCGAGAAGGACTCGCTCGACCCGTCCTTCACTCACGCCATGCAGCTGC TGACGGCAGCTGAAGAAGCATCGGTAGGGAATCCAGAAGGAGCGTTCATGAAGGTGTTACAAGCCCGGAAGAACTACACAAGCACTGAGC TGATTGTTGAGCCAGAGGAGCCCTCAGACAGCAGTGGCATCAACTTGTCAGGCTTTGGGAGTGAGCAGCTAGACACCAATGACGAGAGTG ATTTTATCAGTACACTAAGTTACATCTTGCCTTATTTCTCAGCGGTAAACCTAGATGTGAAATCACTGTTACTACCGTTCATTAAACTGC CAACCACAGGAAACAGCCTGGCAAAGATTCAAACTGTAGGCCAAAACCGGCAGAGAGTGAAGAGAGTCCTCATGGGCCCAAGGAGCATCC AGAAAAGGCACTTCAAAGAGGTAGGAAGGCAGAGCATCAGGAGGGAACAGGGTGCCCAGGCATCTGTGGAGAACGCTGCCGAAGAAAAAA GGCTCACGAGTCCAGCCCCAAGGGAGGTGGAACAGCCCCACACACAGCAGGGGCCTGAGAAGTTAGCGGGAAACGCCGTCTACACCAAGC CTTCGTTCACCCAAGAGCATAAGGCAGCAGTCTCTGTGCTGAAACCCTTCTCCAAGGGCACGCCTTCTACCTCCAGCCCTGCAAAAGCCC TACCACAGGTGAGAGACAGATCGAAAGACTTAACCCACGCTATTTCCATTTTAGAAAGTGCAAAGGCTAGAGTTACAAATACGAAGACGT CTAAACCAATCGTACATGCCAGAAAAAAATACCGCTTTCACAAAACTCGCTCCCACGTGACCCACAGAACAACCAAAGTCAAAAAGAGTC CAAAGGTCAGAAAGAAAAGTTATCTGAGTAGACTGATGCTCGCAAACAGGCTTCCATTCTCTGCAGCGAAGAGCCTCATAAATTCCCCTT CACAAGGGGCTTTTTCATCCTTAGGAGACCTGAGTCCTCAAGAAAACCCTTTTCTGGAAGTATCTGCTCTTTCAGAACATTTTATAGAGA AGAATAATACAAAACACACAACTGCAAGAAATGCCTTTGAAGAAAATGATTTTATGGAAAACACTAACATGCCAGAAGGAACCATCTCTG AAAACACAAACTACAATCATCCTCCTGAGGCAGATTCCGCTGGGACTGCATTCAACTTAGGGCCAACTGTTAAACAAACTGAGACAAAAT GGGAATACAACAACGTGGGCACTGACCTGTCCCCCGAGCCCAAAAGCTTCAATTACCCATTGCTCTCGTCCCCAGGTGATCAGTTTGAAA TTCAGCTAACCCAGCAGCTACAGTCCCTTATCCCCAACAACAATGTGAGAAGGCTCATTGCTCATGTTATCCGGACCTTGAAGATGGACT GCTCTGGGGCCCATGTGCAAGTGACCTGTGCCAAGCTCATCTCCAGGACAGGCCACCTGATGAAGCTTCTCAGTGGGCAGCAGGAAGTAA AGGCATCCAAGATAGAATGGGATACGGACCAATGGAAGATTGAGAACTACATTAATGAGAGCACAGAAGCCCAGAGTGAACAGAAAGAGA AGTCGCTTGAGCTCAAAAAAGAAGTTCCAGGATATGGCTATACTGACAAACTCATCTTGGCATTAATTGTTACTGGAATACTAACGATTT TGATTATACTTTTCTGCCTCATTGTGATATGTTGTCACCGAAGGTCATTACAAGAAGATGAAGAAGGATTCTCAAGGGGCATTTTCAGAT TTCTGCCATGGAGGGGATGCTCTTCGCGAAGGGAGAGTCAGGATGGACTTTCCTCATTTGGACAGCCGCTCTGGTTTAAAGATCTGTACA AACCTCTCAGTGCCACAAGAATAAATAATCATGCATGGAAGCTGCACAAGAAGTCATCTAATGAGGACAAGATCCTCAACAGGGACCCTG GGGACAGCGAAGCCCCAACGGAGGAGGAGGAGAGTGAAGCCCTGCCATAGGAGGAGAACACAGCCCACCTCAGGCCTCCTGCAAAAATAC >41955_41955_3_KHDRBS1-LRRC37A_KHDRBS1_chr1_32479978_ENST00000327300_LRRC37A_chr17_44407816_ENST00000496930_length(amino acids)=824AA_BP=181 MRTRAASFPVLSLAGSLGSASVATAPALPPPPTAARASVAAASLSRSLDRTSSQMQRRDDPAARMSRSSGRSGSMDPSGAHPSVRQTPSR QPPLPHRSRGGGGGSRGGARASPATQPPPLLPPSATGPDATVGGPAPTPLLPPSATASVKMEPENKYLPELMAEKDSLDPSFTHAMQLLT AAEEASVGNPEGAFMKVLQARKNYTSTELIVEPEEPSDSSGINLSGFGSEQLDTNDESDFISTLSYILPYFSAVNLDVKSLLLPFIKLPT TGNSLAKIQTVGQNRQRVKRVLMGPRSIQKRHFKEVGRQSIRREQGAQASVENAAEEKRLTSPAPREVEQPHTQQGPEKLAGNAVYTKPS FTQEHKAAVSVLKPFSKGTPSTSSPAKALPQVRDRSKDLTHAISILESAKARVTNTKTSKPIVHARKKYRFHKTRSHVTHRTTKVKKSPK VRKKSYLSRLMLANRLPFSAAKSLINSPSQGAFSSLGDLSPQENPFLEVSALSEHFIEKNNTKHTTARNAFEENDFMENTNMPEGTISEN TNYNHPPEADSAGTAFNLGPTVKQTETKWEYNNVGTDLSPEPKSFNYPLLSSPGDQFEIQLTQQLQSLIPNNNVRRLIAHVIRTLKMDCS GAHVQVTCAKLISRTGHLMKLLSGQQEVKASKIEWDTDQWKIENYINESTEAQSEQKEKSLELKKEVPGYGYTDKLILALIVTGILTILI ILFCLIVICCHRRSLQEDEEGFSRGIFRFLPWRGCSSRRESQDGLSSFGQPLWFKDLYKPLSATRINNHAWKLHKKSSNEDKILNRDPGD -------------------------------------------------------------- >41955_41955_4_KHDRBS1-LRRC37A_KHDRBS1_chr1_32479978_ENST00000492989_LRRC37A_chr17_44407816_ENST00000320254_length(transcript)=2384nt_BP=382nt ATGCAGCGCCGGGACGACCCCGCCGCGCGCATGAGCCGGTCTTCGGGCCGTAGCGGCTCCATGGACCCCTCCGGTGCCCACCCCTCGGTG CGTCAGACGCCGTCTCGGCAGCCGCCGCTGCCTCACCGGTCCCGGGGAGGCGGAGGGGGATCCCGCGGGGGCGCCCGGGCCTCGCCCGCC ACGCAGCCGCCACCGCTGCTGCCGCCCTCGGCCACGGGTCCCGACGCGACAGTGGGCGGGCCAGCGCCGACCCCGCTGCTGCCCCCCTCG GCCACAGCCTCGGTCAAGATGGAGCCAGAGAACAAGTACCTGCCCGAACTCATGGCCGAGAAGGACTCGCTCGACCCGTCCTTCACTCAC GCCATGCAGCTGCTGACGGCAGCTGAAGAAGCATCGGTAGGGAATCCAGAAGGAGCGTTCATGAAGGTGTTACAAGCCCGGAAGAACTAC ACAAGCACTGAGCTGATTGTTGAGCCAGAGGAGCCCTCAGACAGCAGTGGCATCAACTTGTCAGGCTTTGGGAGTGAGCAGCTAGACACC AATGACGAGAGTGATTTTATCAGTACACTAAGTTACATCTTGCCTTATTTCTCAGCGGTAAACCTAGATGTGAAATCACTGTTACTACCG TTCATTAAACTGCCAACCACAGGAAACAGCCTGGCAAAGATTCAAACTGTAGGCCAAAACCGGCAGAGAGTGAAGAGAGTCCTCATGGGC CCAAGGAGCATCCAGAAAAGGCACTTCAAAGAGGTAGGAAGGCAGAGCATCAGGAGGGAACAGGGTGCCCAGGCATCTGTGGAGAACGCT GCCGAAGAAAAAAGGCTCACGAGTCCAGCCCCAAGGGAGGTGGAACAGCCCCACACACAGCAGGGGCCTGAGAAGTTAGCGGGAAACGCC GTCTACACCAAGCCTTCGTTCACCCAAGAGCATAAGGCAGCAGTCTCTGTGCTGAAACCCTTCTCCAAGGGCACGCCTTCTACCTCCAGC CCTGCAAAAGCCCTACCACAGGTGAGAGACAGATCGAAAGACTTAACCCACGCTATTTCCATTTTAGAAAGTGCAAAGGCTAGAGTTACA AATACGAAGACGTCTAAACCAATCGTACATGCCAGAAAAAAATACCGCTTTCACAAAACTCGCTCCCACGTGACCCACAGAACAACCAAA GTCAAAAAGAGTCCAAAGGTCAGAAAGAAAAGTTATCTGAGTAGACTGATGCTCGCAAACAGGCTTCCATTCTCTGCAGCGAAGAGCCTC ATAAATTCCCCTTCACAAGGGGCTTTTTCATCCTTAGGAGACCTGAGTCCTCAAGAAAACCCTTTTCTGGAAGTATCTGCTCTTTCAGAA CATTTTATAGAGAAGAATAATACAAAACACACAACTGCAAGAAATGCCTTTGAAGAAAATGATTTTATGGAAAACACTAACATGCCAGAA GGAACCATCTCTGAAAACACAAACTACAATCATCCTCCTGAGGCAGATTCCGCTGGGACTGCATTCAACTTAGGGCCAACTGTTAAACAA ACTGAGACAAAATGGGAATACAACAACGTGGGCACTGACCTGTCCCCCGAGCCCAAAAGCTTCAATTACCCATTGCTCTCGTCCCCAGGT GATCAGTTTGAAATTCAGCTAACCCAGCAGCTACAGTCCCTTATCCCCAACAACAATGTGAGAAGGCTCATTGCTCATGTTATCCGGACC TTGAAGATGGACTGCTCTGGGGCCCATGTGCAAGTGACCTGTGCCAAGCTCATCTCCAGGACAGGCCACCTGATGAAGCTTCTCAGTGGG CAGCAGGAAGTAAAGGCATCCAAGATAGAATGGGATACGGACCAATGGAAGATTGAGAACTACATTAATGAGAGCACAGAAGCCCAGAGT GAACAGAAAGAGAAGTCGCTTGAGCTCAAAAAAGAAGTTCCAGGATATGGCTATACTGACAAACTCATCTTGGCATTAATTGTTACTGGA ATACTAACGATTTTGATTATACTTTTCTGCCTCATTGTGATATGTTGTCACCGAAGGTCATTACAAGAAGATGAAGAAGGATTCTCAAGG GGCATTTTCAGATTTCTGCCATGGAGGGGATGCTCTTCGCGAAGGGAGAGTCAGGATGGACTTTCCTCATTTGGACAGCCGCTCTGGTTT AAAGATCTGTACAAACCTCTCAGTGCCACAAGAATAAATAATCATGCATGGAAGCTGCACAAGAAGTCATCTAATGAGGACAAGATCCTC AACAGGGACCCTGGGGACAGCGAAGCCCCAACGGAGGAGGAGGAGAGTGAAGCCCTGCCATAGGAGGAGAACACAGCCCACCTCAGGCCT >41955_41955_4_KHDRBS1-LRRC37A_KHDRBS1_chr1_32479978_ENST00000492989_LRRC37A_chr17_44407816_ENST00000320254_length(amino acids)=770AA_BP=127 MQRRDDPAARMSRSSGRSGSMDPSGAHPSVRQTPSRQPPLPHRSRGGGGGSRGGARASPATQPPPLLPPSATGPDATVGGPAPTPLLPPS ATASVKMEPENKYLPELMAEKDSLDPSFTHAMQLLTAAEEASVGNPEGAFMKVLQARKNYTSTELIVEPEEPSDSSGINLSGFGSEQLDT NDESDFISTLSYILPYFSAVNLDVKSLLLPFIKLPTTGNSLAKIQTVGQNRQRVKRVLMGPRSIQKRHFKEVGRQSIRREQGAQASVENA AEEKRLTSPAPREVEQPHTQQGPEKLAGNAVYTKPSFTQEHKAAVSVLKPFSKGTPSTSSPAKALPQVRDRSKDLTHAISILESAKARVT NTKTSKPIVHARKKYRFHKTRSHVTHRTTKVKKSPKVRKKSYLSRLMLANRLPFSAAKSLINSPSQGAFSSLGDLSPQENPFLEVSALSE HFIEKNNTKHTTARNAFEENDFMENTNMPEGTISENTNYNHPPEADSAGTAFNLGPTVKQTETKWEYNNVGTDLSPEPKSFNYPLLSSPG DQFEIQLTQQLQSLIPNNNVRRLIAHVIRTLKMDCSGAHVQVTCAKLISRTGHLMKLLSGQQEVKASKIEWDTDQWKIENYINESTEAQS EQKEKSLELKKEVPGYGYTDKLILALIVTGILTILIILFCLIVICCHRRSLQEDEEGFSRGIFRFLPWRGCSSRRESQDGLSSFGQPLWF -------------------------------------------------------------- >41955_41955_5_KHDRBS1-LRRC37A_KHDRBS1_chr1_32479978_ENST00000492989_LRRC37A_chr17_44407816_ENST00000393465_length(transcript)=2172nt_BP=382nt ATGCAGCGCCGGGACGACCCCGCCGCGCGCATGAGCCGGTCTTCGGGCCGTAGCGGCTCCATGGACCCCTCCGGTGCCCACCCCTCGGTG CGTCAGACGCCGTCTCGGCAGCCGCCGCTGCCTCACCGGTCCCGGGGAGGCGGAGGGGGATCCCGCGGGGGCGCCCGGGCCTCGCCCGCC ACGCAGCCGCCACCGCTGCTGCCGCCCTCGGCCACGGGTCCCGACGCGACAGTGGGCGGGCCAGCGCCGACCCCGCTGCTGCCCCCCTCG GCCACAGCCTCGGTCAAGATGGAGCCAGAGAACAAGTACCTGCCCGAACTCATGGCCGAGAAGGACTCGCTCGACCCGTCCTTCACTCAC GCCATGCAGCTGCTGACGGCAGCTGAAGAAGCATCGGTAGGGAATCCAGAAGGAGCGTTCATGAAGGTGTTACAAGCCCGGAAGAACTAC ACAAGCACTGAGCTGATTGTTGAGCCAGAGGAGCCCTCAGACAGCAGTGGCATCAACTTGTCAGGCTTTGGGAGTGAGCAGCTAGACACC AATGACGAGAGTGATTTTATCAGTACACTAAGTTACATCTTGCCTTATTTCTCAGCGGTAAACCTAGATGTGAAATCACTGTTACTACCG TTCATTAAACTGCCAACCACAGGAAACAGCCTGGCAAAGATTCAAACTGTAGGCCAAAACCGGCAGAGAGTGAAGAGAGTCCTCATGGGC CCAAGGAGCATCCAGAAAAGGCACTTCAAAGAGGTAGGAAGGCAGAGCATCAGGAGGGAACAGGGTGCCCAGGCATCTGTGGAGAACGCT GCCGAAGAAAAAAGGCTCACGAGTCCAGCCCCAAGGGAGGTGGAACAGCCCCACACACAGCAGGGGCCTGAGAAGTTAGCGGGAAACGCC GTCTACACCAAGCCTTCGTTCACCCAAGAGCATAAGGCAGCAGTCTCTGTGCTGAAACCCTTCTCCAAGGGCACGCCTTCTACCTCCAGC CCTGCAAAAGCCCTACCACAGGTGAGAGACAGATCGAAAGACTTAACCCACGCTATTTCCATTTTAGAAAGTGCAAAGGCTAGAGTTACA AATACGAAGACGTCTAAACCAATCGTACATGCCAGAAAAAAATACCGCTTTCACAAAACTCGCTCCCACGTGACCCACAGAACAACCAAA GTCAAAAAGAGTCCAAAGGTCAGAAAGAAAAGTTATCTGAGTAGACTGATGCTCGCAAACAGGCTTCCATTCTCTGCAGCGAAGAGCCTC ATAAATTCCCCTTCACAAGGGGCTTTTTCATCCTTAGGAGACCTGAGTCCTCAAGAAAACCCTTTTCTGGAAGTATCTGCTCTTTCAGAA CATTTTATAGAGAAGAATAATACAAAACACACAACTGCAAGAAATGCCTTTGAAGAAAATGATTTTATGGAAAACACTAACATGCCAGAA GGAACCATCTCTGAAAACACAAACTACAATCATCCTCCTGAGGCAGATTCCGCTGGGACTGCATTCAACTTAGGGCCAACTGTTAAACAA ACTGAGACAAAATGGGAATACAACAACGTGGGCACTGACCTGTCCCCCGAGCCCAAAAGCTTCAATTACCCATTGCTCTCGTCCCCAGGT GATCAGTTTGAAATTCAGCTAACCCAGCAGCTACAGTCCCTTATCCCCAACAACAATGTGAGAAGGCTCATTGCTCATGTTATCCGGACC TTGAAGATGGACTGCTCTGGGGCCCATGTGCAAGTGACCTGTGCCAAGCTCATCTCCAGGACAGGCCACCTGATGAAGCTTCTCAGTGGG CAGCAGGAAGTAAAGGCATCCAAGATAGAATGGGATACGGACCAATGGAAGATTGAGAACTACATTAATGAGAGCACAGAAGCCCAGAGT GAACAGAAAGAGAAGTCGCTTGAGCTCAAAAAAGAAGTTCCAGGATATGGCTATACTGACAAACTCATCTTGGCATTAATTGTTACTGGA ATACTAACGATTTTGATTATACTTTTCTGCCTCATTGTGATATGTTGTCACCGAAGGTCATTACAAGAAGATGAAGAAGGATTCTCAAGG GACAGCGAAGCCCCAACGGAGGAGGAGGAGAGTGAAGCCCTGCCATAGGAGGAGAACACAGCCCACCTCAGGCCTCCTGCAAAAATACAT >41955_41955_5_KHDRBS1-LRRC37A_KHDRBS1_chr1_32479978_ENST00000492989_LRRC37A_chr17_44407816_ENST00000393465_length(amino acids)=705AA_BP=127 MQRRDDPAARMSRSSGRSGSMDPSGAHPSVRQTPSRQPPLPHRSRGGGGGSRGGARASPATQPPPLLPPSATGPDATVGGPAPTPLLPPS ATASVKMEPENKYLPELMAEKDSLDPSFTHAMQLLTAAEEASVGNPEGAFMKVLQARKNYTSTELIVEPEEPSDSSGINLSGFGSEQLDT NDESDFISTLSYILPYFSAVNLDVKSLLLPFIKLPTTGNSLAKIQTVGQNRQRVKRVLMGPRSIQKRHFKEVGRQSIRREQGAQASVENA AEEKRLTSPAPREVEQPHTQQGPEKLAGNAVYTKPSFTQEHKAAVSVLKPFSKGTPSTSSPAKALPQVRDRSKDLTHAISILESAKARVT NTKTSKPIVHARKKYRFHKTRSHVTHRTTKVKKSPKVRKKSYLSRLMLANRLPFSAAKSLINSPSQGAFSSLGDLSPQENPFLEVSALSE HFIEKNNTKHTTARNAFEENDFMENTNMPEGTISENTNYNHPPEADSAGTAFNLGPTVKQTETKWEYNNVGTDLSPEPKSFNYPLLSSPG DQFEIQLTQQLQSLIPNNNVRRLIAHVIRTLKMDCSGAHVQVTCAKLISRTGHLMKLLSGQQEVKASKIEWDTDQWKIENYINESTEAQS -------------------------------------------------------------- >41955_41955_6_KHDRBS1-LRRC37A_KHDRBS1_chr1_32479978_ENST00000492989_LRRC37A_chr17_44407816_ENST00000496930_length(transcript)=2382nt_BP=382nt ATGCAGCGCCGGGACGACCCCGCCGCGCGCATGAGCCGGTCTTCGGGCCGTAGCGGCTCCATGGACCCCTCCGGTGCCCACCCCTCGGTG CGTCAGACGCCGTCTCGGCAGCCGCCGCTGCCTCACCGGTCCCGGGGAGGCGGAGGGGGATCCCGCGGGGGCGCCCGGGCCTCGCCCGCC ACGCAGCCGCCACCGCTGCTGCCGCCCTCGGCCACGGGTCCCGACGCGACAGTGGGCGGGCCAGCGCCGACCCCGCTGCTGCCCCCCTCG GCCACAGCCTCGGTCAAGATGGAGCCAGAGAACAAGTACCTGCCCGAACTCATGGCCGAGAAGGACTCGCTCGACCCGTCCTTCACTCAC GCCATGCAGCTGCTGACGGCAGCTGAAGAAGCATCGGTAGGGAATCCAGAAGGAGCGTTCATGAAGGTGTTACAAGCCCGGAAGAACTAC ACAAGCACTGAGCTGATTGTTGAGCCAGAGGAGCCCTCAGACAGCAGTGGCATCAACTTGTCAGGCTTTGGGAGTGAGCAGCTAGACACC AATGACGAGAGTGATTTTATCAGTACACTAAGTTACATCTTGCCTTATTTCTCAGCGGTAAACCTAGATGTGAAATCACTGTTACTACCG TTCATTAAACTGCCAACCACAGGAAACAGCCTGGCAAAGATTCAAACTGTAGGCCAAAACCGGCAGAGAGTGAAGAGAGTCCTCATGGGC CCAAGGAGCATCCAGAAAAGGCACTTCAAAGAGGTAGGAAGGCAGAGCATCAGGAGGGAACAGGGTGCCCAGGCATCTGTGGAGAACGCT GCCGAAGAAAAAAGGCTCACGAGTCCAGCCCCAAGGGAGGTGGAACAGCCCCACACACAGCAGGGGCCTGAGAAGTTAGCGGGAAACGCC GTCTACACCAAGCCTTCGTTCACCCAAGAGCATAAGGCAGCAGTCTCTGTGCTGAAACCCTTCTCCAAGGGCACGCCTTCTACCTCCAGC CCTGCAAAAGCCCTACCACAGGTGAGAGACAGATCGAAAGACTTAACCCACGCTATTTCCATTTTAGAAAGTGCAAAGGCTAGAGTTACA AATACGAAGACGTCTAAACCAATCGTACATGCCAGAAAAAAATACCGCTTTCACAAAACTCGCTCCCACGTGACCCACAGAACAACCAAA GTCAAAAAGAGTCCAAAGGTCAGAAAGAAAAGTTATCTGAGTAGACTGATGCTCGCAAACAGGCTTCCATTCTCTGCAGCGAAGAGCCTC ATAAATTCCCCTTCACAAGGGGCTTTTTCATCCTTAGGAGACCTGAGTCCTCAAGAAAACCCTTTTCTGGAAGTATCTGCTCTTTCAGAA CATTTTATAGAGAAGAATAATACAAAACACACAACTGCAAGAAATGCCTTTGAAGAAAATGATTTTATGGAAAACACTAACATGCCAGAA GGAACCATCTCTGAAAACACAAACTACAATCATCCTCCTGAGGCAGATTCCGCTGGGACTGCATTCAACTTAGGGCCAACTGTTAAACAA ACTGAGACAAAATGGGAATACAACAACGTGGGCACTGACCTGTCCCCCGAGCCCAAAAGCTTCAATTACCCATTGCTCTCGTCCCCAGGT GATCAGTTTGAAATTCAGCTAACCCAGCAGCTACAGTCCCTTATCCCCAACAACAATGTGAGAAGGCTCATTGCTCATGTTATCCGGACC TTGAAGATGGACTGCTCTGGGGCCCATGTGCAAGTGACCTGTGCCAAGCTCATCTCCAGGACAGGCCACCTGATGAAGCTTCTCAGTGGG CAGCAGGAAGTAAAGGCATCCAAGATAGAATGGGATACGGACCAATGGAAGATTGAGAACTACATTAATGAGAGCACAGAAGCCCAGAGT GAACAGAAAGAGAAGTCGCTTGAGCTCAAAAAAGAAGTTCCAGGATATGGCTATACTGACAAACTCATCTTGGCATTAATTGTTACTGGA ATACTAACGATTTTGATTATACTTTTCTGCCTCATTGTGATATGTTGTCACCGAAGGTCATTACAAGAAGATGAAGAAGGATTCTCAAGG GGCATTTTCAGATTTCTGCCATGGAGGGGATGCTCTTCGCGAAGGGAGAGTCAGGATGGACTTTCCTCATTTGGACAGCCGCTCTGGTTT AAAGATCTGTACAAACCTCTCAGTGCCACAAGAATAAATAATCATGCATGGAAGCTGCACAAGAAGTCATCTAATGAGGACAAGATCCTC AACAGGGACCCTGGGGACAGCGAAGCCCCAACGGAGGAGGAGGAGAGTGAAGCCCTGCCATAGGAGGAGAACACAGCCCACCTCAGGCCT >41955_41955_6_KHDRBS1-LRRC37A_KHDRBS1_chr1_32479978_ENST00000492989_LRRC37A_chr17_44407816_ENST00000496930_length(amino acids)=770AA_BP=127 MQRRDDPAARMSRSSGRSGSMDPSGAHPSVRQTPSRQPPLPHRSRGGGGGSRGGARASPATQPPPLLPPSATGPDATVGGPAPTPLLPPS ATASVKMEPENKYLPELMAEKDSLDPSFTHAMQLLTAAEEASVGNPEGAFMKVLQARKNYTSTELIVEPEEPSDSSGINLSGFGSEQLDT NDESDFISTLSYILPYFSAVNLDVKSLLLPFIKLPTTGNSLAKIQTVGQNRQRVKRVLMGPRSIQKRHFKEVGRQSIRREQGAQASVENA AEEKRLTSPAPREVEQPHTQQGPEKLAGNAVYTKPSFTQEHKAAVSVLKPFSKGTPSTSSPAKALPQVRDRSKDLTHAISILESAKARVT NTKTSKPIVHARKKYRFHKTRSHVTHRTTKVKKSPKVRKKSYLSRLMLANRLPFSAAKSLINSPSQGAFSSLGDLSPQENPFLEVSALSE HFIEKNNTKHTTARNAFEENDFMENTNMPEGTISENTNYNHPPEADSAGTAFNLGPTVKQTETKWEYNNVGTDLSPEPKSFNYPLLSSPG DQFEIQLTQQLQSLIPNNNVRRLIAHVIRTLKMDCSGAHVQVTCAKLISRTGHLMKLLSGQQEVKASKIEWDTDQWKIENYINESTEAQS EQKEKSLELKKEVPGYGYTDKLILALIVTGILTILIILFCLIVICCHRRSLQEDEEGFSRGIFRFLPWRGCSSRRESQDGLSSFGQPLWF -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for KHDRBS1-LRRC37A |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | KHDRBS1 | chr1:32479978 | chr17:44407816 | ENST00000327300 | + | 1 | 9 | 351_443 | 127.33333333333333 | 444.0 | HNRNPA1 |
| Hgene | KHDRBS1 | chr1:32479978 | chr17:44407816 | ENST00000492989 | + | 1 | 8 | 351_443 | 127.33333333333333 | 405.0 | HNRNPA1 |
| Hgene | KHDRBS1 | chr1:32479978 | chr17:44407816 | ENST00000327300 | + | 1 | 9 | 400_420 | 127.33333333333333 | 444.0 | ZBTB7A |
| Hgene | KHDRBS1 | chr1:32479978 | chr17:44407816 | ENST00000492989 | + | 1 | 8 | 400_420 | 127.33333333333333 | 405.0 | ZBTB7A |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for KHDRBS1-LRRC37A |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for KHDRBS1-LRRC37A |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
