
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:KIAA1328-LIMCH1 (FusionGDB2 ID:42322) |
Fusion Gene Summary for KIAA1328-LIMCH1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: KIAA1328-LIMCH1 | Fusion gene ID: 42322 | Hgene | Tgene | Gene symbol | KIAA1328 | LIMCH1 | Gene ID | 57536 | 22998 |
| Gene name | KIAA1328 | LIM and calponin homology domains 1 | |
| Synonyms | - | LIMCH1A|LMO7B | |
| Cytomap | 18q12.2 | 4p13 | |
| Type of gene | protein-coding | protein-coding | |
| Description | protein hinderinhinderin | LIM and calponin homology domains-containing protein 1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | Q9UPQ0 | |
| Ensembl transtripts involved in fusion gene | ENST00000280020, ENST00000543923, ENST00000591619, ENST00000435985, ENST00000586135, ENST00000586501, ENST00000592521, ENST00000601437, | ENST00000313860, ENST00000381753, ENST00000396595, ENST00000503057, ENST00000508501, ENST00000509277, ENST00000509454, ENST00000509638, ENST00000511496, ENST00000512632, ENST00000512820, ENST00000512946, ENST00000514096, ENST00000515785, ENST00000513024, | |
| Fusion gene scores | * DoF score | 7 X 7 X 5=245 | 18 X 19 X 9=3078 |
| # samples | 7 | 19 | |
| ** MAII score | log2(7/245*10)=-1.8073549220576 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(19/3078*10)=-4.01792190799726 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: KIAA1328 [Title/Abstract] AND LIMCH1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | KIAA1328(34539398)-LIMCH1(41663437), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across KIAA1328 (5'-gene) Fusion gene breakpoints across KIAA1328 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
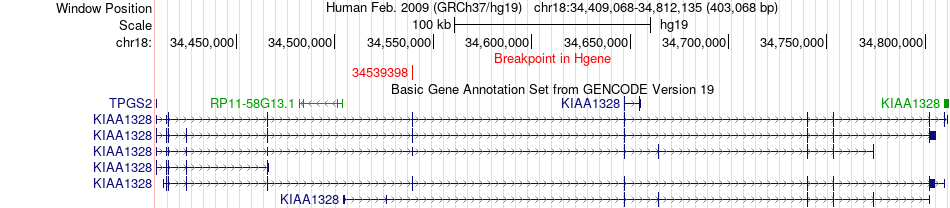 |
 Fusion gene breakpoints across LIMCH1 (3'-gene) Fusion gene breakpoints across LIMCH1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
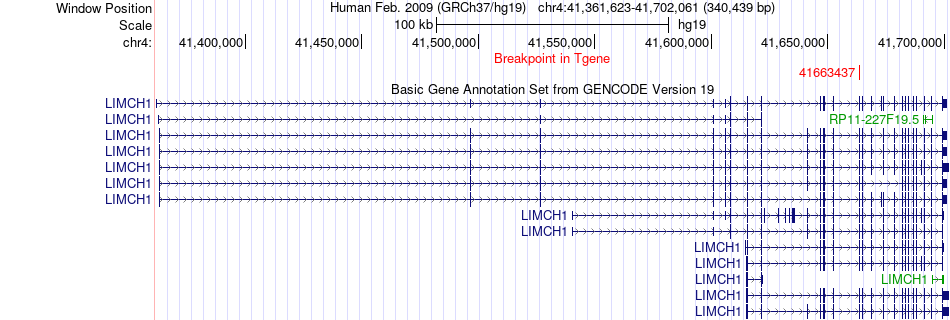 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LGG | TCGA-DU-6407-02B | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
Top |
Fusion Gene ORF analysis for KIAA1328-LIMCH1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000280020 | ENST00000313860 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000280020 | ENST00000381753 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000280020 | ENST00000396595 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000280020 | ENST00000503057 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000280020 | ENST00000508501 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000280020 | ENST00000509277 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000280020 | ENST00000509454 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000280020 | ENST00000509638 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000280020 | ENST00000511496 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000280020 | ENST00000512632 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000280020 | ENST00000512820 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000280020 | ENST00000512946 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000280020 | ENST00000514096 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000280020 | ENST00000515785 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000543923 | ENST00000313860 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000543923 | ENST00000381753 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000543923 | ENST00000396595 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000543923 | ENST00000503057 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000543923 | ENST00000508501 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000543923 | ENST00000509277 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000543923 | ENST00000509454 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000543923 | ENST00000509638 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000543923 | ENST00000511496 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000543923 | ENST00000512632 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000543923 | ENST00000512820 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000543923 | ENST00000512946 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000543923 | ENST00000514096 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000543923 | ENST00000515785 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000591619 | ENST00000313860 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000591619 | ENST00000381753 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000591619 | ENST00000396595 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000591619 | ENST00000503057 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000591619 | ENST00000508501 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000591619 | ENST00000509277 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000591619 | ENST00000509454 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000591619 | ENST00000509638 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000591619 | ENST00000511496 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000591619 | ENST00000512632 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000591619 | ENST00000512820 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000591619 | ENST00000512946 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000591619 | ENST00000514096 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5CDS-intron | ENST00000591619 | ENST00000515785 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5UTR-3CDS | ENST00000435985 | ENST00000513024 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5UTR-intron | ENST00000435985 | ENST00000313860 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5UTR-intron | ENST00000435985 | ENST00000381753 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5UTR-intron | ENST00000435985 | ENST00000396595 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5UTR-intron | ENST00000435985 | ENST00000503057 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5UTR-intron | ENST00000435985 | ENST00000508501 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5UTR-intron | ENST00000435985 | ENST00000509277 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5UTR-intron | ENST00000435985 | ENST00000509454 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5UTR-intron | ENST00000435985 | ENST00000509638 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5UTR-intron | ENST00000435985 | ENST00000511496 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5UTR-intron | ENST00000435985 | ENST00000512632 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5UTR-intron | ENST00000435985 | ENST00000512820 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5UTR-intron | ENST00000435985 | ENST00000512946 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5UTR-intron | ENST00000435985 | ENST00000514096 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| 5UTR-intron | ENST00000435985 | ENST00000515785 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| In-frame | ENST00000280020 | ENST00000513024 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| In-frame | ENST00000543923 | ENST00000513024 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| In-frame | ENST00000591619 | ENST00000513024 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-3CDS | ENST00000586135 | ENST00000513024 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-3CDS | ENST00000586501 | ENST00000513024 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-3CDS | ENST00000592521 | ENST00000513024 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-3CDS | ENST00000601437 | ENST00000513024 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586135 | ENST00000313860 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586135 | ENST00000381753 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586135 | ENST00000396595 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586135 | ENST00000503057 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586135 | ENST00000508501 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586135 | ENST00000509277 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586135 | ENST00000509454 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586135 | ENST00000509638 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586135 | ENST00000511496 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586135 | ENST00000512632 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586135 | ENST00000512820 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586135 | ENST00000512946 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586135 | ENST00000514096 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586135 | ENST00000515785 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586501 | ENST00000313860 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586501 | ENST00000381753 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586501 | ENST00000396595 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586501 | ENST00000503057 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586501 | ENST00000508501 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586501 | ENST00000509277 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586501 | ENST00000509454 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586501 | ENST00000509638 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586501 | ENST00000511496 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586501 | ENST00000512632 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586501 | ENST00000512820 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586501 | ENST00000512946 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586501 | ENST00000514096 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000586501 | ENST00000515785 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000592521 | ENST00000313860 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000592521 | ENST00000381753 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000592521 | ENST00000396595 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000592521 | ENST00000503057 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000592521 | ENST00000508501 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000592521 | ENST00000509277 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000592521 | ENST00000509454 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000592521 | ENST00000509638 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000592521 | ENST00000511496 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000592521 | ENST00000512632 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000592521 | ENST00000512820 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000592521 | ENST00000512946 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000592521 | ENST00000514096 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000592521 | ENST00000515785 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000601437 | ENST00000313860 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000601437 | ENST00000381753 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000601437 | ENST00000396595 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000601437 | ENST00000503057 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000601437 | ENST00000508501 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000601437 | ENST00000509277 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000601437 | ENST00000509454 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000601437 | ENST00000509638 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000601437 | ENST00000511496 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000601437 | ENST00000512632 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000601437 | ENST00000512820 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000601437 | ENST00000512946 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000601437 | ENST00000514096 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
| intron-intron | ENST00000601437 | ENST00000515785 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000543923 | KIAA1328 | chr18 | 34539398 | - | ENST00000513024 | LIMCH1 | chr4 | 41663437 | + | 3697 | 514 | 262 | 1878 | 538 |
| ENST00000280020 | KIAA1328 | chr18 | 34539398 | - | ENST00000513024 | LIMCH1 | chr4 | 41663437 | + | 3781 | 598 | 22 | 1962 | 646 |
| ENST00000591619 | KIAA1328 | chr18 | 34539398 | - | ENST00000513024 | LIMCH1 | chr4 | 41663437 | + | 4533 | 1350 | 786 | 2714 | 642 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000543923 | ENST00000513024 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + | 0.000654475 | 0.9993455 |
| ENST00000280020 | ENST00000513024 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + | 0.000578846 | 0.9994211 |
| ENST00000591619 | ENST00000513024 | KIAA1328 | chr18 | 34539398 | - | LIMCH1 | chr4 | 41663437 | + | 0.001203279 | 0.9987967 |
Top |
Fusion Genomic Features for KIAA1328-LIMCH1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
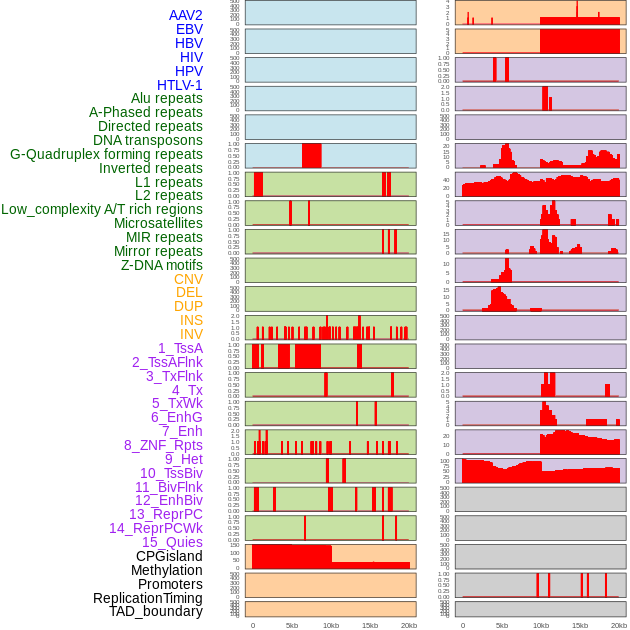 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for KIAA1328-LIMCH1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr18:34539398/chr4:41663437) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | LIMCH1 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Actin stress fibers-associated protein that activates non-muscle myosin IIa. Activates the non-muscle myosin IIa complex by promoting the phosphorylation of its regulatory subunit MRLC/MYL9. Through the activation of non-muscle myosin IIa, positively regulates actin stress fibers assembly and stabilizes focal adhesions. It therefore negatively regulates cell spreading and cell migration. {ECO:0000269|PubMed:28228547}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KIAA1328 | chr18:34539398 | chr4:41663437 | ENST00000280020 | - | 6 | 10 | 91_167 | 192 | 578.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | KIAA1328 | chr18:34539398 | chr4:41663437 | ENST00000591619 | - | 6 | 11 | 91_167 | 188 | 441.3333333333333 | Coiled coil | Ontology_term=ECO:0000255 |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000313860 | 12 | 27 | 782_823 | 627 | 1084.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000381753 | 6 | 20 | 782_823 | 461 | 891.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000396595 | 7 | 21 | 782_823 | 473 | 903.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000508501 | 12 | 26 | 782_823 | 627 | 1057.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000511496 | 9 | 23 | 782_823 | 468 | 898.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000512820 | 11 | 26 | 782_823 | 615 | 1070.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000512946 | 12 | 26 | 782_823 | 627 | 1058.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000513024 | 11 | 26 | 782_823 | 456 | 911.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000313860 | 12 | 27 | 1011_1077 | 627 | 1084.0 | Domain | LIM zinc-binding | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000381753 | 6 | 20 | 1011_1077 | 461 | 891.0 | Domain | LIM zinc-binding | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000396595 | 7 | 21 | 1011_1077 | 473 | 903.0 | Domain | LIM zinc-binding | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000508501 | 12 | 26 | 1011_1077 | 627 | 1057.0 | Domain | LIM zinc-binding | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000511496 | 9 | 23 | 1011_1077 | 468 | 898.0 | Domain | LIM zinc-binding | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000512820 | 11 | 26 | 1011_1077 | 615 | 1070.0 | Domain | LIM zinc-binding | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000512946 | 12 | 26 | 1011_1077 | 627 | 1058.0 | Domain | LIM zinc-binding | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000513024 | 11 | 26 | 1011_1077 | 456 | 911.0 | Domain | LIM zinc-binding |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KIAA1328 | chr18:34539398 | chr4:41663437 | ENST00000280020 | - | 6 | 10 | 358_402 | 192 | 578.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | KIAA1328 | chr18:34539398 | chr4:41663437 | ENST00000435985 | - | 6 | 11 | 358_402 | 0 | 280.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | KIAA1328 | chr18:34539398 | chr4:41663437 | ENST00000435985 | - | 6 | 11 | 91_167 | 0 | 280.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | KIAA1328 | chr18:34539398 | chr4:41663437 | ENST00000586135 | - | 1 | 9 | 358_402 | 0 | 369.3333333333333 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | KIAA1328 | chr18:34539398 | chr4:41663437 | ENST00000586135 | - | 1 | 9 | 91_167 | 0 | 369.3333333333333 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | KIAA1328 | chr18:34539398 | chr4:41663437 | ENST00000586501 | - | 1 | 2 | 358_402 | 0 | 130.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | KIAA1328 | chr18:34539398 | chr4:41663437 | ENST00000586501 | - | 1 | 2 | 91_167 | 0 | 130.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | KIAA1328 | chr18:34539398 | chr4:41663437 | ENST00000591619 | - | 6 | 11 | 358_402 | 188 | 441.3333333333333 | Coiled coil | Ontology_term=ECO:0000255 |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000313860 | 12 | 27 | 354_439 | 627 | 1084.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000381753 | 6 | 20 | 354_439 | 461 | 891.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000396595 | 7 | 21 | 354_439 | 473 | 903.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000508501 | 12 | 26 | 354_439 | 627 | 1057.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000511496 | 9 | 23 | 354_439 | 468 | 898.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000512820 | 11 | 26 | 354_439 | 615 | 1070.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000512946 | 12 | 26 | 354_439 | 627 | 1058.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000513024 | 11 | 26 | 354_439 | 456 | 911.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000313860 | 12 | 27 | 21_125 | 627 | 1084.0 | Domain | Calponin-homology (CH) | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000381753 | 6 | 20 | 21_125 | 461 | 891.0 | Domain | Calponin-homology (CH) | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000396595 | 7 | 21 | 21_125 | 473 | 903.0 | Domain | Calponin-homology (CH) | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000508501 | 12 | 26 | 21_125 | 627 | 1057.0 | Domain | Calponin-homology (CH) | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000511496 | 9 | 23 | 21_125 | 468 | 898.0 | Domain | Calponin-homology (CH) | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000512820 | 11 | 26 | 21_125 | 615 | 1070.0 | Domain | Calponin-homology (CH) | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000512946 | 12 | 26 | 21_125 | 627 | 1058.0 | Domain | Calponin-homology (CH) | |
| Tgene | LIMCH1 | chr18:34539398 | chr4:41663437 | ENST00000513024 | 11 | 26 | 21_125 | 456 | 911.0 | Domain | Calponin-homology (CH) |
Top |
Fusion Gene Sequence for KIAA1328-LIMCH1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >42322_42322_1_KIAA1328-LIMCH1_KIAA1328_chr18_34539398_ENST00000280020_LIMCH1_chr4_41663437_ENST00000513024_length(transcript)=3781nt_BP=598nt CCCGAGTGGCGGTTGTTTCAAGATGGCGGACGTGGCGGGCCCCTCCCGCCCCAGTGCCGCGGCGTTCTGGAGCCGGGACTTTTCTGATGA AGAACAATCAGTAGTATACGTTCCAGGAATTTCTGCTGAAGGAAATGTCAGATCAAGACACAAGCTGATGAGTCCAAAAGCTGATGTTAA ACTTAAGACTTCCAGGGTGACTGATGCTTCAATCTCCATGGAGTCCTTAAAAGGCACAGGAGATTCAGTAGATGAACAGAATTCCTGCAG GGGAGAAATAAAGAGTGCATCATTGAAGGATTTATGTCTTGAAGACAAAAGACGCATTGCAAACTTAATTAAAGAACTGGCCAGAGTAAG TGAGGAAAAGGAAGTGACAGAGGAAAGACTGAAAGCTGAGCAGGAGTCATTTGAGAAGAAGATCAGGCAGTTGGAAGAACAGAATGAACT GATCATCAAAGAAAGGGAAGCTCTTCAGCTACAGTATAGAGAATGCCAAGAACTTCTAAGCCTGTATCAGAAATATTTATCAGAACAACA GGAGAAGCTCACCATGTCTCTCTCAGAACTTGGTGCTGCTAGAATGCAGGAACAGCAGGCAACCACTGAGAAAACGGAACCGAATAGTCA AGAGGACAAGAATGATGGTGGAAAATCAAGAAAAGGGAATATAGAACTTGCCTCATCAGAACCACAGCATTTTACAACAACTGTGACTCG ATGCAGCCCGACCGTGGCCTTTGTGGAATTTCCCTCCAGCCCCCAGCTGAAGAATGATGTGTCGGAAGAAAAAGACCAGAAGAAACCAGA AAATGAAATGAGTGGAAAGGTGGAGTTGGTGCTGTCACAAAAGGTGGTAAAGCCAAAATCTCCAGAACCCGAAGCAACGCTGACATTTCC ATTTCTGGACAAAATGCCTGAAGCCAACCAACTACATTTGCCAAATCTCAATTCTCAAGAGTCGCCTGGCACTGCCAGTGTTCCGCTTAG AGTTCAGAACTCTTGGCGACGATCCCAGTTTTTCTCCCAGTCAGTGGATTCTCCAAGCAGTGAGAAGTCACCTGTTATGACACCTTTTAA GTTCTGGGCATGGGACCCAGAAGAGGAGCGCAGGCGACAGGAAAAATGGCAACAGGAACAGGAACGTTTGCTCCAGGAGAGATACCAGAA GGAGCAGGACAAGCTGAAAGAAGAGTGGGAAAAGGCCCAAAAGGAGGTGGAAGAGGAAGAACGCAGATACTATGAGGAGGAGCGTAAGAT AATTGAAGACACTGTGGTTCCATTTACTGTTTCTTCAAGTTCCGCTGACCAGCTGTCTACCTCTTCCTCCATGACTGAAGGCAGTGGGAC AATGAATAAGATAGACCTGGGAAACTGTCAAGATGAAAAACAAGACAGAAGATGGAAGAAATCATTCCAGGGAGATGACAGTGACTTATT GCTGAAGACTAGGGAAAGTGATCGACTGGAGGAGAAGGGCAGCCTAACTGAAGGGGCCTTGGCTCATTCTGGGAACCCTGTATCAAAAGG AGTCCATGAAGACCATCAGCTGGATACCGAGGCTGGGGCCCCACACTGTGGAACAAACCCACAGCTTGCTCAGGATCCATCCCAGAATCA GCAGACATCAAATCCAACGCACAGTTCAGAAGATGTGAAGCCAAAAACCCTCCCGCTGGATAAAAGCATTAACCATCAGATCGAGTCTCC CAGTGAAAGGCGGAAGTCTATAAGTGGAAAGAAGCTGTGCTCTTCCTGTGGGCTTCCTTTGGGTAAAGGAGCTGCAATGATCATCGAGAC CCTCAATCTCTATTTTCACATCCAGTGTTTCAGGTGTGGAATTTGTAAAGGCCAGCTTGGAGATGCAGTGAGTGGGACGGATGTTAGGAT TCGAAATGGTCTCCTGAACTGTAATGATTGCTACATGCGATCCAGAAGTGCCGGGCAGCCTACAACATTGTGACACGGCTTTCAAGCTTC CGGATCACTCACCATTTCTTTACTGAGAGTGTCCCCTGGCAACTGCTTAACAAAATCCCAAGCTCAGGGGCTTCTCAGCATTTACCTAAT TTCTGAAAGGCTCTTCTGAAAGGTGGTATCTGTTCTTTCGTAGCACAGTGTTTATGTTTTTCCTGTTTATTGTTTTGGTTTTTTTTTTTT TTTTGCATTTGCACAGTATACACAAAAGAATATGGGGTTGTAATGATCCTGAATAGCTCAAAAAAGGTTTTAGCATGGTCAAACAGGCTT ATGGTTTAAAATGTGTTATTCTCTTCTTTGGGAATTAGCTAAATGATGCAATAAACCTGTTTTGTTTTAGAATGTCTAGGAATTAAACAC TTTATGTTTACAGAATTGAGCTGCAGAAAGTGCAAGACATGCCAATTTGAGACACACGGTCTTCTAAGACTGAAGGATAAATTTAATGCA TTTCAGAAACTAAACATCACAGCAAGCTCTATCTCTGAGCTATAATTTGTTTTTAATGCAAAGACACTAGTTTGATAATATATACTGTAA TCCTGAAACATTTGTGTTACTTACCTTTGGAGGTAGAAATTATACCAATAAATTATTGCACCGTTAGTATTAGATTCTGTGTACCTTGGA AGTTATGTCATTAATATAGGCTGGTTCATCAAATAAAGCAAAACCTTGCAATATCAGCTAGATTTACACTCCGGGACGTTGCCCAAAGGT AGGAAGAAAGCAGAGGGAAATATTTCAGTCATCATTTCCAAAGTCATTATCAAAATCTGTGAGGAAGTTTAATCTTCCAAAGAGTCAATG TCAGACATCAGGCCTCTGTTGCCTGCTTCTCTCGAGGCACTAGATTAGGAGTCTTCAATAAGAGACTTAACATGAGGTATATGGAAGATG AGGCACCGAGATAAGTTCATCATTAGGTGTGAGCACTGCTCACCCTTGCTGGCAAGTTCTCCTTAAGGGCCTGAAGCACAGGTGTCCAAA GAAAAGCGTTAAGTCCATCTTAATAGAATCTATGTGGTATATGATGTGGTCAGCCCCTGGTCTGTGATCAGCAAGAACCTACAGCACAGA TTATGCCCTGCCCACTTCAATGAATACCTACTCTCCTCCATTCTCCATCACTTTTTTTGCTATCAAGAACTCCGGACCTTGCCCATGGAG AAGTTTAGAGAGGAACTCTTGTGGAGAGCTGGTTTATTTTCTGCCCTGTGCGACGAGTTTCAGCTGGCCAAGAAAGGAGTCAAGTTATTA AAAAGCATCACAATGTAGATCTCCAGGCTGGTTTTTTGTTTTTTGTTGTTAAGACTGGGGAAAGGGGGACTATTTATTCTGCCTTAAATC AATGGCAAATAAGTCAAGATGACATTTTGTGAATGTAGACTATGGATACACTCCTAATAGATTGATGTAGTCATAAAAGGGGGTCAAGTA GATGTTTTTCTGTTATGTAAGCAATAATTTTTCCGTGTCTTATTGAGTATGGCTAGCGATTATTTATTACATGCTAGATGGGTTCTTTGC ATGTGGGTTCCATATAGGTGCAGAAATTTCCTCAGCCACTGGAGGGATTTCGACCATATTTGTCATTTGGATGAGCTGTTATTAGATTGA AATCTACACATCATTTCATTAAAAATTGTGCCTTAGAAAACGCAAAGCTGTTGCACATGGCGATAAATTATGGATGCAGTACATTGAAGA GAGATGAAGTCACTTCCAAGTTTCCAAGACTTCTCATGGAGGTGTTTGCTGTTTTACAGGAAAAAATAAAAATAAAAAAAGAAAAAAAGA >42322_42322_1_KIAA1328-LIMCH1_KIAA1328_chr18_34539398_ENST00000280020_LIMCH1_chr4_41663437_ENST00000513024_length(amino acids)=646AA_BP=191 MADVAGPSRPSAAAFWSRDFSDEEQSVVYVPGISAEGNVRSRHKLMSPKADVKLKTSRVTDASISMESLKGTGDSVDEQNSCRGEIKSAS LKDLCLEDKRRIANLIKELARVSEEKEVTEERLKAEQESFEKKIRQLEEQNELIIKEREALQLQYRECQELLSLYQKYLSEQQEKLTMSL SELGAARMQEQQATTEKTEPNSQEDKNDGGKSRKGNIELASSEPQHFTTTVTRCSPTVAFVEFPSSPQLKNDVSEEKDQKKPENEMSGKV ELVLSQKVVKPKSPEPEATLTFPFLDKMPEANQLHLPNLNSQESPGTASVPLRVQNSWRRSQFFSQSVDSPSSEKSPVMTPFKFWAWDPE EERRRQEKWQQEQERLLQERYQKEQDKLKEEWEKAQKEVEEEERRYYEEERKIIEDTVVPFTVSSSSADQLSTSSSMTEGSGTMNKIDLG NCQDEKQDRRWKKSFQGDDSDLLLKTRESDRLEEKGSLTEGALAHSGNPVSKGVHEDHQLDTEAGAPHCGTNPQLAQDPSQNQQTSNPTH SSEDVKPKTLPLDKSINHQIESPSERRKSISGKKLCSSCGLPLGKGAAMIIETLNLYFHIQCFRCGICKGQLGDAVSGTDVRIRNGLLNC -------------------------------------------------------------- >42322_42322_2_KIAA1328-LIMCH1_KIAA1328_chr18_34539398_ENST00000543923_LIMCH1_chr4_41663437_ENST00000513024_length(transcript)=3697nt_BP=514nt AGGCGCGACGCCCCGAGTGGCGGTTGTTTCAAGATGGCGGACGTGGCGGGCCCCTCCCGCCCCAGTGCCGCGGCGTTCTGGAGCCGGGAC TTTTCTGATGAAGAACAATCAGTAGTATACGTTCCAGGAATTTCTGCTGAAGGAAATGTCAGATCAAGACACAAGCTGATGAGTCCAAAA GCTGATGTTAAACTTAAGACTTCCAGGGTGACTGATGCTTCAATCTCCATGGAGTCCTTAAAAGGCACAGGAGATTCAGTAGATGAACAG AGTAAGTGAGGAAAAGGAAGTGACAGAGGAAAGACTGAAAGCTGAGCAGGAGTCATTTGAGAAGAAGATCAGGCAGTTGGAAGAACAGAA TGAACTGATCATCAAAGAAAGGGAAGCTCTTCAGCTACAGTATAGAGAATGCCAAGAACTTCTAAGCCTGTATCAGAAATATTTATCAGA ACAACAGGAGAAGCTCACCATGTCTCTCTCAGAACTTGGTGCTGCTAGAATGCAGGAACAGCAGGCAACCACTGAGAAAACGGAACCGAA TAGTCAAGAGGACAAGAATGATGGTGGAAAATCAAGAAAAGGGAATATAGAACTTGCCTCATCAGAACCACAGCATTTTACAACAACTGT GACTCGATGCAGCCCGACCGTGGCCTTTGTGGAATTTCCCTCCAGCCCCCAGCTGAAGAATGATGTGTCGGAAGAAAAAGACCAGAAGAA ACCAGAAAATGAAATGAGTGGAAAGGTGGAGTTGGTGCTGTCACAAAAGGTGGTAAAGCCAAAATCTCCAGAACCCGAAGCAACGCTGAC ATTTCCATTTCTGGACAAAATGCCTGAAGCCAACCAACTACATTTGCCAAATCTCAATTCTCAAGAGTCGCCTGGCACTGCCAGTGTTCC GCTTAGAGTTCAGAACTCTTGGCGACGATCCCAGTTTTTCTCCCAGTCAGTGGATTCTCCAAGCAGTGAGAAGTCACCTGTTATGACACC TTTTAAGTTCTGGGCATGGGACCCAGAAGAGGAGCGCAGGCGACAGGAAAAATGGCAACAGGAACAGGAACGTTTGCTCCAGGAGAGATA CCAGAAGGAGCAGGACAAGCTGAAAGAAGAGTGGGAAAAGGCCCAAAAGGAGGTGGAAGAGGAAGAACGCAGATACTATGAGGAGGAGCG TAAGATAATTGAAGACACTGTGGTTCCATTTACTGTTTCTTCAAGTTCCGCTGACCAGCTGTCTACCTCTTCCTCCATGACTGAAGGCAG TGGGACAATGAATAAGATAGACCTGGGAAACTGTCAAGATGAAAAACAAGACAGAAGATGGAAGAAATCATTCCAGGGAGATGACAGTGA CTTATTGCTGAAGACTAGGGAAAGTGATCGACTGGAGGAGAAGGGCAGCCTAACTGAAGGGGCCTTGGCTCATTCTGGGAACCCTGTATC AAAAGGAGTCCATGAAGACCATCAGCTGGATACCGAGGCTGGGGCCCCACACTGTGGAACAAACCCACAGCTTGCTCAGGATCCATCCCA GAATCAGCAGACATCAAATCCAACGCACAGTTCAGAAGATGTGAAGCCAAAAACCCTCCCGCTGGATAAAAGCATTAACCATCAGATCGA GTCTCCCAGTGAAAGGCGGAAGTCTATAAGTGGAAAGAAGCTGTGCTCTTCCTGTGGGCTTCCTTTGGGTAAAGGAGCTGCAATGATCAT CGAGACCCTCAATCTCTATTTTCACATCCAGTGTTTCAGGTGTGGAATTTGTAAAGGCCAGCTTGGAGATGCAGTGAGTGGGACGGATGT TAGGATTCGAAATGGTCTCCTGAACTGTAATGATTGCTACATGCGATCCAGAAGTGCCGGGCAGCCTACAACATTGTGACACGGCTTTCA AGCTTCCGGATCACTCACCATTTCTTTACTGAGAGTGTCCCCTGGCAACTGCTTAACAAAATCCCAAGCTCAGGGGCTTCTCAGCATTTA CCTAATTTCTGAAAGGCTCTTCTGAAAGGTGGTATCTGTTCTTTCGTAGCACAGTGTTTATGTTTTTCCTGTTTATTGTTTTGGTTTTTT TTTTTTTTTTGCATTTGCACAGTATACACAAAAGAATATGGGGTTGTAATGATCCTGAATAGCTCAAAAAAGGTTTTAGCATGGTCAAAC AGGCTTATGGTTTAAAATGTGTTATTCTCTTCTTTGGGAATTAGCTAAATGATGCAATAAACCTGTTTTGTTTTAGAATGTCTAGGAATT AAACACTTTATGTTTACAGAATTGAGCTGCAGAAAGTGCAAGACATGCCAATTTGAGACACACGGTCTTCTAAGACTGAAGGATAAATTT AATGCATTTCAGAAACTAAACATCACAGCAAGCTCTATCTCTGAGCTATAATTTGTTTTTAATGCAAAGACACTAGTTTGATAATATATA CTGTAATCCTGAAACATTTGTGTTACTTACCTTTGGAGGTAGAAATTATACCAATAAATTATTGCACCGTTAGTATTAGATTCTGTGTAC CTTGGAAGTTATGTCATTAATATAGGCTGGTTCATCAAATAAAGCAAAACCTTGCAATATCAGCTAGATTTACACTCCGGGACGTTGCCC AAAGGTAGGAAGAAAGCAGAGGGAAATATTTCAGTCATCATTTCCAAAGTCATTATCAAAATCTGTGAGGAAGTTTAATCTTCCAAAGAG TCAATGTCAGACATCAGGCCTCTGTTGCCTGCTTCTCTCGAGGCACTAGATTAGGAGTCTTCAATAAGAGACTTAACATGAGGTATATGG AAGATGAGGCACCGAGATAAGTTCATCATTAGGTGTGAGCACTGCTCACCCTTGCTGGCAAGTTCTCCTTAAGGGCCTGAAGCACAGGTG TCCAAAGAAAAGCGTTAAGTCCATCTTAATAGAATCTATGTGGTATATGATGTGGTCAGCCCCTGGTCTGTGATCAGCAAGAACCTACAG CACAGATTATGCCCTGCCCACTTCAATGAATACCTACTCTCCTCCATTCTCCATCACTTTTTTTGCTATCAAGAACTCCGGACCTTGCCC ATGGAGAAGTTTAGAGAGGAACTCTTGTGGAGAGCTGGTTTATTTTCTGCCCTGTGCGACGAGTTTCAGCTGGCCAAGAAAGGAGTCAAG TTATTAAAAAGCATCACAATGTAGATCTCCAGGCTGGTTTTTTGTTTTTTGTTGTTAAGACTGGGGAAAGGGGGACTATTTATTCTGCCT TAAATCAATGGCAAATAAGTCAAGATGACATTTTGTGAATGTAGACTATGGATACACTCCTAATAGATTGATGTAGTCATAAAAGGGGGT CAAGTAGATGTTTTTCTGTTATGTAAGCAATAATTTTTCCGTGTCTTATTGAGTATGGCTAGCGATTATTTATTACATGCTAGATGGGTT CTTTGCATGTGGGTTCCATATAGGTGCAGAAATTTCCTCAGCCACTGGAGGGATTTCGACCATATTTGTCATTTGGATGAGCTGTTATTA GATTGAAATCTACACATCATTTCATTAAAAATTGTGCCTTAGAAAACGCAAAGCTGTTGCACATGGCGATAAATTATGGATGCAGTACAT TGAAGAGAGATGAAGTCACTTCCAAGTTTCCAAGACTTCTCATGGAGGTGTTTGCTGTTTTACAGGAAAAAATAAAAATAAAAAAAGAAA >42322_42322_2_KIAA1328-LIMCH1_KIAA1328_chr18_34539398_ENST00000543923_LIMCH1_chr4_41663437_ENST00000513024_length(amino acids)=538AA_BP=83 MNRVSEEKEVTEERLKAEQESFEKKIRQLEEQNELIIKEREALQLQYRECQELLSLYQKYLSEQQEKLTMSLSELGAARMQEQQATTEKT EPNSQEDKNDGGKSRKGNIELASSEPQHFTTTVTRCSPTVAFVEFPSSPQLKNDVSEEKDQKKPENEMSGKVELVLSQKVVKPKSPEPEA TLTFPFLDKMPEANQLHLPNLNSQESPGTASVPLRVQNSWRRSQFFSQSVDSPSSEKSPVMTPFKFWAWDPEEERRRQEKWQQEQERLLQ ERYQKEQDKLKEEWEKAQKEVEEEERRYYEEERKIIEDTVVPFTVSSSSADQLSTSSSMTEGSGTMNKIDLGNCQDEKQDRRWKKSFQGD DSDLLLKTRESDRLEEKGSLTEGALAHSGNPVSKGVHEDHQLDTEAGAPHCGTNPQLAQDPSQNQQTSNPTHSSEDVKPKTLPLDKSINH -------------------------------------------------------------- >42322_42322_3_KIAA1328-LIMCH1_KIAA1328_chr18_34539398_ENST00000591619_LIMCH1_chr4_41663437_ENST00000513024_length(transcript)=4533nt_BP=1350nt TTAAATCAATACCAATTTTATTCTTTCATAGATGGAGCCAGACATTCTTTTATGTTTGAGGGTACAACACTGAACAAAATAAAACAAAAA TCATTACCCTCTTGGTGCCTACATTTTAGTGAGATGAGATCAGGTCATGAATAAGGAAAATATATGTTCAATGATTTTTTTTTTTTTTTT TGAGTCAGAGTCTCGGTCTGTTGCCCAGGCTGGAGTGCAGTGGTGCCATCTCAGCTCACTGCAAGCTCCGCCTTCTGGGTTCACACCATT CTCCTGCCTCAACCTCTGGAGTAGCTGGGACTACAGGCACCCGCCACCATGCCCGGCTAATTTTTTTTTTTTACATTTTTAGTAGAGACA GGGTTTCACCGTGTTAGCCAGGATGATCTCGATTTTCTGACCTCGTGATATGCCCACCTCGGCCTCCCAAAGTGCTGGGATTACAGACGT GAGCCACTGCACCTGGCCTATGTTCAATGATATTGGTGCTGTGGAGAAAAAATAAAGCAGGAAAGAGGCAGATAGTAGCGGGAGGTAGGT AGGTTGTGGGAAGAGAGAGTTTTGCAATTTCAAGTACCTATATGGTTAGGAAGTGAAAGCTTCAATCAGAAGGTGACATGACATGACATA TGACATAAGTCCTAAAGTGAGCCATGAAGATATTTGAGGAAAGAGCTTCCTGGGTAGAAGAAATCATACCTGCAGAAGCTATAAGTTAGT TACAAATTCCCAGGTCAAAAGAAATTATGAATTATAAGAGGTATACAGAACAGAAGCAGCATTTGGATGCCGGATAATATTATTGTATTT TCCTTCATGTTCTCCTGCGTAGTTTCTGATGAAGAACAATCAGTAGTATACGTTCCAGGAATTTCTGCTGAAGGAAATGTCAGATCAAGA CACAAGCTGATGAGTCCAAAAGCTGATGTTAAACTTAAGACTTCCAGGGTGACTGATGCTTCAATCTCCATGGAGTCCTTAAAAGGCACA GGAGATTCAGTAGATGAACAGAATTCCTGCAGGGGAGAAATAAAGAGTGCATCATTGAAGGATTTATGTCTTGAAGACAAAAGACGCATT GCAAACTTAATTAAAGAACTGGCCAGAGTAAGTGAGGAAAAGGAAGTGACAGAGGAAAGACTGAAAGCTGAGCAGGAGTCATTTGAGAAG AAGATCAGGCAGTTGGAAGAACAGAATGAACTGATCATCAAAGAAAGGGAAGCTCTTCAGCTACAGTATAGAGAATGCCAAGAACTTCTA AGCCTGTATCAGAAATATTTATCAGAACAACAGGAGAAGCTCACCATGTCTCTCTCAGAACTTGGTGCTGCTAGAATGCAGGAACAGCAG GCAACCACTGAGAAAACGGAACCGAATAGTCAAGAGGACAAGAATGATGGTGGAAAATCAAGAAAAGGGAATATAGAACTTGCCTCATCA GAACCACAGCATTTTACAACAACTGTGACTCGATGCAGCCCGACCGTGGCCTTTGTGGAATTTCCCTCCAGCCCCCAGCTGAAGAATGAT GTGTCGGAAGAAAAAGACCAGAAGAAACCAGAAAATGAAATGAGTGGAAAGGTGGAGTTGGTGCTGTCACAAAAGGTGGTAAAGCCAAAA TCTCCAGAACCCGAAGCAACGCTGACATTTCCATTTCTGGACAAAATGCCTGAAGCCAACCAACTACATTTGCCAAATCTCAATTCTCAA GAGTCGCCTGGCACTGCCAGTGTTCCGCTTAGAGTTCAGAACTCTTGGCGACGATCCCAGTTTTTCTCCCAGTCAGTGGATTCTCCAAGC AGTGAGAAGTCACCTGTTATGACACCTTTTAAGTTCTGGGCATGGGACCCAGAAGAGGAGCGCAGGCGACAGGAAAAATGGCAACAGGAA CAGGAACGTTTGCTCCAGGAGAGATACCAGAAGGAGCAGGACAAGCTGAAAGAAGAGTGGGAAAAGGCCCAAAAGGAGGTGGAAGAGGAA GAACGCAGATACTATGAGGAGGAGCGTAAGATAATTGAAGACACTGTGGTTCCATTTACTGTTTCTTCAAGTTCCGCTGACCAGCTGTCT ACCTCTTCCTCCATGACTGAAGGCAGTGGGACAATGAATAAGATAGACCTGGGAAACTGTCAAGATGAAAAACAAGACAGAAGATGGAAG AAATCATTCCAGGGAGATGACAGTGACTTATTGCTGAAGACTAGGGAAAGTGATCGACTGGAGGAGAAGGGCAGCCTAACTGAAGGGGCC TTGGCTCATTCTGGGAACCCTGTATCAAAAGGAGTCCATGAAGACCATCAGCTGGATACCGAGGCTGGGGCCCCACACTGTGGAACAAAC CCACAGCTTGCTCAGGATCCATCCCAGAATCAGCAGACATCAAATCCAACGCACAGTTCAGAAGATGTGAAGCCAAAAACCCTCCCGCTG GATAAAAGCATTAACCATCAGATCGAGTCTCCCAGTGAAAGGCGGAAGTCTATAAGTGGAAAGAAGCTGTGCTCTTCCTGTGGGCTTCCT TTGGGTAAAGGAGCTGCAATGATCATCGAGACCCTCAATCTCTATTTTCACATCCAGTGTTTCAGGTGTGGAATTTGTAAAGGCCAGCTT GGAGATGCAGTGAGTGGGACGGATGTTAGGATTCGAAATGGTCTCCTGAACTGTAATGATTGCTACATGCGATCCAGAAGTGCCGGGCAG CCTACAACATTGTGACACGGCTTTCAAGCTTCCGGATCACTCACCATTTCTTTACTGAGAGTGTCCCCTGGCAACTGCTTAACAAAATCC CAAGCTCAGGGGCTTCTCAGCATTTACCTAATTTCTGAAAGGCTCTTCTGAAAGGTGGTATCTGTTCTTTCGTAGCACAGTGTTTATGTT TTTCCTGTTTATTGTTTTGGTTTTTTTTTTTTTTTTGCATTTGCACAGTATACACAAAAGAATATGGGGTTGTAATGATCCTGAATAGCT CAAAAAAGGTTTTAGCATGGTCAAACAGGCTTATGGTTTAAAATGTGTTATTCTCTTCTTTGGGAATTAGCTAAATGATGCAATAAACCT GTTTTGTTTTAGAATGTCTAGGAATTAAACACTTTATGTTTACAGAATTGAGCTGCAGAAAGTGCAAGACATGCCAATTTGAGACACACG GTCTTCTAAGACTGAAGGATAAATTTAATGCATTTCAGAAACTAAACATCACAGCAAGCTCTATCTCTGAGCTATAATTTGTTTTTAATG CAAAGACACTAGTTTGATAATATATACTGTAATCCTGAAACATTTGTGTTACTTACCTTTGGAGGTAGAAATTATACCAATAAATTATTG CACCGTTAGTATTAGATTCTGTGTACCTTGGAAGTTATGTCATTAATATAGGCTGGTTCATCAAATAAAGCAAAACCTTGCAATATCAGC TAGATTTACACTCCGGGACGTTGCCCAAAGGTAGGAAGAAAGCAGAGGGAAATATTTCAGTCATCATTTCCAAAGTCATTATCAAAATCT GTGAGGAAGTTTAATCTTCCAAAGAGTCAATGTCAGACATCAGGCCTCTGTTGCCTGCTTCTCTCGAGGCACTAGATTAGGAGTCTTCAA TAAGAGACTTAACATGAGGTATATGGAAGATGAGGCACCGAGATAAGTTCATCATTAGGTGTGAGCACTGCTCACCCTTGCTGGCAAGTT CTCCTTAAGGGCCTGAAGCACAGGTGTCCAAAGAAAAGCGTTAAGTCCATCTTAATAGAATCTATGTGGTATATGATGTGGTCAGCCCCT GGTCTGTGATCAGCAAGAACCTACAGCACAGATTATGCCCTGCCCACTTCAATGAATACCTACTCTCCTCCATTCTCCATCACTTTTTTT GCTATCAAGAACTCCGGACCTTGCCCATGGAGAAGTTTAGAGAGGAACTCTTGTGGAGAGCTGGTTTATTTTCTGCCCTGTGCGACGAGT TTCAGCTGGCCAAGAAAGGAGTCAAGTTATTAAAAAGCATCACAATGTAGATCTCCAGGCTGGTTTTTTGTTTTTTGTTGTTAAGACTGG GGAAAGGGGGACTATTTATTCTGCCTTAAATCAATGGCAAATAAGTCAAGATGACATTTTGTGAATGTAGACTATGGATACACTCCTAAT AGATTGATGTAGTCATAAAAGGGGGTCAAGTAGATGTTTTTCTGTTATGTAAGCAATAATTTTTCCGTGTCTTATTGAGTATGGCTAGCG ATTATTTATTACATGCTAGATGGGTTCTTTGCATGTGGGTTCCATATAGGTGCAGAAATTTCCTCAGCCACTGGAGGGATTTCGACCATA TTTGTCATTTGGATGAGCTGTTATTAGATTGAAATCTACACATCATTTCATTAAAAATTGTGCCTTAGAAAACGCAAAGCTGTTGCACAT GGCGATAAATTATGGATGCAGTACATTGAAGAGAGATGAAGTCACTTCCAAGTTTCCAAGACTTCTCATGGAGGTGTTTGCTGTTTTACA >42322_42322_3_KIAA1328-LIMCH1_KIAA1328_chr18_34539398_ENST00000591619_LIMCH1_chr4_41663437_ENST00000513024_length(amino acids)=642AA_BP=187 MPDNIIVFSFMFSCVVSDEEQSVVYVPGISAEGNVRSRHKLMSPKADVKLKTSRVTDASISMESLKGTGDSVDEQNSCRGEIKSASLKDL CLEDKRRIANLIKELARVSEEKEVTEERLKAEQESFEKKIRQLEEQNELIIKEREALQLQYRECQELLSLYQKYLSEQQEKLTMSLSELG AARMQEQQATTEKTEPNSQEDKNDGGKSRKGNIELASSEPQHFTTTVTRCSPTVAFVEFPSSPQLKNDVSEEKDQKKPENEMSGKVELVL SQKVVKPKSPEPEATLTFPFLDKMPEANQLHLPNLNSQESPGTASVPLRVQNSWRRSQFFSQSVDSPSSEKSPVMTPFKFWAWDPEEERR RQEKWQQEQERLLQERYQKEQDKLKEEWEKAQKEVEEEERRYYEEERKIIEDTVVPFTVSSSSADQLSTSSSMTEGSGTMNKIDLGNCQD EKQDRRWKKSFQGDDSDLLLKTRESDRLEEKGSLTEGALAHSGNPVSKGVHEDHQLDTEAGAPHCGTNPQLAQDPSQNQQTSNPTHSSED VKPKTLPLDKSINHQIESPSERRKSISGKKLCSSCGLPLGKGAAMIIETLNLYFHIQCFRCGICKGQLGDAVSGTDVRIRNGLLNCNDCY -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for KIAA1328-LIMCH1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for KIAA1328-LIMCH1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for KIAA1328-LIMCH1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
