
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:KIF13B-INTS9 (FusionGDB2 ID:42554) |
Fusion Gene Summary for KIF13B-INTS9 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: KIF13B-INTS9 | Fusion gene ID: 42554 | Hgene | Tgene | Gene symbol | KIF13B | INTS9 | Gene ID | 23303 | 55756 |
| Gene name | kinesin family member 13B | integrator complex subunit 9 | |
| Synonyms | GAKIN | CPSF2L|INT9|RC74 | |
| Cytomap | 8p12 | 8p21.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | kinesin-like protein KIF13Bguanylate kinase associated kinesinkinesin 13Bkinesin-like protein GAKIN | integrator complex subunit 9protein related to CPSF subunits of 74 kDa | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9NQT8 | Q9NV88 | |
| Ensembl transtripts involved in fusion gene | ENST00000524189, ENST00000404075, ENST00000521515, | ENST00000521777, ENST00000397363, ENST00000521070, ENST00000416984, ENST00000521022, | |
| Fusion gene scores | * DoF score | 23 X 22 X 14=7084 | 11 X 10 X 5=550 |
| # samples | 27 | 11 | |
| ** MAII score | log2(27/7084*10)=-4.71353289970108 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(11/550*10)=-2.32192809488736 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: KIF13B [Title/Abstract] AND INTS9 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | KIF13B(28988050)-INTS9(28717080), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | KIF13B | GO:0050770 | regulation of axonogenesis | 20194617 |
| Tgene | INTS9 | GO:0016180 | snRNA processing | 16239144 |
 Fusion gene breakpoints across KIF13B (5'-gene) Fusion gene breakpoints across KIF13B (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
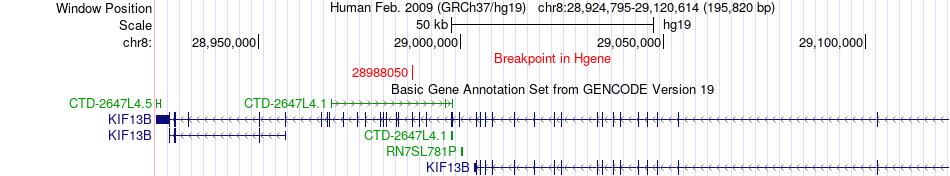 |
 Fusion gene breakpoints across INTS9 (3'-gene) Fusion gene breakpoints across INTS9 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
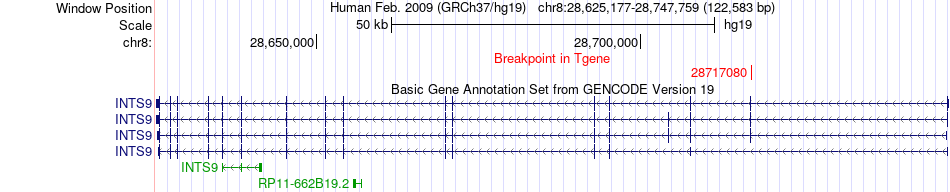 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-BH-A1F6-01A | KIF13B | chr8 | 28988050 | - | INTS9 | chr8 | 28717080 | - |
Top |
Fusion Gene ORF analysis for KIF13B-INTS9 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000524189 | ENST00000521777 | KIF13B | chr8 | 28988050 | - | INTS9 | chr8 | 28717080 | - |
| 5CDS-intron | ENST00000524189 | ENST00000397363 | KIF13B | chr8 | 28988050 | - | INTS9 | chr8 | 28717080 | - |
| 5CDS-intron | ENST00000524189 | ENST00000521070 | KIF13B | chr8 | 28988050 | - | INTS9 | chr8 | 28717080 | - |
| In-frame | ENST00000524189 | ENST00000416984 | KIF13B | chr8 | 28988050 | - | INTS9 | chr8 | 28717080 | - |
| In-frame | ENST00000524189 | ENST00000521022 | KIF13B | chr8 | 28988050 | - | INTS9 | chr8 | 28717080 | - |
| intron-3CDS | ENST00000404075 | ENST00000416984 | KIF13B | chr8 | 28988050 | - | INTS9 | chr8 | 28717080 | - |
| intron-3CDS | ENST00000404075 | ENST00000521022 | KIF13B | chr8 | 28988050 | - | INTS9 | chr8 | 28717080 | - |
| intron-3CDS | ENST00000521515 | ENST00000416984 | KIF13B | chr8 | 28988050 | - | INTS9 | chr8 | 28717080 | - |
| intron-3CDS | ENST00000521515 | ENST00000521022 | KIF13B | chr8 | 28988050 | - | INTS9 | chr8 | 28717080 | - |
| intron-5UTR | ENST00000404075 | ENST00000521777 | KIF13B | chr8 | 28988050 | - | INTS9 | chr8 | 28717080 | - |
| intron-5UTR | ENST00000521515 | ENST00000521777 | KIF13B | chr8 | 28988050 | - | INTS9 | chr8 | 28717080 | - |
| intron-intron | ENST00000404075 | ENST00000397363 | KIF13B | chr8 | 28988050 | - | INTS9 | chr8 | 28717080 | - |
| intron-intron | ENST00000404075 | ENST00000521070 | KIF13B | chr8 | 28988050 | - | INTS9 | chr8 | 28717080 | - |
| intron-intron | ENST00000521515 | ENST00000397363 | KIF13B | chr8 | 28988050 | - | INTS9 | chr8 | 28717080 | - |
| intron-intron | ENST00000521515 | ENST00000521070 | KIF13B | chr8 | 28988050 | - | INTS9 | chr8 | 28717080 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000524189 | KIF13B | chr8 | 28988050 | - | ENST00000521022 | INTS9 | chr8 | 28717080 | - | 5567 | 3114 | 39 | 5081 | 1680 |
| ENST00000524189 | KIF13B | chr8 | 28988050 | - | ENST00000416984 | INTS9 | chr8 | 28717080 | - | 5504 | 3114 | 39 | 5018 | 1659 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000524189 | ENST00000521022 | KIF13B | chr8 | 28988050 | - | INTS9 | chr8 | 28717080 | - | 0.000517548 | 0.9994824 |
| ENST00000524189 | ENST00000416984 | KIF13B | chr8 | 28988050 | - | INTS9 | chr8 | 28717080 | - | 0.000677082 | 0.9993229 |
Top |
Fusion Genomic Features for KIF13B-INTS9 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
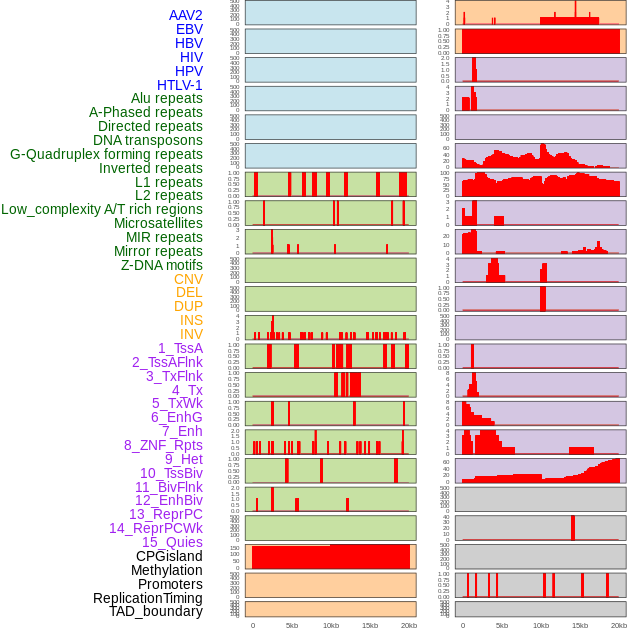 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for KIF13B-INTS9 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr8:28988050/chr8:28717080) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| KIF13B | INTS9 |
| FUNCTION: Involved in reorganization of the cortical cytoskeleton. Regulates axon formation by promoting the formation of extra axons. May be functionally important for the intracellular trafficking of MAGUKs and associated protein complexes. {ECO:0000269|PubMed:20194617}. | FUNCTION: Component of the Integrator (INT) complex, a complex involved in the small nuclear RNAs (snRNA) U1 and U2 transcription and in their 3'-box-dependent processing. The Integrator complex is associated with the C-terminal domain (CTD) of RNA polymerase II largest subunit (POLR2A) and is recruited to the U1 and U2 snRNAs genes (Probable). Mediates recruitment of cytoplasmic dynein to the nuclear envelope, probably as component of the INT complex (PubMed:23904267). {ECO:0000269|PubMed:23904267, ECO:0000305|PubMed:16239144}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KIF13B | chr8:28988050 | chr8:28717080 | ENST00000524189 | - | 24 | 40 | 364_439 | 1025 | 1827.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | KIF13B | chr8:28988050 | chr8:28717080 | ENST00000524189 | - | 24 | 40 | 607_710 | 1025 | 1827.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | KIF13B | chr8:28988050 | chr8:28717080 | ENST00000524189 | - | 24 | 40 | 752_772 | 1025 | 1827.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | KIF13B | chr8:28988050 | chr8:28717080 | ENST00000524189 | - | 24 | 40 | 471_535 | 1025 | 1827.0 | Domain | Note=FHA |
| Hgene | KIF13B | chr8:28988050 | chr8:28717080 | ENST00000524189 | - | 24 | 40 | 5_353 | 1025 | 1827.0 | Domain | Kinesin motor |
| Hgene | KIF13B | chr8:28988050 | chr8:28717080 | ENST00000524189 | - | 24 | 40 | 103_110 | 1025 | 1827.0 | Nucleotide binding | ATP |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KIF13B | chr8:28988050 | chr8:28717080 | ENST00000524189 | - | 24 | 40 | 1096_1143 | 1025 | 1827.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | KIF13B | chr8:28988050 | chr8:28717080 | ENST00000524189 | - | 24 | 40 | 1721_1763 | 1025 | 1827.0 | Domain | CAP-Gly |
Top |
Fusion Gene Sequence for KIF13B-INTS9 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >42554_42554_1_KIF13B-INTS9_KIF13B_chr8_28988050_ENST00000524189_INTS9_chr8_28717080_ENST00000416984_length(transcript)=5504nt_BP=3114nt GGCAGACGGAAGCCGAACGAGTTCCTCGGCGGCTGCAGGATGGGGGACTCCAAAGTGAAAGTGGCGGTGCGGATACGACCCATGAACCGG CGAGAGACTGACTTGCATACCAAATGTGTGGTGGATGTGGATGCAAACAAGGTTATTCTTAATCCTGTAAATACGAATCTTTCCAAAGGA GATGCCCGGGGCCAGCCGAAGGTGTTTGCTTATGATCATTGTTTCTGGTCTATGGATGAATCTGTCAAAGAAAAGTATGCAGGTCAAGAT ATTGTTTTCAAGTGCCTTGGAGAGAATATCCTGCAGAATGCTTTTGATGGCTACAATGCATGTATCTTTGCCTATGGACAGACTGGCTCT GGAAAATCTTATACCATGATGGGCACAGCTGACCAACCTGGATTAATCCCAAGACTTTGCAGTGGACTCTTTGAACGAACTCAGAAAGAG GAAAATGAAGAACAGAGTTTTAAAGTAGAAGTGTCCTACATGGAAATTTATAATGAAAAAGTTCGAGACCTTCTTGATCCCAAAGGAAGC CGTCAGACGTTGAAAGTCAGAGAGCATAGTGTGTTGGGACCTTATGTCGACGGACTTTCTAAACTGGCTGTCACAAGCTACAAGGATATT GAGTCGTTGATGTCTGAGGGTAACAAATCTCGCACAGTTGCTGCAACCAACATGAACGAGGAGAGTAGCCGATCCCATGCAGTTTTCAAA ATCACCCTCACACATACTCTCTACGATGTGAAGTCTGGGACATCTGGAGAGAAAGTGGGCAAACTCAGCCTGGTGGATTTAGCTGGCAGT GAACGAGCAACGAAGACAGGCGCTGCAGGGGACAGGCTGAAGGAAGGGAGCAACATTAACAAGTCCCTCACAACCCTCGGTCTGGTTATC TCAGCTCTTGCAGATCAGAGTGCTGGCAAAAACAAGAATAAATTTGTTCCATATCGTGACTCAGTTCTCACTTGGCTGCTCAAAGACAGC CTCGGGGGTAACAGCAAGACCGCCATGGTGGCTACTGTGAGTCCTGCAGCTGATAACTATGATGAAACCCTCTCAACTCTGCGGTATGCA GATCGAGCCAAGCACATTGTAAACCACGCTGTGGTGAATGAGGACCCTAATGCCCGAATTATCCGGGATCTCCGGGAAGAAGTTGAGAAA CTCCGGGAGCAGCTGACCAAAGCAGAGGCAATGAAATCTCCAGAGCTAAAGGACCGGCTGGAAGAATCTGAGAAGCTAATCCAGGAAATG ACTGTGACCTGGGAGGAGAAATTAAGGAAAACGGAGGAGATTGCACAGGAACGACAGAAACAGCTTGAGAGTCTTGGAATATCTCTTCAG TCTTCGGGAATCAAAGTTGGGGATGATAAATGCTTCCTTGTGAATCTGAATGCTGACCCAGCTCTGAATGAGCTTCTGGTGTACTATTTA AAGGAACATACATTGATAGGGTCAGCAAATTCCCAAGATATCCAACTGTGCGGCATGGGAATTCTTCCTGAACACTGTATTATAGACATC ACGTCAGAAGGCCAGGTTATGCTGACTCCTCAGAAGAACACCAGAACATTTGTAAATGGGTCATCTGTCTCCAGTCCAATACAGCTACAC CATGGGGACAGGATATTATGGGGAAACAATCATTTCTTCAGACTCAATTTGCCTAAAAAGAAAAAGAAAGCAGAACGAGAGGATGAGGAC CAGGATCCCTCCATGAAGAACGAGAATAGTTCTGAGCAGCTGGATGTAGACGGAGACTCCTCCAGCGAGGTGTCCAGTGAAGTTAACTTT AATTACGAATACGCACAGATGGAGGTCACCATGAAGGCCCTGGGCAGCAATGATCCGATGCAGTCCATATTAAACAGCTTAGAACAACAG CATGAAGAAGAAAAACGATCTGCACTGGAGCGCCAGAGGCTTATGTATGAGCACGAATTGGAGCAGCTCCGGAGAAGGCTGTCTCCTGAG AAGCAGAACTGCCGGAGCATGGACAGGTTTTCTTTCCACTCGCCCAGCGCTCAGCAACGCTTAAGACAGTGGGCTGAGGAGAGAGAAGCA ACGTTGAATAACAGCCTGATGAGGCTGAGGGAACAAATTGTTAAGGCCAATCTATTGGTGAGAGAAGCTAATTACATTGCTGAGGAGCTG GATAAAAGAACAGAATACAAAGTTACCCTACAGATTCCAGCCTCCAGCCTGGATGCCAACAGGAAGCGAGGCTCTCTTCTTAGTGAGCCT GCAATCCAGGTGAGAAGAAAAGGAAAAGGAAAGCAGATTTGGTCTTTGGAAAAACTGGACAACAGGCTGTTGGATATGAGAGACCTTTAT CAGGAGTGGAAAGAGTGTGAAGAAGATAACCCAGTAATACGATCATACTTCAAACGTGCTGATCCATTCTATGATGAGCAGGAAAATCAC AGTCTCATTGGGGTGGCCAATGTCTTCCTCGAGTCACTTTTCTATGATGTGAAGTTACAATACGCTGTTCCCATCATCAACCAGAAAGGA GAGGTGGCAGGTCGGCTGCACGTGGAGGTGATGCGACTCAGTGGTGATGTTGGGGAGAGGATCGCAGGAGGCGATGAGGTGGCAGAGGTC TCCTTTGAGAAGGAGACCCAGGAGAACAAACTGGTGTGCATGGTTAAAATCCTGCAAGCTACTGGGTTGCCACAGCATCTGTCCCACTTT GTGTTCTGCAAATACAGCTTCTGGGATCAACAGGAGCCGGTGATTGTCGCTCCTGAAGTGGACACCTCCTCCTCTTCCGTCAGCAAGGAG CCGCACTGCATGGTTGTCTTTGATCATTGCAATGAGTTTTCTGTTAACATCACCGAAGACTTTATCGAGCATCTTTCCGAAGGAGCATTG GCAATTGAAGTATATGGACATAAAATAAACGATCCCCGGAAAAACCCCGCCCTGTGGGATTTGGGAATCATCCAAGCAAAGACACGTAGT CTTCGGGACAGATGGAGTGAAGTGACCAGGAAATTGGAATTCTGGGTTCAAATCTTGGAACAGAATGAGAATGGTGAATACTGCCCTGTA GAAGTGATTTCTGCAAAGGATGTCCCAACAGGAGGAATCTTCCAGCTCCGGCAGTATTGCCTGTCAGGGCACCCAACCTTACCATGCAAT GTGCTCAAATTCAAATCAACCACCATTATGTTGGACTGCGGACTGGACATGACTTCTACCCTCAATTTCCTTCCTTTGCCACTTGTTCAA AGTCCCAGGCTGTCCAATCTTCCTGGCTGGTCCCTGAAGGATGGAAATGCTTTCTTGGACAAGACGGAGCTAATAGATCTGTCTACAGTA GATGTGATTCTCATCTCTAACTATCACTGTATGATGGCGCTGCCATACATCACCGAGCACACCGGCTTCACAGGCACAGTGTATGCCACG GAACCCACCGTCCAGATCGGCAGGCTTCTCATGGAAGAGCTGGTGAATTTCATTGAAAGAGTGCCAAAGGCTCAGTCTGCCTCCTTGTGG AAGAATAAGGACATTCAGAGGCTGTTACCTTCTCCTCTCAAGGATGCAGTGGAAGTCTCAACCTGGAGAAGATGCTATACAATGCAAGAG GTGAACTCTGCCCTTAGTAAAATCCAGCTGGTGGGATATTCTCAGAAAATTGAGCTTTTTGGTGCGGTCCAGGTGACTCCTCTGAGCTCT GGCTATGCCCTTGGGAGCTCCAACTGGATCATCCAGTCTCATTACGAGAAAGTGTCTTATGTCTCTGGATCCTCCTTGCTTACCACACAC CCCCAGCCCATGGACCAAGCTTCTCTCAAAAACAGCGATGTTCTTGTTCTGACAGGGCTTACCCAGATCCCCACTGCAAACCCAGATGGA ATGGTGGGAGAGTTCTGCAGCAACCTAGCTCTGACAGTCCGGAATGGAGGAAACGTGTTGGTTCCCTGCTACCCTTCTGGAGTGATCTAT GACCTCCTGGAGTGCCTATATCAGTACATCGACTCAGCCGGGCTTTCCAGCGTCCCCCTCTACTTCATCTCCCCTGTGGCCAACAGTTCA CTGGAGTTTTCCCAGATCTTTGCTGAGTGGCTTTGTCACAACAAACAGAGTAAGGTGTATCTTCCAGAACCACCTTTTCCTCATGCAGAG CTCATTCAGACCAATAAGCTGAAGCACTACCCCAGCATCCACGGAGACTTCAGCAACGACTTTAGACAGCCCTGTGTGGTGTTCACCGGG CACCCTTCCCTCCGCTTCGGGGACGTGGTCCACTTCATGGAGCTCTGGGGAAAATCTAGTCTCAATACCGTCATATTCACGGAACCAGAC TTCTCCTACCTGGAAGCCCTGGCTCCTTACCAGCCGCTGGCCATGAAATGCATCTACTGCCCCATCGACACCCGGCTGAACTTCATCCAG GTGTCAAAGCTGCTTAAAGAAGTGCAGCCCCTGCACGTGGTGTGTCCTGAGCAGTACACTCAGCCGCCCCCAGCCCAGTCCCACAGGATG GACCTCATGATCGACTGCCAGCCCCCCGCCATGTCCTATCGGCGGGCTGAGGTTCTCGCCCTGCCCTTCAAACGTCGGTACGAGAAGATC GAGATCATGCCAGAGCTCGCAGATTCACTGGTGCCCATGGAGATCAAGCCTGGCATCTCCTTGGCAACTGTCTCGGCCGTGCTGCACACC AAAGATAACAAGCACTTGCTTCAGCCCCCTCCTCGGCCCGCCCAGCCCACGAGCGGGAAGAAGAGAAAGCGGGTGAGCGATGACGTACCA GACTGCAAAGTCCTGAAGCCTTTGTTGAGCGGTTCCATCCCTGTGGAGCAGTTCGTGCAGACCCTGGAGAAGCATGGCTTCAGTGATATT AAGGTGGAGGACACAGCCAAGGGCCATATCGTCCTGCTCCAGGAGGCTGAGACGCTCATCCAGATTGAAGAAGACTCGACCCATATCATC TGCGACAATGACGAGATGCTCAGAGTGCGACTGCGGGACCTTGTCCTCAAATTCTTACAGAAGTTCTGAGTGGGCCATCTGAGCTACTTC CCTGAAATCCTGCAGTCCCTCACTGGCTGCCCTCACAAGCCACCTGAGGAGTGGCATGAGAGGCCATTAACTGTGTCTTTGTGGTGTCCT CTGGCTTAAGGAGTGAAGAGGTGGCTCTTGAGGGAAATGGTCTGGACTTATTCCCAGCACTGTTTCAGGCAAGAACTTTCCCTTTCAACT TCAGGCTCATTTTCTTCTCAACTCTGGCTCTCTCAAGGAGCTGGAGGGTGGCAGAAGTGGGACAGGAGAAGTTTTCCAAGAGGTTCATGG GAGGCGGAGGTGACTGGCTGGCTGTCTTGCATCAGTCCCAGGCCTCGGCCAGGGGAGCCAGCCTTTGGTTTCGTTTACTTGCCTACAGTG CTGTACGCAATAAGATGATGATCCCAAAATATGGTAAAGTGAACCCATCTGTCTGCATTTTCTACTCTGAGCCCATTTGTTAATAAACAC >42554_42554_1_KIF13B-INTS9_KIF13B_chr8_28988050_ENST00000524189_INTS9_chr8_28717080_ENST00000416984_length(amino acids)=1659AA_BP=0 MGDSKVKVAVRIRPMNRRETDLHTKCVVDVDANKVILNPVNTNLSKGDARGQPKVFAYDHCFWSMDESVKEKYAGQDIVFKCLGENILQN AFDGYNACIFAYGQTGSGKSYTMMGTADQPGLIPRLCSGLFERTQKEENEEQSFKVEVSYMEIYNEKVRDLLDPKGSRQTLKVREHSVLG PYVDGLSKLAVTSYKDIESLMSEGNKSRTVAATNMNEESSRSHAVFKITLTHTLYDVKSGTSGEKVGKLSLVDLAGSERATKTGAAGDRL KEGSNINKSLTTLGLVISALADQSAGKNKNKFVPYRDSVLTWLLKDSLGGNSKTAMVATVSPAADNYDETLSTLRYADRAKHIVNHAVVN EDPNARIIRDLREEVEKLREQLTKAEAMKSPELKDRLEESEKLIQEMTVTWEEKLRKTEEIAQERQKQLESLGISLQSSGIKVGDDKCFL VNLNADPALNELLVYYLKEHTLIGSANSQDIQLCGMGILPEHCIIDITSEGQVMLTPQKNTRTFVNGSSVSSPIQLHHGDRILWGNNHFF RLNLPKKKKKAEREDEDQDPSMKNENSSEQLDVDGDSSSEVSSEVNFNYEYAQMEVTMKALGSNDPMQSILNSLEQQHEEEKRSALERQR LMYEHELEQLRRRLSPEKQNCRSMDRFSFHSPSAQQRLRQWAEEREATLNNSLMRLREQIVKANLLVREANYIAEELDKRTEYKVTLQIP ASSLDANRKRGSLLSEPAIQVRRKGKGKQIWSLEKLDNRLLDMRDLYQEWKECEEDNPVIRSYFKRADPFYDEQENHSLIGVANVFLESL FYDVKLQYAVPIINQKGEVAGRLHVEVMRLSGDVGERIAGGDEVAEVSFEKETQENKLVCMVKILQATGLPQHLSHFVFCKYSFWDQQEP VIVAPEVDTSSSSVSKEPHCMVVFDHCNEFSVNITEDFIEHLSEGALAIEVYGHKINDPRKNPALWDLGIIQAKTRSLRDRWSEVTRKLE FWVQILEQNENGEYCPVEVISAKDVPTGGIFQLRQYCLSGHPTLPCNVLKFKSTTIMLDCGLDMTSTLNFLPLPLVQSPRLSNLPGWSLK DGNAFLDKTELIDLSTVDVILISNYHCMMALPYITEHTGFTGTVYATEPTVQIGRLLMEELVNFIERVPKAQSASLWKNKDIQRLLPSPL KDAVEVSTWRRCYTMQEVNSALSKIQLVGYSQKIELFGAVQVTPLSSGYALGSSNWIIQSHYEKVSYVSGSSLLTTHPQPMDQASLKNSD VLVLTGLTQIPTANPDGMVGEFCSNLALTVRNGGNVLVPCYPSGVIYDLLECLYQYIDSAGLSSVPLYFISPVANSSLEFSQIFAEWLCH NKQSKVYLPEPPFPHAELIQTNKLKHYPSIHGDFSNDFRQPCVVFTGHPSLRFGDVVHFMELWGKSSLNTVIFTEPDFSYLEALAPYQPL AMKCIYCPIDTRLNFIQVSKLLKEVQPLHVVCPEQYTQPPPAQSHRMDLMIDCQPPAMSYRRAEVLALPFKRRYEKIEIMPELADSLVPM EIKPGISLATVSAVLHTKDNKHLLQPPPRPAQPTSGKKRKRVSDDVPDCKVLKPLLSGSIPVEQFVQTLEKHGFSDIKVEDTAKGHIVLL -------------------------------------------------------------- >42554_42554_2_KIF13B-INTS9_KIF13B_chr8_28988050_ENST00000524189_INTS9_chr8_28717080_ENST00000521022_length(transcript)=5567nt_BP=3114nt GGCAGACGGAAGCCGAACGAGTTCCTCGGCGGCTGCAGGATGGGGGACTCCAAAGTGAAAGTGGCGGTGCGGATACGACCCATGAACCGG CGAGAGACTGACTTGCATACCAAATGTGTGGTGGATGTGGATGCAAACAAGGTTATTCTTAATCCTGTAAATACGAATCTTTCCAAAGGA GATGCCCGGGGCCAGCCGAAGGTGTTTGCTTATGATCATTGTTTCTGGTCTATGGATGAATCTGTCAAAGAAAAGTATGCAGGTCAAGAT ATTGTTTTCAAGTGCCTTGGAGAGAATATCCTGCAGAATGCTTTTGATGGCTACAATGCATGTATCTTTGCCTATGGACAGACTGGCTCT GGAAAATCTTATACCATGATGGGCACAGCTGACCAACCTGGATTAATCCCAAGACTTTGCAGTGGACTCTTTGAACGAACTCAGAAAGAG GAAAATGAAGAACAGAGTTTTAAAGTAGAAGTGTCCTACATGGAAATTTATAATGAAAAAGTTCGAGACCTTCTTGATCCCAAAGGAAGC CGTCAGACGTTGAAAGTCAGAGAGCATAGTGTGTTGGGACCTTATGTCGACGGACTTTCTAAACTGGCTGTCACAAGCTACAAGGATATT GAGTCGTTGATGTCTGAGGGTAACAAATCTCGCACAGTTGCTGCAACCAACATGAACGAGGAGAGTAGCCGATCCCATGCAGTTTTCAAA ATCACCCTCACACATACTCTCTACGATGTGAAGTCTGGGACATCTGGAGAGAAAGTGGGCAAACTCAGCCTGGTGGATTTAGCTGGCAGT GAACGAGCAACGAAGACAGGCGCTGCAGGGGACAGGCTGAAGGAAGGGAGCAACATTAACAAGTCCCTCACAACCCTCGGTCTGGTTATC TCAGCTCTTGCAGATCAGAGTGCTGGCAAAAACAAGAATAAATTTGTTCCATATCGTGACTCAGTTCTCACTTGGCTGCTCAAAGACAGC CTCGGGGGTAACAGCAAGACCGCCATGGTGGCTACTGTGAGTCCTGCAGCTGATAACTATGATGAAACCCTCTCAACTCTGCGGTATGCA GATCGAGCCAAGCACATTGTAAACCACGCTGTGGTGAATGAGGACCCTAATGCCCGAATTATCCGGGATCTCCGGGAAGAAGTTGAGAAA CTCCGGGAGCAGCTGACCAAAGCAGAGGCAATGAAATCTCCAGAGCTAAAGGACCGGCTGGAAGAATCTGAGAAGCTAATCCAGGAAATG ACTGTGACCTGGGAGGAGAAATTAAGGAAAACGGAGGAGATTGCACAGGAACGACAGAAACAGCTTGAGAGTCTTGGAATATCTCTTCAG TCTTCGGGAATCAAAGTTGGGGATGATAAATGCTTCCTTGTGAATCTGAATGCTGACCCAGCTCTGAATGAGCTTCTGGTGTACTATTTA AAGGAACATACATTGATAGGGTCAGCAAATTCCCAAGATATCCAACTGTGCGGCATGGGAATTCTTCCTGAACACTGTATTATAGACATC ACGTCAGAAGGCCAGGTTATGCTGACTCCTCAGAAGAACACCAGAACATTTGTAAATGGGTCATCTGTCTCCAGTCCAATACAGCTACAC CATGGGGACAGGATATTATGGGGAAACAATCATTTCTTCAGACTCAATTTGCCTAAAAAGAAAAAGAAAGCAGAACGAGAGGATGAGGAC CAGGATCCCTCCATGAAGAACGAGAATAGTTCTGAGCAGCTGGATGTAGACGGAGACTCCTCCAGCGAGGTGTCCAGTGAAGTTAACTTT AATTACGAATACGCACAGATGGAGGTCACCATGAAGGCCCTGGGCAGCAATGATCCGATGCAGTCCATATTAAACAGCTTAGAACAACAG CATGAAGAAGAAAAACGATCTGCACTGGAGCGCCAGAGGCTTATGTATGAGCACGAATTGGAGCAGCTCCGGAGAAGGCTGTCTCCTGAG AAGCAGAACTGCCGGAGCATGGACAGGTTTTCTTTCCACTCGCCCAGCGCTCAGCAACGCTTAAGACAGTGGGCTGAGGAGAGAGAAGCA ACGTTGAATAACAGCCTGATGAGGCTGAGGGAACAAATTGTTAAGGCCAATCTATTGGTGAGAGAAGCTAATTACATTGCTGAGGAGCTG GATAAAAGAACAGAATACAAAGTTACCCTACAGATTCCAGCCTCCAGCCTGGATGCCAACAGGAAGCGAGGCTCTCTTCTTAGTGAGCCT GCAATCCAGGTGAGAAGAAAAGGAAAAGGAAAGCAGATTTGGTCTTTGGAAAAACTGGACAACAGGCTGTTGGATATGAGAGACCTTTAT CAGGAGTGGAAAGAGTGTGAAGAAGATAACCCAGTAATACGATCATACTTCAAACGTGCTGATCCATTCTATGATGAGCAGGAAAATCAC AGTCTCATTGGGGTGGCCAATGTCTTCCTCGAGTCACTTTTCTATGATGTGAAGTTACAATACGCTGTTCCCATCATCAACCAGAAAGGA GAGGTGGCAGGTCGGCTGCACGTGGAGGTGATGCGACTCAGTGGTGATGTTGGGGAGAGGATCGCAGGAGGCGATGAGGTGGCAGAGGTC TCCTTTGAGAAGGAGACCCAGGAGAACAAACTGGTGTGCATGGTTAAAATCCTGCAAGCTACTGGGTTGCCACAGCATCTGTCCCACTTT GTGTTCTGCAAATACAGCTTCTGGGATCAACAGGAGCCGGTGATTGTCGCTCCTGAAGTGGACACCTCCTCCTCTTCCGTCAGCAAGGAG CCGCACTGCATGGTTGTCTTTGATCATTGCAATGAGTTTTCTGTTAACATCACCGAAGACTTTATCGAGCATCTTTCCGAAGGAGCATTG GCAATTGAAGTATATGGACATAAAATAAACGATCCCCGGAAAAACCCCGCCCTGTGGGATTTGGGAATCATCCAAGCAAAGACACGTAGT CTTCGGGACAGATGGAGTGAAGTGACCAGGAAATTGGAATTCTGGGTTCAAATCTTGGAACAGAATGAGAATGGTGAATACTGCCCTGTA GAAGTGATTTCTGCAAAGGATGTCCCAACAGGAGGAATCTTCCAGCTCCGGCAGTATTGCCTGTCAGGGCACCCAACCTTACCATGCAAT GTGCTCAAATTCAAATCAACCACCATTATGTTGGACTGCGGACTGGACATGACTTCTACCCTCAATTTCCTTCCTTTGCCACTTGTTCAA AGTCCCAGGCTGTCCAATCTTCCTGGCTGGTCCCTGAAGGATGGAAATGCTTTCTTGGACAAGGAGCTAAAGGAGTGCTCGGGTCATGTA TTTGTGGATTCTGTGCCGGAATTCTGTTTACCAGAGACGGAGCTAATAGATCTGTCTACAGTAGATGTGATTCTCATCTCTAACTATCAC TGTATGATGGCGCTGCCATACATCACCGAGCACACCGGCTTCACAGGCACAGTGTATGCCACGGAACCCACCGTCCAGATCGGCAGGCTT CTCATGGAAGAGCTGGTGAATTTCATTGAAAGAGTGCCAAAGGCTCAGTCTGCCTCCTTGTGGAAGAATAAGGACATTCAGAGGCTGTTA CCTTCTCCTCTCAAGGATGCAGTGGAAGTCTCAACCTGGAGAAGATGCTATACAATGCAAGAGGTGAACTCTGCCCTTAGTAAAATCCAG CTGGTGGGATATTCTCAGAAAATTGAGCTTTTTGGTGCGGTCCAGGTGACTCCTCTGAGCTCTGGCTATGCCCTTGGGAGCTCCAACTGG ATCATCCAGTCTCATTACGAGAAAGTGTCTTATGTCTCTGGATCCTCCTTGCTTACCACACACCCCCAGCCCATGGACCAAGCTTCTCTC AAAAACAGCGATGTTCTTGTTCTGACAGGGCTTACCCAGATCCCCACTGCAAACCCAGATGGAATGGTGGGAGAGTTCTGCAGCAACCTA GCTCTGACAGTCCGGAATGGAGGAAACGTGTTGGTTCCCTGCTACCCTTCTGGAGTGATCTATGACCTCCTGGAGTGCCTATATCAGTAC ATCGACTCAGCCGGGCTTTCCAGCGTCCCCCTCTACTTCATCTCCCCTGTGGCCAACAGTTCACTGGAGTTTTCCCAGATCTTTGCTGAG TGGCTTTGTCACAACAAACAGAGTAAGGTGTATCTTCCAGAACCACCTTTTCCTCATGCAGAGCTCATTCAGACCAATAAGCTGAAGCAC TACCCCAGCATCCACGGAGACTTCAGCAACGACTTTAGACAGCCCTGTGTGGTGTTCACCGGGCACCCTTCCCTCCGCTTCGGGGACGTG GTCCACTTCATGGAGCTCTGGGGAAAATCTAGTCTCAATACCGTCATATTCACGGAACCAGACTTCTCCTACCTGGAAGCCCTGGCTCCT TACCAGCCGCTGGCCATGAAATGCATCTACTGCCCCATCGACACCCGGCTGAACTTCATCCAGGTGTCAAAGCTGCTTAAAGAAGTGCAG CCCCTGCACGTGGTGTGTCCTGAGCAGTACACTCAGCCGCCCCCAGCCCAGTCCCACAGGATGGACCTCATGATCGACTGCCAGCCCCCC GCCATGTCCTATCGGCGGGCTGAGGTTCTCGCCCTGCCCTTCAAACGTCGGTACGAGAAGATCGAGATCATGCCAGAGCTCGCAGATTCA CTGGTGCCCATGGAGATCAAGCCTGGCATCTCCTTGGCAACTGTCTCGGCCGTGCTGCACACCAAAGATAACAAGCACTTGCTTCAGCCC CCTCCTCGGCCCGCCCAGCCCACGAGCGGGAAGAAGAGAAAGCGGGTGAGCGATGACGTACCAGACTGCAAAGTCCTGAAGCCTTTGTTG AGCGGTTCCATCCCTGTGGAGCAGTTCGTGCAGACCCTGGAGAAGCATGGCTTCAGTGATATTAAGGTGGAGGACACAGCCAAGGGCCAT ATCGTCCTGCTCCAGGAGGCTGAGACGCTCATCCAGATTGAAGAAGACTCGACCCATATCATCTGCGACAATGACGAGATGCTCAGAGTG CGACTGCGGGACCTTGTCCTCAAATTCTTACAGAAGTTCTGAGTGGGCCATCTGAGCTACTTCCCTGAAATCCTGCAGTCCCTCACTGGC TGCCCTCACAAGCCACCTGAGGAGTGGCATGAGAGGCCATTAACTGTGTCTTTGTGGTGTCCTCTGGCTTAAGGAGTGAAGAGGTGGCTC TTGAGGGAAATGGTCTGGACTTATTCCCAGCACTGTTTCAGGCAAGAACTTTCCCTTTCAACTTCAGGCTCATTTTCTTCTCAACTCTGG CTCTCTCAAGGAGCTGGAGGGTGGCAGAAGTGGGACAGGAGAAGTTTTCCAAGAGGTTCATGGGAGGCGGAGGTGACTGGCTGGCTGTCT TGCATCAGTCCCAGGCCTCGGCCAGGGGAGCCAGCCTTTGGTTTCGTTTACTTGCCTACAGTGCTGTACGCAATAAGATGATGATCCCAA >42554_42554_2_KIF13B-INTS9_KIF13B_chr8_28988050_ENST00000524189_INTS9_chr8_28717080_ENST00000521022_length(amino acids)=1680AA_BP=0 MGDSKVKVAVRIRPMNRRETDLHTKCVVDVDANKVILNPVNTNLSKGDARGQPKVFAYDHCFWSMDESVKEKYAGQDIVFKCLGENILQN AFDGYNACIFAYGQTGSGKSYTMMGTADQPGLIPRLCSGLFERTQKEENEEQSFKVEVSYMEIYNEKVRDLLDPKGSRQTLKVREHSVLG PYVDGLSKLAVTSYKDIESLMSEGNKSRTVAATNMNEESSRSHAVFKITLTHTLYDVKSGTSGEKVGKLSLVDLAGSERATKTGAAGDRL KEGSNINKSLTTLGLVISALADQSAGKNKNKFVPYRDSVLTWLLKDSLGGNSKTAMVATVSPAADNYDETLSTLRYADRAKHIVNHAVVN EDPNARIIRDLREEVEKLREQLTKAEAMKSPELKDRLEESEKLIQEMTVTWEEKLRKTEEIAQERQKQLESLGISLQSSGIKVGDDKCFL VNLNADPALNELLVYYLKEHTLIGSANSQDIQLCGMGILPEHCIIDITSEGQVMLTPQKNTRTFVNGSSVSSPIQLHHGDRILWGNNHFF RLNLPKKKKKAEREDEDQDPSMKNENSSEQLDVDGDSSSEVSSEVNFNYEYAQMEVTMKALGSNDPMQSILNSLEQQHEEEKRSALERQR LMYEHELEQLRRRLSPEKQNCRSMDRFSFHSPSAQQRLRQWAEEREATLNNSLMRLREQIVKANLLVREANYIAEELDKRTEYKVTLQIP ASSLDANRKRGSLLSEPAIQVRRKGKGKQIWSLEKLDNRLLDMRDLYQEWKECEEDNPVIRSYFKRADPFYDEQENHSLIGVANVFLESL FYDVKLQYAVPIINQKGEVAGRLHVEVMRLSGDVGERIAGGDEVAEVSFEKETQENKLVCMVKILQATGLPQHLSHFVFCKYSFWDQQEP VIVAPEVDTSSSSVSKEPHCMVVFDHCNEFSVNITEDFIEHLSEGALAIEVYGHKINDPRKNPALWDLGIIQAKTRSLRDRWSEVTRKLE FWVQILEQNENGEYCPVEVISAKDVPTGGIFQLRQYCLSGHPTLPCNVLKFKSTTIMLDCGLDMTSTLNFLPLPLVQSPRLSNLPGWSLK DGNAFLDKELKECSGHVFVDSVPEFCLPETELIDLSTVDVILISNYHCMMALPYITEHTGFTGTVYATEPTVQIGRLLMEELVNFIERVP KAQSASLWKNKDIQRLLPSPLKDAVEVSTWRRCYTMQEVNSALSKIQLVGYSQKIELFGAVQVTPLSSGYALGSSNWIIQSHYEKVSYVS GSSLLTTHPQPMDQASLKNSDVLVLTGLTQIPTANPDGMVGEFCSNLALTVRNGGNVLVPCYPSGVIYDLLECLYQYIDSAGLSSVPLYF ISPVANSSLEFSQIFAEWLCHNKQSKVYLPEPPFPHAELIQTNKLKHYPSIHGDFSNDFRQPCVVFTGHPSLRFGDVVHFMELWGKSSLN TVIFTEPDFSYLEALAPYQPLAMKCIYCPIDTRLNFIQVSKLLKEVQPLHVVCPEQYTQPPPAQSHRMDLMIDCQPPAMSYRRAEVLALP FKRRYEKIEIMPELADSLVPMEIKPGISLATVSAVLHTKDNKHLLQPPPRPAQPTSGKKRKRVSDDVPDCKVLKPLLSGSIPVEQFVQTL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for KIF13B-INTS9 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for KIF13B-INTS9 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for KIF13B-INTS9 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
