
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:KIF5B-HERC4 (FusionGDB2 ID:42728) |
Fusion Gene Summary for KIF5B-HERC4 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: KIF5B-HERC4 | Fusion gene ID: 42728 | Hgene | Tgene | Gene symbol | KIF5B | HERC4 | Gene ID | 3799 | 26091 |
| Gene name | kinesin family member 5B | HECT and RLD domain containing E3 ubiquitin protein ligase 4 | |
| Synonyms | HEL-S-61|KINH|KNS|KNS1|UKHC | - | |
| Cytomap | 10p11.22 | 10q21.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | kinesin-1 heavy chainconventional kinesin heavy chainepididymis secretory protein Li 61kinesin 1 (110-120kD)kinesin heavy chainubiquitous kinesin heavy chain | probable E3 ubiquitin-protein ligase HERC4HECT domain and RCC1-like domain-containing protein 4HECT-type E3 ubiquitin transferase HERC4hect domain and RLD 4 | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | P33176 | Q5GLZ8 | |
| Ensembl transtripts involved in fusion gene | ENST00000302418, ENST00000493889, | ENST00000395187, ENST00000480158, ENST00000492996, ENST00000277817, ENST00000373700, ENST00000395198, ENST00000412272, | |
| Fusion gene scores | * DoF score | 12 X 17 X 10=2040 | 5 X 5 X 3=75 |
| # samples | 22 | 5 | |
| ** MAII score | log2(22/2040*10)=-3.2129937233342 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/75*10)=-0.584962500721156 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: KIF5B [Title/Abstract] AND HERC4 [Title/Abstract] AND fusion [Title/Abstract] | ||
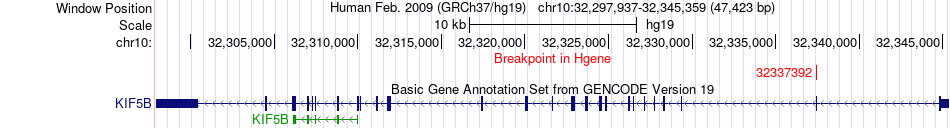
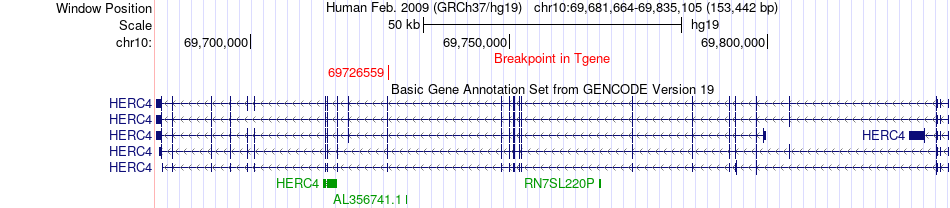
| Most frequent breakpoint | KIF5B(32337392)-HERC4(69726559), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | KIF5B | GO:0042391 | regulation of membrane potential | 19675065 |
| Hgene | KIF5B | GO:0043268 | positive regulation of potassium ion transport | 19675065 |
| Hgene | KIF5B | GO:0047496 | vesicle transport along microtubule | 28426968 |
| Hgene | KIF5B | GO:1903078 | positive regulation of protein localization to plasma membrane | 19675065 |
 Fusion gene breakpoints across KIF5B (5'-gene) Fusion gene breakpoints across KIF5B (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across HERC4 (3'-gene) Fusion gene breakpoints across HERC4 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-HU-A4HB-01A | KIF5B | chr10 | 32337392 | - | HERC4 | chr10 | 69726559 | - |
Top |
Fusion Gene ORF analysis for KIF5B-HERC4 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000302418 | ENST00000395187 | KIF5B | chr10 | 32337392 | - | HERC4 | chr10 | 69726559 | - |
| 5CDS-intron | ENST00000302418 | ENST00000480158 | KIF5B | chr10 | 32337392 | - | HERC4 | chr10 | 69726559 | - |
| 5CDS-intron | ENST00000302418 | ENST00000492996 | KIF5B | chr10 | 32337392 | - | HERC4 | chr10 | 69726559 | - |
| In-frame | ENST00000302418 | ENST00000277817 | KIF5B | chr10 | 32337392 | - | HERC4 | chr10 | 69726559 | - |
| In-frame | ENST00000302418 | ENST00000373700 | KIF5B | chr10 | 32337392 | - | HERC4 | chr10 | 69726559 | - |
| In-frame | ENST00000302418 | ENST00000395198 | KIF5B | chr10 | 32337392 | - | HERC4 | chr10 | 69726559 | - |
| In-frame | ENST00000302418 | ENST00000412272 | KIF5B | chr10 | 32337392 | - | HERC4 | chr10 | 69726559 | - |
| intron-3CDS | ENST00000493889 | ENST00000277817 | KIF5B | chr10 | 32337392 | - | HERC4 | chr10 | 69726559 | - |
| intron-3CDS | ENST00000493889 | ENST00000373700 | KIF5B | chr10 | 32337392 | - | HERC4 | chr10 | 69726559 | - |
| intron-3CDS | ENST00000493889 | ENST00000395198 | KIF5B | chr10 | 32337392 | - | HERC4 | chr10 | 69726559 | - |
| intron-3CDS | ENST00000493889 | ENST00000412272 | KIF5B | chr10 | 32337392 | - | HERC4 | chr10 | 69726559 | - |
| intron-3UTR | ENST00000493889 | ENST00000395187 | KIF5B | chr10 | 32337392 | - | HERC4 | chr10 | 69726559 | - |
| intron-intron | ENST00000493889 | ENST00000480158 | KIF5B | chr10 | 32337392 | - | HERC4 | chr10 | 69726559 | - |
| intron-intron | ENST00000493889 | ENST00000492996 | KIF5B | chr10 | 32337392 | - | HERC4 | chr10 | 69726559 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000302418 | KIF5B | chr10 | 32337392 | - | ENST00000277817 | HERC4 | chr10 | 69726559 | - | 3063 | 672 | 594 | 2039 | 481 |
| ENST00000302418 | KIF5B | chr10 | 32337392 | - | ENST00000412272 | HERC4 | chr10 | 69726559 | - | 2829 | 672 | 594 | 1805 | 403 |
| ENST00000302418 | KIF5B | chr10 | 32337392 | - | ENST00000395198 | HERC4 | chr10 | 69726559 | - | 3063 | 672 | 594 | 2039 | 481 |
| ENST00000302418 | KIF5B | chr10 | 32337392 | - | ENST00000373700 | HERC4 | chr10 | 69726559 | - | 2458 | 672 | 594 | 2015 | 473 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000302418 | ENST00000277817 | KIF5B | chr10 | 32337392 | - | HERC4 | chr10 | 69726559 | - | 0.000254382 | 0.99974567 |
| ENST00000302418 | ENST00000412272 | KIF5B | chr10 | 32337392 | - | HERC4 | chr10 | 69726559 | - | 0.000235208 | 0.99976474 |
| ENST00000302418 | ENST00000395198 | KIF5B | chr10 | 32337392 | - | HERC4 | chr10 | 69726559 | - | 0.000254382 | 0.99974567 |
| ENST00000302418 | ENST00000373700 | KIF5B | chr10 | 32337392 | - | HERC4 | chr10 | 69726559 | - | 0.000277872 | 0.99972206 |
Top |
Fusion Genomic Features for KIF5B-HERC4 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
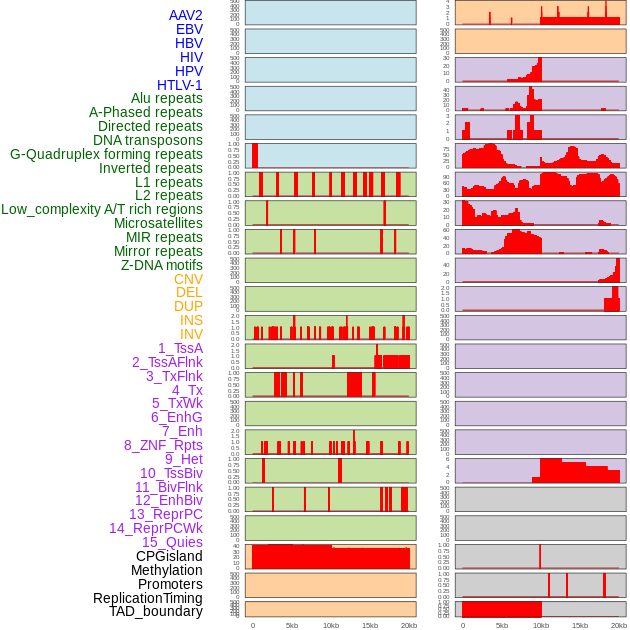 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for KIF5B-HERC4 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr10:32337392/chr10:69726559) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| KIF5B | HERC4 |
| FUNCTION: Microtubule-dependent motor required for normal distribution of mitochondria and lysosomes. Can induce formation of neurite-like membrane protrusions in non-neuronal cells in a ZFYVE27-dependent manner (By similarity). Regulates centrosome and nuclear positioning during mitotic entry. During the G2 phase of the cell cycle in a BICD2-dependent manner, antagonizes dynein function and drives the separation of nuclei and centrosomes (PubMed:20386726). Required for anterograde axonal transportation of MAPK8IP3/JIP3 which is essential for MAPK8IP3/JIP3 function in axon elongation (By similarity). Through binding with PLEKHM2 and ARL8B, directs lysosome movement toward microtubule plus ends (Probable). Involved in NK cell-mediated cytotoxicity. Drives the polarization of cytolytic granules and microtubule-organizing centers (MTOCs) toward the immune synapse between effector NK lymphocytes and target cells (PubMed:24088571). {ECO:0000250|UniProtKB:Q2PQA9, ECO:0000250|UniProtKB:Q61768, ECO:0000269|PubMed:20386726, ECO:0000269|PubMed:24088571, ECO:0000305|PubMed:22172677, ECO:0000305|PubMed:24088571}. | FUNCTION: Probable E3 ubiquitin-protein ligase involved in either protein trafficking or in the distribution of cellular structures. Required for spermatozoon maturation and fertility, and for the removal of the cytoplasmic droplet of the spermatozoon. E3 ubiquitin-protein ligases accept ubiquitin from an E2 ubiquitin-conjugating enzyme in the form of a thioester and then directly transfer it to targeted substrates. {ECO:0000250|UniProtKB:Q6PAV2}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000277817 | 11 | 23 | 730_1057 | 492 | 948.0 | Domain | HECT | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000373700 | 14 | 25 | 730_1057 | 602 | 1050.0 | Domain | HECT | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000395187 | 13 | 24 | 730_1057 | 548 | 123.66666666666667 | Domain | HECT | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000395198 | 14 | 26 | 730_1057 | 602 | 1058.0 | Domain | HECT | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000412272 | 14 | 24 | 730_1057 | 602 | 980.0 | Domain | HECT | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000492996 | 0 | 4 | 730_1057 | 0 | 111.0 | Domain | HECT | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000492996 | 0 | 4 | 102_154 | 0 | 111.0 | Repeat | Note=RCC1 3 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000492996 | 0 | 4 | 156_207 | 0 | 111.0 | Repeat | Note=RCC1 4 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000492996 | 0 | 4 | 1_51 | 0 | 111.0 | Repeat | Note=RCC1 1 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000492996 | 0 | 4 | 208_259 | 0 | 111.0 | Repeat | Note=RCC1 5 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000492996 | 0 | 4 | 261_311 | 0 | 111.0 | Repeat | Note=RCC1 6 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000492996 | 0 | 4 | 313_368 | 0 | 111.0 | Repeat | Note=RCC1 7 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000492996 | 0 | 4 | 52_101 | 0 | 111.0 | Repeat | Note=RCC1 2 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KIF5B | chr10:32337392 | chr10:69726559 | ENST00000302418 | - | 2 | 26 | 329_914 | 71 | 1626.0 | Coiled coil | . |
| Hgene | KIF5B | chr10:32337392 | chr10:69726559 | ENST00000302418 | - | 2 | 26 | 8_325 | 71 | 1626.0 | Domain | Kinesin motor |
| Hgene | KIF5B | chr10:32337392 | chr10:69726559 | ENST00000302418 | - | 2 | 26 | 85_92 | 71 | 1626.0 | Nucleotide binding | Note=ATP |
| Hgene | KIF5B | chr10:32337392 | chr10:69726559 | ENST00000302418 | - | 2 | 26 | 915_963 | 71 | 1626.0 | Region | Note=Globular |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000277817 | 11 | 23 | 102_154 | 492 | 948.0 | Repeat | Note=RCC1 3 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000277817 | 11 | 23 | 156_207 | 492 | 948.0 | Repeat | Note=RCC1 4 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000277817 | 11 | 23 | 1_51 | 492 | 948.0 | Repeat | Note=RCC1 1 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000277817 | 11 | 23 | 208_259 | 492 | 948.0 | Repeat | Note=RCC1 5 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000277817 | 11 | 23 | 261_311 | 492 | 948.0 | Repeat | Note=RCC1 6 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000277817 | 11 | 23 | 313_368 | 492 | 948.0 | Repeat | Note=RCC1 7 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000277817 | 11 | 23 | 52_101 | 492 | 948.0 | Repeat | Note=RCC1 2 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000373700 | 14 | 25 | 102_154 | 602 | 1050.0 | Repeat | Note=RCC1 3 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000373700 | 14 | 25 | 156_207 | 602 | 1050.0 | Repeat | Note=RCC1 4 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000373700 | 14 | 25 | 1_51 | 602 | 1050.0 | Repeat | Note=RCC1 1 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000373700 | 14 | 25 | 208_259 | 602 | 1050.0 | Repeat | Note=RCC1 5 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000373700 | 14 | 25 | 261_311 | 602 | 1050.0 | Repeat | Note=RCC1 6 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000373700 | 14 | 25 | 313_368 | 602 | 1050.0 | Repeat | Note=RCC1 7 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000373700 | 14 | 25 | 52_101 | 602 | 1050.0 | Repeat | Note=RCC1 2 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000395187 | 13 | 24 | 102_154 | 548 | 123.66666666666667 | Repeat | Note=RCC1 3 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000395187 | 13 | 24 | 156_207 | 548 | 123.66666666666667 | Repeat | Note=RCC1 4 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000395187 | 13 | 24 | 1_51 | 548 | 123.66666666666667 | Repeat | Note=RCC1 1 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000395187 | 13 | 24 | 208_259 | 548 | 123.66666666666667 | Repeat | Note=RCC1 5 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000395187 | 13 | 24 | 261_311 | 548 | 123.66666666666667 | Repeat | Note=RCC1 6 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000395187 | 13 | 24 | 313_368 | 548 | 123.66666666666667 | Repeat | Note=RCC1 7 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000395187 | 13 | 24 | 52_101 | 548 | 123.66666666666667 | Repeat | Note=RCC1 2 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000395198 | 14 | 26 | 102_154 | 602 | 1058.0 | Repeat | Note=RCC1 3 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000395198 | 14 | 26 | 156_207 | 602 | 1058.0 | Repeat | Note=RCC1 4 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000395198 | 14 | 26 | 1_51 | 602 | 1058.0 | Repeat | Note=RCC1 1 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000395198 | 14 | 26 | 208_259 | 602 | 1058.0 | Repeat | Note=RCC1 5 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000395198 | 14 | 26 | 261_311 | 602 | 1058.0 | Repeat | Note=RCC1 6 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000395198 | 14 | 26 | 313_368 | 602 | 1058.0 | Repeat | Note=RCC1 7 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000395198 | 14 | 26 | 52_101 | 602 | 1058.0 | Repeat | Note=RCC1 2 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000412272 | 14 | 24 | 102_154 | 602 | 980.0 | Repeat | Note=RCC1 3 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000412272 | 14 | 24 | 156_207 | 602 | 980.0 | Repeat | Note=RCC1 4 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000412272 | 14 | 24 | 1_51 | 602 | 980.0 | Repeat | Note=RCC1 1 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000412272 | 14 | 24 | 208_259 | 602 | 980.0 | Repeat | Note=RCC1 5 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000412272 | 14 | 24 | 261_311 | 602 | 980.0 | Repeat | Note=RCC1 6 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000412272 | 14 | 24 | 313_368 | 602 | 980.0 | Repeat | Note=RCC1 7 | |
| Tgene | HERC4 | chr10:32337392 | chr10:69726559 | ENST00000412272 | 14 | 24 | 52_101 | 602 | 980.0 | Repeat | Note=RCC1 2 |
Top |
Fusion Gene Sequence for KIF5B-HERC4 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >42728_42728_1_KIF5B-HERC4_KIF5B_chr10_32337392_ENST00000302418_HERC4_chr10_69726559_ENST00000277817_length(transcript)=3063nt_BP=672nt CGCCCTGTCGCCCAACGGCGGCCTCAGGAGTGATCGGGCAGCAGTCGGCCGGCCAGCGGACGGCAGAGCGGGCGGACGGGTAGGCCCGGC CTGCTCTTCGCGAGGAGGAAGAAGGTGGCCACTCTCCCGGTCCCCAGAACCTCCCCAGCCCCCGCAGTCCGCCCAGACCGTAAAGGGGGA CGCTGAGGAGCCGCGGACGCTCTCCCCGGTGCCGCCGCCGCTGCCGCCGCCATGGCTGCCATGATGGATCGGAAGTGAGCATTAGGGTTA ACGGCTGCCGGCGCCGGCTCTTCAAGTCCCGGCTCCCCGGCCGCCTCCACCCGGGGAAGCGCAGCGCGGCGCAGCTGACTGCTGCCTCTC ACGGCCCTCGCGACCACAAGCCCTCAGGTCCGGCGCGTTCCCTGCAAGACTGAGCGGCGGGGAGTGGCTCCCGGCCGCCGGCCCCGGCTG CGAGAAAGATGGCGGACCTGGCCGAGTGCAACATCAAAGTGATGTGTCGCTTCAGACCTCTCAACGAGTCTGAAGTGAACCGCGGCGACA AGTACATCGCCAAGTTTCAGGGAGAAGACACGGTCGTGATCGCGTCCAAGCCTTATGCATTTGATCGGGTGTTCCAGTCAAGCACATCTC AAGAGCAAGTGTATAATGACTGTGCAAAGAAGATTGTTAAAGGTAAATGAGAAAATGGGACAGATTATACAGTATGATAAATTTTATATA CATGAAGTACAAGAATTGATAGACATAAGAAATGATTATATCAACTGGGTCCAACAGCAGGCCTATGGAATGGATGTCAACCATGGATTA ACTGAGTTGGCAGATATCCCTGTTACAATCTGTACATATCCATTTGTATTTGATGCCCAAGCAAAAACTACTCTGTTACAGACCGATGCA GTCTTACAGATGCAGATGGCTATTGATCAGGCCCACAGGCAGAATGTCTCCTCTCTTTTTCTCCCAGTGATTGAATCTGTGAATCCCTGC TTAATTCTAGTGGTGCGTAGAGAAAATATTGTAGGAGATGCAATGGAAGTCCTTAGGAAAACAAAGAACATAGATTACAAGAAGCCACTC AAGGTTATATTTGTTGGAGAAGATGCTGTGGATGCAGGAGGGGTGCGCAAAGAATTTTTCTTGCTCATCATGAGGGAATTATTGGATCCT AAATACGGCATGTTTAGGTATTATGAAGATTCCAGGCTCATTTGGTTTTCTGATAAGACATTTGAAGACAGTGATTTGTTCCATTTGATT GGTGTTATCTGTGGCTTAGCAATTTATAATTGTACCATTGTGGACCTCCATTTTCCTTTGGCTTTATATAAGAAACTACTGAAAAAGAAG CCATCCTTGGATGATTTGAAAGAACTAATGCCTGATGTTGGGAGAAGCATGCAACAGTTACTGGATTATCCAGAAGATGACATAGAGGAA ACATTTTGTCTTAATTTTACGATCACAGTTGAAAACTTTGGTGCAACAGAAGTGAAAGAGCTGGTTCTAAATGGTGCAGACACAGCTGTT AACAAACAAAATCGGCAAGAGTTTGTCGATGCTTATGTGGATTACATATTCAATAAATCAGTGGCTTCCTTATTTGATGCTTTTCATGCG GGCTTTCATAAGGTCTGTGGAGGAAAAGTCCTTCTGCTCTTTCAGCCTAATGAACTACAAGCAATGGTCATTGGAAATACAAATTATGAT TGGAAGGAACTGGAAAAGAATACAGAATACAAAGGGGAATATTGGGCAGAACATCCTACGATAAAAATTTTTTGGGAAGTATTTCACGAA TTACCATTGGAAAAGAAGAAACAGTTTCTGTTATTTTTGACAGGTAGTGATCGCATTCCTATTCTTGGTATGAAGAGTCTGAAACTAGTC ATCCAGTCCACAGGAGGTGGTGAGGAGTATCTCCCAGTTTCCCATACTTGTTTTAATCTTCTGGATCTTCCAAAATATACAGAAAAAGAA ACTCTACGCTCTAAACTGATCCAAGCTATTGATCACAATGAAGGCTTCAGTTTAATATAACTTTGGAGTTATAACTATTCAGTTTAGTGC AAAAGCATTAAACTATTTGTGTTTTTCTTGTGGTGATGAATTCAGCAAGGTGACAGAGGTACTATTATAATTCTTACTTGCAGAATGTTC AATCTACGAGTGTTCATGGAAGCCAAAAAATATTAAAGGAAAATGAACAAACTGTTAATATTATTGTACAGAACCATGGATTTTTTTTGA CCATCTTCTAATAAACATAGCAAGTATTATGAATACATTAAAGTTTTACTAACATGAATTTTAAGAGTTTGCATATTTCAGAAATGATCT GGTGTGAGTGCATGGAAATATTGCTTAATTTTTCTTCAATCATTGAGTGAAAAACCTTTAACTTTGGCCTGCAATAGTCATTTGATTATT TTTTCATTTTGTAAATAATGTTAAGTTTTGTAATAAAATAGTTATGTTCTGATACCAGTACAGTTTCTATGGTTGTAATTGAACTTGAAC AGACTTTTAAAGGTTAAAAATTATGATTTAAATCTTACTCTGAGACTCTATAAAAAGAAAAAAAAGGTAGCATGGTGGAAACACTATCTT TTCTTTTTTGCTAGAATAAGTGTCTTTGTGCAAACCTAAATCACAGATAGGGAATATACTACATAACCTGCAGTTCTTTTCTTTGTGAAT CCTGACACACTGATAGATGGGGGATTGTCGATCAGAGAACTTATTAATATTTAGTACTGGAAGAACTCTGTCTCCACAGTTGCCAGTAAT AAAAAGAAACATTGGCTACTATGAGCACCAATCACTGGGTTATAGCTTTCAAAATTATTGATGCTGCAGTGCTTTAGAGCTATTTCCTTG AATTTAAGAAACAAATCTTAACAGTTTTATGGTGCTGATGCTTAGTTGTCTCATGCCATTAAATTGTAAAAGTGAGTTGATGCAATACAT GTGACTTTCTGCTATAATGCAAATATTTATTTTTTAAATTTATTTTAAAATGCCGTGAAAACTGTTTAATAAAGATTTATTGTTTTAATA >42728_42728_1_KIF5B-HERC4_KIF5B_chr10_32337392_ENST00000302418_HERC4_chr10_69726559_ENST00000277817_length(amino acids)=481AA_BP=23 MHLIGCSSQAHLKSKCIMTVQRRLLKVNEKMGQIIQYDKFYIHEVQELIDIRNDYINWVQQQAYGMDVNHGLTELADIPVTICTYPFVFD AQAKTTLLQTDAVLQMQMAIDQAHRQNVSSLFLPVIESVNPCLILVVRRENIVGDAMEVLRKTKNIDYKKPLKVIFVGEDAVDAGGVRKE FFLLIMRELLDPKYGMFRYYEDSRLIWFSDKTFEDSDLFHLIGVICGLAIYNCTIVDLHFPLALYKKLLKKKPSLDDLKELMPDVGRSMQ QLLDYPEDDIEETFCLNFTITVENFGATEVKELVLNGADTAVNKQNRQEFVDAYVDYIFNKSVASLFDAFHAGFHKVCGGKVLLLFQPNE LQAMVIGNTNYDWKELEKNTEYKGEYWAEHPTIKIFWEVFHELPLEKKKQFLLFLTGSDRIPILGMKSLKLVIQSTGGGEEYLPVSHTCF -------------------------------------------------------------- >42728_42728_2_KIF5B-HERC4_KIF5B_chr10_32337392_ENST00000302418_HERC4_chr10_69726559_ENST00000373700_length(transcript)=2458nt_BP=672nt CGCCCTGTCGCCCAACGGCGGCCTCAGGAGTGATCGGGCAGCAGTCGGCCGGCCAGCGGACGGCAGAGCGGGCGGACGGGTAGGCCCGGC CTGCTCTTCGCGAGGAGGAAGAAGGTGGCCACTCTCCCGGTCCCCAGAACCTCCCCAGCCCCCGCAGTCCGCCCAGACCGTAAAGGGGGA CGCTGAGGAGCCGCGGACGCTCTCCCCGGTGCCGCCGCCGCTGCCGCCGCCATGGCTGCCATGATGGATCGGAAGTGAGCATTAGGGTTA ACGGCTGCCGGCGCCGGCTCTTCAAGTCCCGGCTCCCCGGCCGCCTCCACCCGGGGAAGCGCAGCGCGGCGCAGCTGACTGCTGCCTCTC ACGGCCCTCGCGACCACAAGCCCTCAGGTCCGGCGCGTTCCCTGCAAGACTGAGCGGCGGGGAGTGGCTCCCGGCCGCCGGCCCCGGCTG CGAGAAAGATGGCGGACCTGGCCGAGTGCAACATCAAAGTGATGTGTCGCTTCAGACCTCTCAACGAGTCTGAAGTGAACCGCGGCGACA AGTACATCGCCAAGTTTCAGGGAGAAGACACGGTCGTGATCGCGTCCAAGCCTTATGCATTTGATCGGGTGTTCCAGTCAAGCACATCTC AAGAGCAAGTGTATAATGACTGTGCAAAGAAGATTGTTAAAGGTAAATGAGAAAATGGGACAGATTATACAGTATGATAAATTTTATATA CATGAAGTACAAGAATTGATAGACATAAGAAATGATTATATCAACTGGGTCCAACAGCAGGCCTATGGAATGTTGGCAGATATCCCTGTT ACAATCTGTACATATCCATTTGTATTTGATGCCCAAGCAAAAACTACTCTGTTACAGACCGATGCAGTCTTACAGATGCAGATGGCTATT GATCAGGCCCACAGGCAGAATGTCTCCTCTCTTTTTCTCCCAGTGATTGAATCTGTGAATCCCTGCTTAATTCTAGTGGTGCGTAGAGAA AATATTGTAGGAGATGCAATGGAAGTCCTTAGGAAAACAAAGAACATAGATTACAAGAAGCCACTCAAGGTTATATTTGTTGGAGAAGAT GCTGTGGATGCAGGAGGGGTGCGCAAAGAATTTTTCTTGCTCATCATGAGGGAATTATTGGATCCTAAATACGGCATGTTTAGGTATTAT GAAGATTCCAGGCTCATTTGGTTTTCTGATAAGACATTTGAAGACAGTGATTTGTTCCATTTGATTGGTGTTATCTGTGGCTTAGCAATT TATAATTGTACCATTGTGGACCTCCATTTTCCTTTGGCTTTATATAAGAAACTACTGAAAAAGAAGCCATCCTTGGATGATTTGAAAGAA CTAATGCCTGATGTTGGGAGAAGCATGCAACAGTTACTGGATTATCCAGAAGATGACATAGAGGAAACATTTTGTCTTAATTTTACGATC ACAGTTGAAAACTTTGGTGCAACAGAAGTGAAAGAGCTGGTTCTAAATGGTGCAGACACAGCTGTTAACAAACAAAATCGGCAAGAGTTT GTCGATGCTTATGTGGATTACATATTCAATAAATCAGTGGCTTCCTTATTTGATGCTTTTCATGCGGGCTTTCATAAGGTCTGTGGAGGA AAAGTCCTTCTGCTCTTTCAGCCTAATGAACTACAAGCAATGGTCATTGGAAATACAAATTATGATTGGAAGGAACTGGAAAAGAATACA GAATACAAAGGGGAATATTGGGCAGAACATCCTACGATAAAAATTTTTTGGGAAGTATTTCACGAATTACCATTGGAAAAGAAGAAACAG TTTCTGTTATTTTTGACAGGTAGTGATCGCATTCCTATTCTTGGTATGAAGAGTCTGAAACTAGTCATCCAGTCCACAGGAGGTGGTGAG GAGTATCTCCCAGTTTCCCATACTTGTTTTAATCTTCTGGATCTTCCAAAATATACAGAAAAAGAAACTCTACGCTCTAAACTGATCCAA GCTATTGATCACAATGAAGGCTTCAGTTTAATATAACTTTGGAGTTATAACTATTCAGTTTAGTGCAAAAGCATTAAACTATTTGTGTTT TTCTTGTGGTGATGAATTCAGCAAGGTGACAGAGGTACTATTATAATTCTTACTTGCAGAATGTTCAATCTACGAGTGTTCATGGAAGCC AAAAAATATTAAAGGAAAATGAACAAACTGTTAATATTATTGTACAGAACCATGGATTTTTTTTGACCATCTTCTAATAAACATAGCAAG TATTATGAATACATTAAAGTTTTACTAACATGAATTTTAAGAGTTTGCATATTTCAGAAATGATCTGGTGTGAGTGCATGGAAATATTGC TTAATTTTTCTTCAATCATTGAGTGAAAAACCTTTAACTTTGGCCTGCAATAGTCATTTGATTATTTTTTCATTTTGTAAATAATGTTAA >42728_42728_2_KIF5B-HERC4_KIF5B_chr10_32337392_ENST00000302418_HERC4_chr10_69726559_ENST00000373700_length(amino acids)=473AA_BP=23 MHLIGCSSQAHLKSKCIMTVQRRLLKVNEKMGQIIQYDKFYIHEVQELIDIRNDYINWVQQQAYGMLADIPVTICTYPFVFDAQAKTTLL QTDAVLQMQMAIDQAHRQNVSSLFLPVIESVNPCLILVVRRENIVGDAMEVLRKTKNIDYKKPLKVIFVGEDAVDAGGVRKEFFLLIMRE LLDPKYGMFRYYEDSRLIWFSDKTFEDSDLFHLIGVICGLAIYNCTIVDLHFPLALYKKLLKKKPSLDDLKELMPDVGRSMQQLLDYPED DIEETFCLNFTITVENFGATEVKELVLNGADTAVNKQNRQEFVDAYVDYIFNKSVASLFDAFHAGFHKVCGGKVLLLFQPNELQAMVIGN TNYDWKELEKNTEYKGEYWAEHPTIKIFWEVFHELPLEKKKQFLLFLTGSDRIPILGMKSLKLVIQSTGGGEEYLPVSHTCFNLLDLPKY -------------------------------------------------------------- >42728_42728_3_KIF5B-HERC4_KIF5B_chr10_32337392_ENST00000302418_HERC4_chr10_69726559_ENST00000395198_length(transcript)=3063nt_BP=672nt CGCCCTGTCGCCCAACGGCGGCCTCAGGAGTGATCGGGCAGCAGTCGGCCGGCCAGCGGACGGCAGAGCGGGCGGACGGGTAGGCCCGGC CTGCTCTTCGCGAGGAGGAAGAAGGTGGCCACTCTCCCGGTCCCCAGAACCTCCCCAGCCCCCGCAGTCCGCCCAGACCGTAAAGGGGGA CGCTGAGGAGCCGCGGACGCTCTCCCCGGTGCCGCCGCCGCTGCCGCCGCCATGGCTGCCATGATGGATCGGAAGTGAGCATTAGGGTTA ACGGCTGCCGGCGCCGGCTCTTCAAGTCCCGGCTCCCCGGCCGCCTCCACCCGGGGAAGCGCAGCGCGGCGCAGCTGACTGCTGCCTCTC ACGGCCCTCGCGACCACAAGCCCTCAGGTCCGGCGCGTTCCCTGCAAGACTGAGCGGCGGGGAGTGGCTCCCGGCCGCCGGCCCCGGCTG CGAGAAAGATGGCGGACCTGGCCGAGTGCAACATCAAAGTGATGTGTCGCTTCAGACCTCTCAACGAGTCTGAAGTGAACCGCGGCGACA AGTACATCGCCAAGTTTCAGGGAGAAGACACGGTCGTGATCGCGTCCAAGCCTTATGCATTTGATCGGGTGTTCCAGTCAAGCACATCTC AAGAGCAAGTGTATAATGACTGTGCAAAGAAGATTGTTAAAGGTAAATGAGAAAATGGGACAGATTATACAGTATGATAAATTTTATATA CATGAAGTACAAGAATTGATAGACATAAGAAATGATTATATCAACTGGGTCCAACAGCAGGCCTATGGAATGGATGTCAACCATGGATTA ACTGAGTTGGCAGATATCCCTGTTACAATCTGTACATATCCATTTGTATTTGATGCCCAAGCAAAAACTACTCTGTTACAGACCGATGCA GTCTTACAGATGCAGATGGCTATTGATCAGGCCCACAGGCAGAATGTCTCCTCTCTTTTTCTCCCAGTGATTGAATCTGTGAATCCCTGC TTAATTCTAGTGGTGCGTAGAGAAAATATTGTAGGAGATGCAATGGAAGTCCTTAGGAAAACAAAGAACATAGATTACAAGAAGCCACTC AAGGTTATATTTGTTGGAGAAGATGCTGTGGATGCAGGAGGGGTGCGCAAAGAATTTTTCTTGCTCATCATGAGGGAATTATTGGATCCT AAATACGGCATGTTTAGGTATTATGAAGATTCCAGGCTCATTTGGTTTTCTGATAAGACATTTGAAGACAGTGATTTGTTCCATTTGATT GGTGTTATCTGTGGCTTAGCAATTTATAATTGTACCATTGTGGACCTCCATTTTCCTTTGGCTTTATATAAGAAACTACTGAAAAAGAAG CCATCCTTGGATGATTTGAAAGAACTAATGCCTGATGTTGGGAGAAGCATGCAACAGTTACTGGATTATCCAGAAGATGACATAGAGGAA ACATTTTGTCTTAATTTTACGATCACAGTTGAAAACTTTGGTGCAACAGAAGTGAAAGAGCTGGTTCTAAATGGTGCAGACACAGCTGTT AACAAACAAAATCGGCAAGAGTTTGTCGATGCTTATGTGGATTACATATTCAATAAATCAGTGGCTTCCTTATTTGATGCTTTTCATGCG GGCTTTCATAAGGTCTGTGGAGGAAAAGTCCTTCTGCTCTTTCAGCCTAATGAACTACAAGCAATGGTCATTGGAAATACAAATTATGAT TGGAAGGAACTGGAAAAGAATACAGAATACAAAGGGGAATATTGGGCAGAACATCCTACGATAAAAATTTTTTGGGAAGTATTTCACGAA TTACCATTGGAAAAGAAGAAACAGTTTCTGTTATTTTTGACAGGTAGTGATCGCATTCCTATTCTTGGTATGAAGAGTCTGAAACTAGTC ATCCAGTCCACAGGAGGTGGTGAGGAGTATCTCCCAGTTTCCCATACTTGTTTTAATCTTCTGGATCTTCCAAAATATACAGAAAAAGAA ACTCTACGCTCTAAACTGATCCAAGCTATTGATCACAATGAAGGCTTCAGTTTAATATAACTTTGGAGTTATAACTATTCAGTTTAGTGC AAAAGCATTAAACTATTTGTGTTTTTCTTGTGGTGATGAATTCAGCAAGGTGACAGAGGTACTATTATAATTCTTACTTGCAGAATGTTC AATCTACGAGTGTTCATGGAAGCCAAAAAATATTAAAGGAAAATGAACAAACTGTTAATATTATTGTACAGAACCATGGATTTTTTTTGA CCATCTTCTAATAAACATAGCAAGTATTATGAATACATTAAAGTTTTACTAACATGAATTTTAAGAGTTTGCATATTTCAGAAATGATCT GGTGTGAGTGCATGGAAATATTGCTTAATTTTTCTTCAATCATTGAGTGAAAAACCTTTAACTTTGGCCTGCAATAGTCATTTGATTATT TTTTCATTTTGTAAATAATGTTAAGTTTTGTAATAAAATAGTTATGTTCTGATACCAGTACAGTTTCTATGGTTGTAATTGAACTTGAAC AGACTTTTAAAGGTTAAAAATTATGATTTAAATCTTACTCTGAGACTCTATAAAAAGAAAAAAAAGGTAGCATGGTGGAAACACTATCTT TTCTTTTTTGCTAGAATAAGTGTCTTTGTGCAAACCTAAATCACAGATAGGGAATATACTACATAACCTGCAGTTCTTTTCTTTGTGAAT CCTGACACACTGATAGATGGGGGATTGTCGATCAGAGAACTTATTAATATTTAGTACTGGAAGAACTCTGTCTCCACAGTTGCCAGTAAT AAAAAGAAACATTGGCTACTATGAGCACCAATCACTGGGTTATAGCTTTCAAAATTATTGATGCTGCAGTGCTTTAGAGCTATTTCCTTG AATTTAAGAAACAAATCTTAACAGTTTTATGGTGCTGATGCTTAGTTGTCTCATGCCATTAAATTGTAAAAGTGAGTTGATGCAATACAT GTGACTTTCTGCTATAATGCAAATATTTATTTTTTAAATTTATTTTAAAATGCCGTGAAAACTGTTTAATAAAGATTTATTGTTTTAATA >42728_42728_3_KIF5B-HERC4_KIF5B_chr10_32337392_ENST00000302418_HERC4_chr10_69726559_ENST00000395198_length(amino acids)=481AA_BP=23 MHLIGCSSQAHLKSKCIMTVQRRLLKVNEKMGQIIQYDKFYIHEVQELIDIRNDYINWVQQQAYGMDVNHGLTELADIPVTICTYPFVFD AQAKTTLLQTDAVLQMQMAIDQAHRQNVSSLFLPVIESVNPCLILVVRRENIVGDAMEVLRKTKNIDYKKPLKVIFVGEDAVDAGGVRKE FFLLIMRELLDPKYGMFRYYEDSRLIWFSDKTFEDSDLFHLIGVICGLAIYNCTIVDLHFPLALYKKLLKKKPSLDDLKELMPDVGRSMQ QLLDYPEDDIEETFCLNFTITVENFGATEVKELVLNGADTAVNKQNRQEFVDAYVDYIFNKSVASLFDAFHAGFHKVCGGKVLLLFQPNE LQAMVIGNTNYDWKELEKNTEYKGEYWAEHPTIKIFWEVFHELPLEKKKQFLLFLTGSDRIPILGMKSLKLVIQSTGGGEEYLPVSHTCF -------------------------------------------------------------- >42728_42728_4_KIF5B-HERC4_KIF5B_chr10_32337392_ENST00000302418_HERC4_chr10_69726559_ENST00000412272_length(transcript)=2829nt_BP=672nt CGCCCTGTCGCCCAACGGCGGCCTCAGGAGTGATCGGGCAGCAGTCGGCCGGCCAGCGGACGGCAGAGCGGGCGGACGGGTAGGCCCGGC CTGCTCTTCGCGAGGAGGAAGAAGGTGGCCACTCTCCCGGTCCCCAGAACCTCCCCAGCCCCCGCAGTCCGCCCAGACCGTAAAGGGGGA CGCTGAGGAGCCGCGGACGCTCTCCCCGGTGCCGCCGCCGCTGCCGCCGCCATGGCTGCCATGATGGATCGGAAGTGAGCATTAGGGTTA ACGGCTGCCGGCGCCGGCTCTTCAAGTCCCGGCTCCCCGGCCGCCTCCACCCGGGGAAGCGCAGCGCGGCGCAGCTGACTGCTGCCTCTC ACGGCCCTCGCGACCACAAGCCCTCAGGTCCGGCGCGTTCCCTGCAAGACTGAGCGGCGGGGAGTGGCTCCCGGCCGCCGGCCCCGGCTG CGAGAAAGATGGCGGACCTGGCCGAGTGCAACATCAAAGTGATGTGTCGCTTCAGACCTCTCAACGAGTCTGAAGTGAACCGCGGCGACA AGTACATCGCCAAGTTTCAGGGAGAAGACACGGTCGTGATCGCGTCCAAGCCTTATGCATTTGATCGGGTGTTCCAGTCAAGCACATCTC AAGAGCAAGTGTATAATGACTGTGCAAAGAAGATTGTTAAAGGTAAATGAGAAAATGGGACAGATTATACAGTATGATAAATTTTATATA CATGAAGTACAAGAATTGATAGACATAAGAAATGATTATATCAACTGGGTCCAACAGCAGGCCTATGGAATGGATGTCAACCATGGATTA ACTGAGTTGGCAGATATCCCTGTTACAATCTGTACATATCCATTTGTATTTGATGCCCAAGCAAAAACTACTCTGTTACAGACCGATGCA GTCTTACAGATGCAGATGGCTATTGATCAGGCCCACAGGCAGAATGTCTCCTCTCTTTTTCTCCCAGTGATTGAATCTGTGAATCCCTGC TTAATTCTAGTGGTGCGTAGAGAAAATATTGTAGGAGATGCAATGGAAGTCCTTAGGAAAACAAAGAACATAGATTACAAGAAGCCACTC AAGGTTATATTTGTTGGAGAAGATGCTGTGGATGCAGGAGGGGTGCGCAAAGAATTTTTCTTGCTCATCATGAGGGAATTATTGGATCCT AAATACGGCATGTTTAGGTATTATGAAGATTCCAGGCTCATTTGGTTTTCTGATAAGATCACAGTTGAAAACTTTGGTGCAACAGAAGTG AAAGAGCTGGTTCTAAATGGTGCAGACACAGCTGTTAACAAACAAAATCGGCAAGAGTTTGTCGATGCTTATGTGGATTACATATTCAAT AAATCAGTGGCTTCCTTATTTGATGCTTTTCATGCGGGCTTTCATAAGGTCTGTGGAGGAAAAGTCCTTCTGCTCTTTCAGCCTAATGAA CTACAAGCAATGGTCATTGGAAATACAAATTATGATTGGAAGGAACTGGAAAAGAATACAGAATACAAAGGGGAATATTGGGCAGAACAT CCTACGATAAAAATTTTTTGGGAAGTATTTCACGAATTACCATTGGAAAAGAAGAAACAGTTTCTGTTATTTTTGACAGGTAGTGATCGC ATTCCTATTCTTGGTATGAAGAGTCTGAAACTAGTCATCCAGTCCACAGGAGGTGGTGAGGAGTATCTCCCAGTTTCCCATACTTGTTTT AATCTTCTGGATCTTCCAAAATATACAGAAAAAGAAACTCTACGCTCTAAACTGATCCAAGCTATTGATCACAATGAAGGCTTCAGTTTA ATATAACTTTGGAGTTATAACTATTCAGTTTAGTGCAAAAGCATTAAACTATTTGTGTTTTTCTTGTGGTGATGAATTCAGCAAGGTGAC AGAGGTACTATTATAATTCTTACTTGCAGAATGTTCAATCTACGAGTGTTCATGGAAGCCAAAAAATATTAAAGGAAAATGAACAAACTG TTAATATTATTGTACAGAACCATGGATTTTTTTTGACCATCTTCTAATAAACATAGCAAGTATTATGAATACATTAAAGTTTTACTAACA TGAATTTTAAGAGTTTGCATATTTCAGAAATGATCTGGTGTGAGTGCATGGAAATATTGCTTAATTTTTCTTCAATCATTGAGTGAAAAA CCTTTAACTTTGGCCTGCAATAGTCATTTGATTATTTTTTCATTTTGTAAATAATGTTAAGTTTTGTAATAAAATAGTTATGTTCTGATA CCAGTACAGTTTCTATGGTTGTAATTGAACTTGAACAGACTTTTAAAGGTTAAAAATTATGATTTAAATCTTACTCTGAGACTCTATAAA AAGAAAAAAAAGGTAGCATGGTGGAAACACTATCTTTTCTTTTTTGCTAGAATAAGTGTCTTTGTGCAAACCTAAATCACAGATAGGGAA TATACTACATAACCTGCAGTTCTTTTCTTTGTGAATCCTGACACACTGATAGATGGGGGATTGTCGATCAGAGAACTTATTAATATTTAG TACTGGAAGAACTCTGTCTCCACAGTTGCCAGTAATAAAAAGAAACATTGGCTACTATGAGCACCAATCACTGGGTTATAGCTTTCAAAA TTATTGATGCTGCAGTGCTTTAGAGCTATTTCCTTGAATTTAAGAAACAAATCTTAACAGTTTTATGGTGCTGATGCTTAGTTGTCTCAT GCCATTAAATTGTAAAAGTGAGTTGATGCAATACATGTGACTTTCTGCTATAATGCAAATATTTATTTTTTAAATTTATTTTAAAATGCC >42728_42728_4_KIF5B-HERC4_KIF5B_chr10_32337392_ENST00000302418_HERC4_chr10_69726559_ENST00000412272_length(amino acids)=403AA_BP=23 MHLIGCSSQAHLKSKCIMTVQRRLLKVNEKMGQIIQYDKFYIHEVQELIDIRNDYINWVQQQAYGMDVNHGLTELADIPVTICTYPFVFD AQAKTTLLQTDAVLQMQMAIDQAHRQNVSSLFLPVIESVNPCLILVVRRENIVGDAMEVLRKTKNIDYKKPLKVIFVGEDAVDAGGVRKE FFLLIMRELLDPKYGMFRYYEDSRLIWFSDKITVENFGATEVKELVLNGADTAVNKQNRQEFVDAYVDYIFNKSVASLFDAFHAGFHKVC GGKVLLLFQPNELQAMVIGNTNYDWKELEKNTEYKGEYWAEHPTIKIFWEVFHELPLEKKKQFLLFLTGSDRIPILGMKSLKLVIQSTGG -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for KIF5B-HERC4 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for KIF5B-HERC4 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for KIF5B-HERC4 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
