
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:KLHL24-CEP89 (FusionGDB2 ID:42971) |
Fusion Gene Summary for KLHL24-CEP89 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: KLHL24-CEP89 | Fusion gene ID: 42971 | Hgene | Tgene | Gene symbol | KLHL24 | CEP89 | Gene ID | 54800 | 84902 |
| Gene name | kelch like family member 24 | centrosomal protein 89 | |
| Synonyms | DRE1|EBSSH|KRIP6 | CCDC123|CEP123 | |
| Cytomap | 3q27.1 | 19q13.11 | |
| Type of gene | protein-coding | protein-coding | |
| Description | kelch-like protein 24kainate receptor interacting protein for GluR6kelch-like 24 | centrosomal protein of 89 kDacentrosomal protein 123centrosomal protein 89kDacoiled-coil domain containing 123coiled-coil domain-containing protein 123, mitochondrial | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q6TFL4 | Q96ST8 | |
| Ensembl transtripts involved in fusion gene | ENST00000242810, ENST00000454652, ENST00000476808, ENST00000461813, | ENST00000590597, ENST00000591863, ENST00000305768, | |
| Fusion gene scores | * DoF score | 11 X 11 X 5=605 | 11 X 11 X 7=847 |
| # samples | 14 | 15 | |
| ** MAII score | log2(14/605*10)=-2.11150831521699 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(15/847*10)=-2.49739946883632 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: KLHL24 [Title/Abstract] AND CEP89 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | KLHL24(183369064)-CEP89(33392318), # samples:2 KLHL24(183381430)-CEP89(33392318), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | KLHL24-CEP89 seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | KLHL24 | GO:0016567 | protein ubiquitination | 27798626 |
 Fusion gene breakpoints across KLHL24 (5'-gene) Fusion gene breakpoints across KLHL24 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
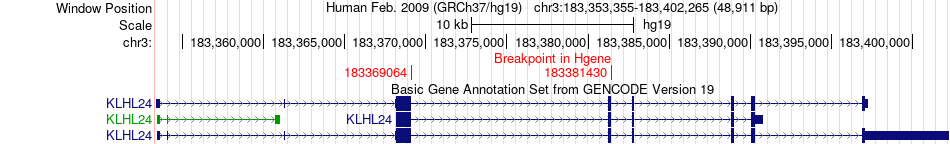 |
 Fusion gene breakpoints across CEP89 (3'-gene) Fusion gene breakpoints across CEP89 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
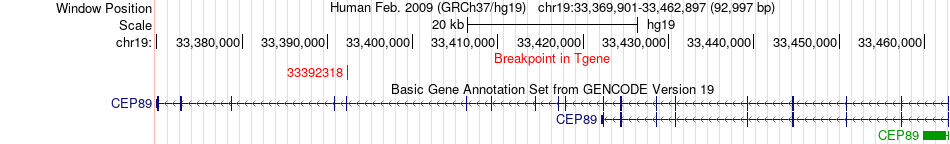 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUSC | TCGA-21-1071-01A | KLHL24 | chr3 | 183369064 | + | CEP89 | chr19 | 33392318 | - |
| ChimerDB4 | LUSC | TCGA-21-1071-01A | KLHL24 | chr3 | 183381430 | - | CEP89 | chr19 | 33392318 | - |
| ChimerDB4 | LUSC | TCGA-21-1071 | KLHL24 | chr3 | 183369064 | + | CEP89 | chr19 | 33392318 | - |
| ChimerDB4 | LUSC | TCGA-21-1071 | KLHL24 | chr3 | 183381430 | + | CEP89 | chr19 | 33392318 | - |
Top |
Fusion Gene ORF analysis for KLHL24-CEP89 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000242810 | ENST00000590597 | KLHL24 | chr3 | 183369064 | + | CEP89 | chr19 | 33392318 | - |
| 5CDS-intron | ENST00000242810 | ENST00000590597 | KLHL24 | chr3 | 183381430 | + | CEP89 | chr19 | 33392318 | - |
| 5CDS-intron | ENST00000242810 | ENST00000591863 | KLHL24 | chr3 | 183369064 | + | CEP89 | chr19 | 33392318 | - |
| 5CDS-intron | ENST00000242810 | ENST00000591863 | KLHL24 | chr3 | 183381430 | + | CEP89 | chr19 | 33392318 | - |
| 5CDS-intron | ENST00000454652 | ENST00000590597 | KLHL24 | chr3 | 183369064 | + | CEP89 | chr19 | 33392318 | - |
| 5CDS-intron | ENST00000454652 | ENST00000590597 | KLHL24 | chr3 | 183381430 | + | CEP89 | chr19 | 33392318 | - |
| 5CDS-intron | ENST00000454652 | ENST00000591863 | KLHL24 | chr3 | 183369064 | + | CEP89 | chr19 | 33392318 | - |
| 5CDS-intron | ENST00000454652 | ENST00000591863 | KLHL24 | chr3 | 183381430 | + | CEP89 | chr19 | 33392318 | - |
| 5CDS-intron | ENST00000476808 | ENST00000590597 | KLHL24 | chr3 | 183369064 | + | CEP89 | chr19 | 33392318 | - |
| 5CDS-intron | ENST00000476808 | ENST00000590597 | KLHL24 | chr3 | 183381430 | + | CEP89 | chr19 | 33392318 | - |
| 5CDS-intron | ENST00000476808 | ENST00000591863 | KLHL24 | chr3 | 183369064 | + | CEP89 | chr19 | 33392318 | - |
| 5CDS-intron | ENST00000476808 | ENST00000591863 | KLHL24 | chr3 | 183381430 | + | CEP89 | chr19 | 33392318 | - |
| Frame-shift | ENST00000242810 | ENST00000305768 | KLHL24 | chr3 | 183381430 | + | CEP89 | chr19 | 33392318 | - |
| Frame-shift | ENST00000454652 | ENST00000305768 | KLHL24 | chr3 | 183381430 | + | CEP89 | chr19 | 33392318 | - |
| Frame-shift | ENST00000476808 | ENST00000305768 | KLHL24 | chr3 | 183381430 | + | CEP89 | chr19 | 33392318 | - |
| In-frame | ENST00000242810 | ENST00000305768 | KLHL24 | chr3 | 183369064 | + | CEP89 | chr19 | 33392318 | - |
| In-frame | ENST00000454652 | ENST00000305768 | KLHL24 | chr3 | 183369064 | + | CEP89 | chr19 | 33392318 | - |
| In-frame | ENST00000476808 | ENST00000305768 | KLHL24 | chr3 | 183369064 | + | CEP89 | chr19 | 33392318 | - |
| intron-3CDS | ENST00000461813 | ENST00000305768 | KLHL24 | chr3 | 183369064 | + | CEP89 | chr19 | 33392318 | - |
| intron-3CDS | ENST00000461813 | ENST00000305768 | KLHL24 | chr3 | 183381430 | + | CEP89 | chr19 | 33392318 | - |
| intron-intron | ENST00000461813 | ENST00000590597 | KLHL24 | chr3 | 183369064 | + | CEP89 | chr19 | 33392318 | - |
| intron-intron | ENST00000461813 | ENST00000590597 | KLHL24 | chr3 | 183381430 | + | CEP89 | chr19 | 33392318 | - |
| intron-intron | ENST00000461813 | ENST00000591863 | KLHL24 | chr3 | 183369064 | + | CEP89 | chr19 | 33392318 | - |
| intron-intron | ENST00000461813 | ENST00000591863 | KLHL24 | chr3 | 183381430 | + | CEP89 | chr19 | 33392318 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000242810 | KLHL24 | chr3 | 183369064 | + | ENST00000305768 | CEP89 | chr19 | 33392318 | - | 2223 | 1270 | 350 | 2056 | 568 |
| ENST00000454652 | KLHL24 | chr3 | 183369064 | + | ENST00000305768 | CEP89 | chr19 | 33392318 | - | 2259 | 1306 | 386 | 2092 | 568 |
| ENST00000476808 | KLHL24 | chr3 | 183369064 | + | ENST00000305768 | CEP89 | chr19 | 33392318 | - | 1873 | 920 | 0 | 1706 | 568 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000242810 | ENST00000305768 | KLHL24 | chr3 | 183369064 | + | CEP89 | chr19 | 33392318 | - | 0.001155328 | 0.9988446 |
| ENST00000454652 | ENST00000305768 | KLHL24 | chr3 | 183369064 | + | CEP89 | chr19 | 33392318 | - | 0.001226793 | 0.99877316 |
| ENST00000476808 | ENST00000305768 | KLHL24 | chr3 | 183369064 | + | CEP89 | chr19 | 33392318 | - | 0.001715493 | 0.9982845 |
Top |
Fusion Genomic Features for KLHL24-CEP89 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
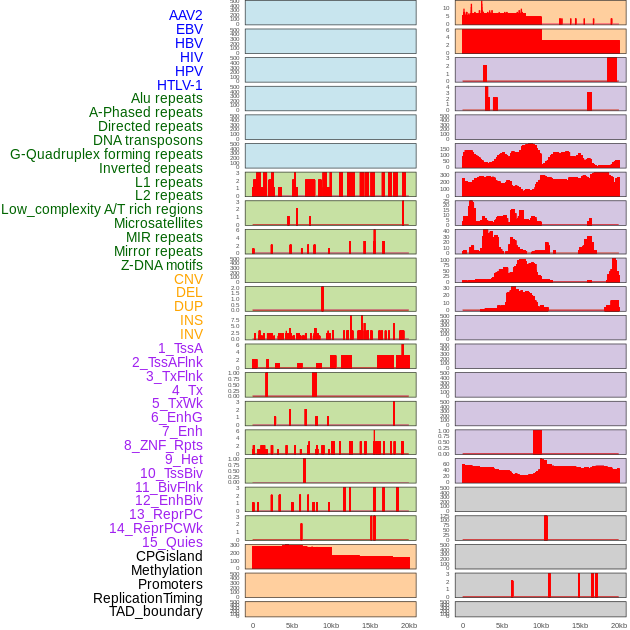 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for KLHL24-CEP89 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:183369064/chr19:33392318) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| KLHL24 | CEP89 |
| FUNCTION: Necessary to maintain the balance between intermediate filament stability and degradation, a process that is essential for skin integrity (PubMed:27889062). As part of the BCR(KLHL24) E3 ubiquitin ligase complex, mediates ubiquitination of KRT14 and controls its levels during keratinocytes differentiation (PubMed:27798626). Specifically reduces kainate receptor-mediated currents in hippocampal neurons, most probably by modulating channel properties (By similarity). {ECO:0000250|UniProtKB:Q56A24, ECO:0000269|PubMed:27798626, ECO:0000269|PubMed:27889062}. | FUNCTION: Required for ciliogenesis. Also plays a role in mitochondrial metabolism where it may modulate complex IV activity. {ECO:0000269|PubMed:23348840, ECO:0000269|PubMed:23575228}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KLHL24 | chr3:183369064 | chr19:33392318 | ENST00000242810 | + | 3 | 8 | 168_270 | 306 | 601.0 | Domain | Note=BACK |
| Hgene | KLHL24 | chr3:183369064 | chr19:33392318 | ENST00000242810 | + | 3 | 8 | 66_133 | 306 | 601.0 | Domain | BTB |
| Hgene | KLHL24 | chr3:183369064 | chr19:33392318 | ENST00000454652 | + | 4 | 9 | 168_270 | 306 | 601.0 | Domain | Note=BACK |
| Hgene | KLHL24 | chr3:183369064 | chr19:33392318 | ENST00000454652 | + | 4 | 9 | 66_133 | 306 | 601.0 | Domain | BTB |
| Hgene | KLHL24 | chr3:183369064 | chr19:33392318 | ENST00000476808 | + | 1 | 5 | 168_270 | 306 | 538.0 | Domain | Note=BACK |
| Hgene | KLHL24 | chr3:183369064 | chr19:33392318 | ENST00000476808 | + | 1 | 5 | 66_133 | 306 | 538.0 | Domain | BTB |
| Tgene | CEP89 | chr3:183369064 | chr19:33392318 | ENST00000590597 | 0 | 9 | 234_333 | 0 | 357.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | CEP89 | chr3:183369064 | chr19:33392318 | ENST00000590597 | 0 | 9 | 369_719 | 0 | 357.0 | Coiled coil | Ontology_term=ECO:0000255 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KLHL24 | chr3:183369064 | chr19:33392318 | ENST00000242810 | + | 3 | 8 | 314_363 | 306 | 601.0 | Repeat | Note=Kelch 1 |
| Hgene | KLHL24 | chr3:183369064 | chr19:33392318 | ENST00000242810 | + | 3 | 8 | 365_407 | 306 | 601.0 | Repeat | Note=Kelch 2 |
| Hgene | KLHL24 | chr3:183369064 | chr19:33392318 | ENST00000242810 | + | 3 | 8 | 408_454 | 306 | 601.0 | Repeat | Note=Kelch 3 |
| Hgene | KLHL24 | chr3:183369064 | chr19:33392318 | ENST00000242810 | + | 3 | 8 | 456_502 | 306 | 601.0 | Repeat | Note=Kelch 4 |
| Hgene | KLHL24 | chr3:183369064 | chr19:33392318 | ENST00000242810 | + | 3 | 8 | 504_544 | 306 | 601.0 | Repeat | Note=Kelch 5 |
| Hgene | KLHL24 | chr3:183369064 | chr19:33392318 | ENST00000242810 | + | 3 | 8 | 546_592 | 306 | 601.0 | Repeat | Note=Kelch 6 |
| Hgene | KLHL24 | chr3:183369064 | chr19:33392318 | ENST00000454652 | + | 4 | 9 | 314_363 | 306 | 601.0 | Repeat | Note=Kelch 1 |
| Hgene | KLHL24 | chr3:183369064 | chr19:33392318 | ENST00000454652 | + | 4 | 9 | 365_407 | 306 | 601.0 | Repeat | Note=Kelch 2 |
| Hgene | KLHL24 | chr3:183369064 | chr19:33392318 | ENST00000454652 | + | 4 | 9 | 408_454 | 306 | 601.0 | Repeat | Note=Kelch 3 |
| Hgene | KLHL24 | chr3:183369064 | chr19:33392318 | ENST00000454652 | + | 4 | 9 | 456_502 | 306 | 601.0 | Repeat | Note=Kelch 4 |
| Hgene | KLHL24 | chr3:183369064 | chr19:33392318 | ENST00000454652 | + | 4 | 9 | 504_544 | 306 | 601.0 | Repeat | Note=Kelch 5 |
| Hgene | KLHL24 | chr3:183369064 | chr19:33392318 | ENST00000454652 | + | 4 | 9 | 546_592 | 306 | 601.0 | Repeat | Note=Kelch 6 |
| Hgene | KLHL24 | chr3:183369064 | chr19:33392318 | ENST00000476808 | + | 1 | 5 | 314_363 | 306 | 538.0 | Repeat | Note=Kelch 1 |
| Hgene | KLHL24 | chr3:183369064 | chr19:33392318 | ENST00000476808 | + | 1 | 5 | 365_407 | 306 | 538.0 | Repeat | Note=Kelch 2 |
| Hgene | KLHL24 | chr3:183369064 | chr19:33392318 | ENST00000476808 | + | 1 | 5 | 408_454 | 306 | 538.0 | Repeat | Note=Kelch 3 |
| Hgene | KLHL24 | chr3:183369064 | chr19:33392318 | ENST00000476808 | + | 1 | 5 | 456_502 | 306 | 538.0 | Repeat | Note=Kelch 4 |
| Hgene | KLHL24 | chr3:183369064 | chr19:33392318 | ENST00000476808 | + | 1 | 5 | 504_544 | 306 | 538.0 | Repeat | Note=Kelch 5 |
| Hgene | KLHL24 | chr3:183369064 | chr19:33392318 | ENST00000476808 | + | 1 | 5 | 546_592 | 306 | 538.0 | Repeat | Note=Kelch 6 |
| Tgene | CEP89 | chr3:183369064 | chr19:33392318 | ENST00000305768 | 13 | 19 | 234_333 | 521 | 784.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | CEP89 | chr3:183369064 | chr19:33392318 | ENST00000305768 | 13 | 19 | 369_719 | 521 | 784.0 | Coiled coil | Ontology_term=ECO:0000255 |
Top |
Fusion Gene Sequence for KLHL24-CEP89 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >42971_42971_1_KLHL24-CEP89_KLHL24_chr3_183369064_ENST00000242810_CEP89_chr19_33392318_ENST00000305768_length(transcript)=2223nt_BP=1270nt GGCCAGGGTTCGCTGACGCTCAGTGTTTTGGCCCGGACGGTCACATGTTTCCTTTGTTGTGAGCTGCGGCAGAGACTGGTGGCTGGAGGA GACGCCGGCGCTGGAGAGTGCGCTGCGCCGCCCGCCGCTGAGGGACCGCGGGGTTAGCCACTGCTGGCTGCTTCCAGTGTTCGCCGAGAG GTACCGGGGGTGACAGCTCCGGGACCGGCCGAAAGGCGAGGAACCGGTGTGGAAATTAAAAGAACACACATATTTTGACTGGGGCTTTGA TCAACCAAATGCTAAAAAGCCACATAAAGAAGATCCCTAATAGTCATTTCTCAACAATTATATAGTCAACTGATGTAACAATGGTACTAA TATTGGGACGCAGACTAAACAGAGAGGATCTTGGGGTGCGTGATTCCCCAGCAACTAAGCGAAAAGTTTTTGAAATGGACCCCAAATCTC TGACAGGTCATGAGTTTTTTGACTTCTCTTCAGGATCATCCCATGCCGAAAACATACTCCAGATATTTAATGAATTTCGTGATAGCCGCT TATTCACAGATGTTATCATTTGTGTGGAAGGAAAAGAATTTCCTTGCCATAGAGCTGTGCTCTCAGCCTGTAGCAGCTACTTCAGAGCTA TGTTTTGTAATGACCACAGGGAAAGCCGAGAAATGTTGGTTGAGATCAATGGTATTTTAGCTGAAGCTATGGAATGTTTTTTGCAGTATG TTTATACTGGAAAGGTGAAGATCACTACAGAGAATGTACAGTATCTCTTTGAGACATCAAGCCTCTTTCAGATTAGTGTTCTCCGTGATG CATGTGCCAAGTTCTTGGAGGAGCAACTTGATCCTTGTAATTGCTTAGGAATCCAGCGCTTTGCTGATACCCATTCACTCAAAACACTCT TCACAAAATGCAAAAATTTTGCGTTACAGACTTTTGAGGATGTATCCCAGCACGAAGAATTTCTTGAGCTTGACAAAGATGAACTTATTG ATTATATTTGTAGTGATGAACTTGTTATTGGTAAAGAGGAGATGGTTTTTGAAGCCGTCATGCGTTGGGTCTATCGTGCCGTTGATCTGA GAAGACCACTGTTACACGAGCTCCTGACACATGTGAGACTCCCTCTGTTGCATCCCAACTACTTTGTTCAAACAGTTGAAGTGGACCAAT TGATCCAGAATTCTCCTGAGTGTTATCAGTTGTTGCATGAAGCAAGACGGTACCACATACTTGGGAATGAAATGATGTCCCCAAGGACTA GGCCACGCAGCCAATTACAGAAGGAAGAAGAGAAAGAAAGGGCTGAGATGGAGGAGTTGATGGAGAAGCTGACAGTCCTGCAAGCGCAGA AGAAGAGCCTGCTGTTAGAGAAGAACAGTTTGACAGAGCAAAACAAAGCACTGGAAGCCGAACTTGAACGAGCACAGAAAATCAATAGGA AATCTCAAAAGAAAATTGAGGTCCTCAAAAAGCAGGTGGAAAAAGCCATGGGGAACGAAATGTCTGCTCATCAGTACCTGGCAAACCTTG TTGGCCTGGCAGAAAACATAACCCAGGAACGTGACAGTCTTATGTGTTTGGCAAAATGTTTAGAAAGTGAGAAGGATGGAGTGCTTAATA AAGTCATAAAAAGCAACATTCGCCTGGGAAAGTTAGAGGAAAAAGTCAAGGGCTACAAGAAGCAGGCAGCACTGAAGCTGGGGGACATCA GTCACCGTCTGCTGGAGCAGCAGGAGGACTTCGCCGGCAAGACAGCCCAGTACCGGCAGGAGATGCGGCACCTGCACCAGGTGCTGAAGG ACAAGCAGGAGGTGCTGGACCAGGCGCTGCAGCAGAACAGAGAAATGGAAGGTGAACTTGAAGTTATTTGGGAATCTACCTTCAGGGAAA ACCGAAGAATCCGAGAACTTCTCCAGGACACACTCACGAGGACAGGCGTGCAGGACAACCCCAGAGCTCTGGTTGCCCCCAGCCTCAATG GCGTCTCTCAGGCAGACCTGCTGGACGGCTGCGATGTCTGCTCCTATGACCTGAAGTCTCATGCCCCCACCTGCTAGAATCTGCGGGAGC CCGTGGTGTAGCCTCCTCTGGCCTGAAAGGTCTGCCTCACACAGGCCTTCCCTGCAGGGCAGCCCATGGTGGCAGCCGGAGGTGCGTAAG >42971_42971_1_KLHL24-CEP89_KLHL24_chr3_183369064_ENST00000242810_CEP89_chr19_33392318_ENST00000305768_length(amino acids)=568AA_BP=306 MVLILGRRLNREDLGVRDSPATKRKVFEMDPKSLTGHEFFDFSSGSSHAENILQIFNEFRDSRLFTDVIICVEGKEFPCHRAVLSACSSY FRAMFCNDHRESREMLVEINGILAEAMECFLQYVYTGKVKITTENVQYLFETSSLFQISVLRDACAKFLEEQLDPCNCLGIQRFADTHSL KTLFTKCKNFALQTFEDVSQHEEFLELDKDELIDYICSDELVIGKEEMVFEAVMRWVYRAVDLRRPLLHELLTHVRLPLLHPNYFVQTVE VDQLIQNSPECYQLLHEARRYHILGNEMMSPRTRPRSQLQKEEEKERAEMEELMEKLTVLQAQKKSLLLEKNSLTEQNKALEAELERAQK INRKSQKKIEVLKKQVEKAMGNEMSAHQYLANLVGLAENITQERDSLMCLAKCLESEKDGVLNKVIKSNIRLGKLEEKVKGYKKQAALKL GDISHRLLEQQEDFAGKTAQYRQEMRHLHQVLKDKQEVLDQALQQNREMEGELEVIWESTFRENRRIRELLQDTLTRTGVQDNPRALVAP -------------------------------------------------------------- >42971_42971_2_KLHL24-CEP89_KLHL24_chr3_183369064_ENST00000454652_CEP89_chr19_33392318_ENST00000305768_length(transcript)=2259nt_BP=1306nt GTTGTGAGCTGCGGCAGAGACTGGTGGCTGGAGGAGACGCCGGCGCTGGAGAGTGCGCTGCGCCGCCCGCCGCTGAGGGACCGCGGGGTT AGCCACTGCTGGCTGCTTCCAGTGTTCGCCGAGAGGTACCGGGGGTGACAGCTCCGGGACCGGCCGAAAGGCGAGGAACCGGAAGCCCGG GGGAGTGCGACCGCCTCAAATCCAGCCTTCTTGTTTGGTGGGACGACCGTCGGTTCTGTCCGGGAGGAATGACGGGGTCAAGGTGTGGAA ATTAAAAGAACACACATATTTTGACTGGGGCTTTGATCAACCAAATGCTAAAAAGCCACATAAAGAAGATCCCTAATAGTCATTTCTCAA CAATTATATAGTCAACTGATGTAACAATGGTACTAATATTGGGACGCAGACTAAACAGAGAGGATCTTGGGGTGCGTGATTCCCCAGCAA CTAAGCGAAAAGTTTTTGAAATGGACCCCAAATCTCTGACAGGTCATGAGTTTTTTGACTTCTCTTCAGGATCATCCCATGCCGAAAACA TACTCCAGATATTTAATGAATTTCGTGATAGCCGCTTATTCACAGATGTTATCATTTGTGTGGAAGGAAAAGAATTTCCTTGCCATAGAG CTGTGCTCTCAGCCTGTAGCAGCTACTTCAGAGCTATGTTTTGTAATGACCACAGGGAAAGCCGAGAAATGTTGGTTGAGATCAATGGTA TTTTAGCTGAAGCTATGGAATGTTTTTTGCAGTATGTTTATACTGGAAAGGTGAAGATCACTACAGAGAATGTACAGTATCTCTTTGAGA CATCAAGCCTCTTTCAGATTAGTGTTCTCCGTGATGCATGTGCCAAGTTCTTGGAGGAGCAACTTGATCCTTGTAATTGCTTAGGAATCC AGCGCTTTGCTGATACCCATTCACTCAAAACACTCTTCACAAAATGCAAAAATTTTGCGTTACAGACTTTTGAGGATGTATCCCAGCACG AAGAATTTCTTGAGCTTGACAAAGATGAACTTATTGATTATATTTGTAGTGATGAACTTGTTATTGGTAAAGAGGAGATGGTTTTTGAAG CCGTCATGCGTTGGGTCTATCGTGCCGTTGATCTGAGAAGACCACTGTTACACGAGCTCCTGACACATGTGAGACTCCCTCTGTTGCATC CCAACTACTTTGTTCAAACAGTTGAAGTGGACCAATTGATCCAGAATTCTCCTGAGTGTTATCAGTTGTTGCATGAAGCAAGACGGTACC ACATACTTGGGAATGAAATGATGTCCCCAAGGACTAGGCCACGCAGCCAATTACAGAAGGAAGAAGAGAAAGAAAGGGCTGAGATGGAGG AGTTGATGGAGAAGCTGACAGTCCTGCAAGCGCAGAAGAAGAGCCTGCTGTTAGAGAAGAACAGTTTGACAGAGCAAAACAAAGCACTGG AAGCCGAACTTGAACGAGCACAGAAAATCAATAGGAAATCTCAAAAGAAAATTGAGGTCCTCAAAAAGCAGGTGGAAAAAGCCATGGGGA ACGAAATGTCTGCTCATCAGTACCTGGCAAACCTTGTTGGCCTGGCAGAAAACATAACCCAGGAACGTGACAGTCTTATGTGTTTGGCAA AATGTTTAGAAAGTGAGAAGGATGGAGTGCTTAATAAAGTCATAAAAAGCAACATTCGCCTGGGAAAGTTAGAGGAAAAAGTCAAGGGCT ACAAGAAGCAGGCAGCACTGAAGCTGGGGGACATCAGTCACCGTCTGCTGGAGCAGCAGGAGGACTTCGCCGGCAAGACAGCCCAGTACC GGCAGGAGATGCGGCACCTGCACCAGGTGCTGAAGGACAAGCAGGAGGTGCTGGACCAGGCGCTGCAGCAGAACAGAGAAATGGAAGGTG AACTTGAAGTTATTTGGGAATCTACCTTCAGGGAAAACCGAAGAATCCGAGAACTTCTCCAGGACACACTCACGAGGACAGGCGTGCAGG ACAACCCCAGAGCTCTGGTTGCCCCCAGCCTCAATGGCGTCTCTCAGGCAGACCTGCTGGACGGCTGCGATGTCTGCTCCTATGACCTGA AGTCTCATGCCCCCACCTGCTAGAATCTGCGGGAGCCCGTGGTGTAGCCTCCTCTGGCCTGAAAGGTCTGCCTCACACAGGCCTTCCCTG CAGGGCAGCCCATGGTGGCAGCCGGAGGTGCGTAAGCCCACTGAACGCTGCAGGGCAGGGAAGAAATAAATGGTCTCATAGTTTAATCCA >42971_42971_2_KLHL24-CEP89_KLHL24_chr3_183369064_ENST00000454652_CEP89_chr19_33392318_ENST00000305768_length(amino acids)=568AA_BP=306 MVLILGRRLNREDLGVRDSPATKRKVFEMDPKSLTGHEFFDFSSGSSHAENILQIFNEFRDSRLFTDVIICVEGKEFPCHRAVLSACSSY FRAMFCNDHRESREMLVEINGILAEAMECFLQYVYTGKVKITTENVQYLFETSSLFQISVLRDACAKFLEEQLDPCNCLGIQRFADTHSL KTLFTKCKNFALQTFEDVSQHEEFLELDKDELIDYICSDELVIGKEEMVFEAVMRWVYRAVDLRRPLLHELLTHVRLPLLHPNYFVQTVE VDQLIQNSPECYQLLHEARRYHILGNEMMSPRTRPRSQLQKEEEKERAEMEELMEKLTVLQAQKKSLLLEKNSLTEQNKALEAELERAQK INRKSQKKIEVLKKQVEKAMGNEMSAHQYLANLVGLAENITQERDSLMCLAKCLESEKDGVLNKVIKSNIRLGKLEEKVKGYKKQAALKL GDISHRLLEQQEDFAGKTAQYRQEMRHLHQVLKDKQEVLDQALQQNREMEGELEVIWESTFRENRRIRELLQDTLTRTGVQDNPRALVAP -------------------------------------------------------------- >42971_42971_3_KLHL24-CEP89_KLHL24_chr3_183369064_ENST00000476808_CEP89_chr19_33392318_ENST00000305768_length(transcript)=1873nt_BP=920nt ATGGTACTAATATTGGGACGCAGACTAAACAGAGAGGATCTTGGGGTGCGTGATTCCCCAGCAACTAAGCGAAAAGTTTTTGAAATGGAC CCCAAATCTCTGACAGGTCATGAGTTTTTTGACTTCTCTTCAGGATCATCCCATGCCGAAAACATACTCCAGATATTTAATGAATTTCGT GATAGCCGCTTATTCACAGATGTTATCATTTGTGTGGAAGGAAAAGAATTTCCTTGCCATAGAGCTGTGCTCTCAGCCTGTAGCAGCTAC TTCAGAGCTATGTTTTGTAATGACCACAGGGAAAGCCGAGAAATGTTGGTTGAGATCAATGGTATTTTAGCTGAAGCTATGGAATGTTTT TTGCAGTATGTTTATACTGGAAAGGTGAAGATCACTACAGAGAATGTACAGTATCTCTTTGAGACATCAAGCCTCTTTCAGATTAGTGTT CTCCGTGATGCATGTGCCAAGTTCTTGGAGGAGCAACTTGATCCTTGTAATTGCTTAGGAATCCAGCGCTTTGCTGATACCCATTCACTC AAAACACTCTTCACAAAATGCAAAAATTTTGCGTTACAGACTTTTGAGGATGTATCCCAGCACGAAGAATTTCTTGAGCTTGACAAAGAT GAACTTATTGATTATATTTGTAGTGATGAACTTGTTATTGGTAAAGAGGAGATGGTTTTTGAAGCCGTCATGCGTTGGGTCTATCGTGCC GTTGATCTGAGAAGACCACTGTTACACGAGCTCCTGACACATGTGAGACTCCCTCTGTTGCATCCCAACTACTTTGTTCAAACAGTTGAA GTGGACCAATTGATCCAGAATTCTCCTGAGTGTTATCAGTTGTTGCATGAAGCAAGACGGTACCACATACTTGGGAATGAAATGATGTCC CCAAGGACTAGGCCACGCAGCCAATTACAGAAGGAAGAAGAGAAAGAAAGGGCTGAGATGGAGGAGTTGATGGAGAAGCTGACAGTCCTG CAAGCGCAGAAGAAGAGCCTGCTGTTAGAGAAGAACAGTTTGACAGAGCAAAACAAAGCACTGGAAGCCGAACTTGAACGAGCACAGAAA ATCAATAGGAAATCTCAAAAGAAAATTGAGGTCCTCAAAAAGCAGGTGGAAAAAGCCATGGGGAACGAAATGTCTGCTCATCAGTACCTG GCAAACCTTGTTGGCCTGGCAGAAAACATAACCCAGGAACGTGACAGTCTTATGTGTTTGGCAAAATGTTTAGAAAGTGAGAAGGATGGA GTGCTTAATAAAGTCATAAAAAGCAACATTCGCCTGGGAAAGTTAGAGGAAAAAGTCAAGGGCTACAAGAAGCAGGCAGCACTGAAGCTG GGGGACATCAGTCACCGTCTGCTGGAGCAGCAGGAGGACTTCGCCGGCAAGACAGCCCAGTACCGGCAGGAGATGCGGCACCTGCACCAG GTGCTGAAGGACAAGCAGGAGGTGCTGGACCAGGCGCTGCAGCAGAACAGAGAAATGGAAGGTGAACTTGAAGTTATTTGGGAATCTACC TTCAGGGAAAACCGAAGAATCCGAGAACTTCTCCAGGACACACTCACGAGGACAGGCGTGCAGGACAACCCCAGAGCTCTGGTTGCCCCC AGCCTCAATGGCGTCTCTCAGGCAGACCTGCTGGACGGCTGCGATGTCTGCTCCTATGACCTGAAGTCTCATGCCCCCACCTGCTAGAAT CTGCGGGAGCCCGTGGTGTAGCCTCCTCTGGCCTGAAAGGTCTGCCTCACACAGGCCTTCCCTGCAGGGCAGCCCATGGTGGCAGCCGGA >42971_42971_3_KLHL24-CEP89_KLHL24_chr3_183369064_ENST00000476808_CEP89_chr19_33392318_ENST00000305768_length(amino acids)=568AA_BP=306 MVLILGRRLNREDLGVRDSPATKRKVFEMDPKSLTGHEFFDFSSGSSHAENILQIFNEFRDSRLFTDVIICVEGKEFPCHRAVLSACSSY FRAMFCNDHRESREMLVEINGILAEAMECFLQYVYTGKVKITTENVQYLFETSSLFQISVLRDACAKFLEEQLDPCNCLGIQRFADTHSL KTLFTKCKNFALQTFEDVSQHEEFLELDKDELIDYICSDELVIGKEEMVFEAVMRWVYRAVDLRRPLLHELLTHVRLPLLHPNYFVQTVE VDQLIQNSPECYQLLHEARRYHILGNEMMSPRTRPRSQLQKEEEKERAEMEELMEKLTVLQAQKKSLLLEKNSLTEQNKALEAELERAQK INRKSQKKIEVLKKQVEKAMGNEMSAHQYLANLVGLAENITQERDSLMCLAKCLESEKDGVLNKVIKSNIRLGKLEEKVKGYKKQAALKL GDISHRLLEQQEDFAGKTAQYRQEMRHLHQVLKDKQEVLDQALQQNREMEGELEVIWESTFRENRRIRELLQDTLTRTGVQDNPRALVAP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for KLHL24-CEP89 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for KLHL24-CEP89 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for KLHL24-CEP89 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
