
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:KLK3-CDC42EP4 (FusionGDB2 ID:43076) |
Fusion Gene Summary for KLK3-CDC42EP4 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: KLK3-CDC42EP4 | Fusion gene ID: 43076 | Hgene | Tgene | Gene symbol | KLK3 | CDC42EP4 | Gene ID | 3818 | 23580 |
| Gene name | kallikrein B1 | CDC42 effector protein 4 | |
| Synonyms | KLK3|PKK|PKKD|PPK | BORG4|CEP4|KAIA1777 | |
| Cytomap | 4q35.2 | 17q25.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | plasma kallikreinkallikrein B, plasma (Fletcher factor) 1kininogeninplasma prekallikrein | cdc42 effector protein 4CDC42 effector protein (Rho GTPase binding) 4binder of Rho GTPases 4 | |
| Modification date | 20200320 | 20200313 | |
| UniProtAcc | P07288 | Q9H3Q1 | |
| Ensembl transtripts involved in fusion gene | ENST00000326003, ENST00000360617, ENST00000593997, ENST00000595952, ENST00000597483, | ENST00000581014, ENST00000582303, ENST00000335793, ENST00000439510, | |
| Fusion gene scores | * DoF score | 24 X 31 X 2=1488 | 7 X 3 X 6=126 |
| # samples | 31 | 10 | |
| ** MAII score | log2(31/1488*10)=-2.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(10/126*10)=-0.333423733725192 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: KLK3 [Title/Abstract] AND CDC42EP4 [Title/Abstract] AND fusion [Title/Abstract] | ||
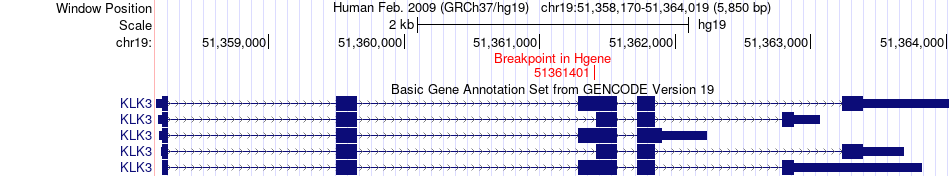
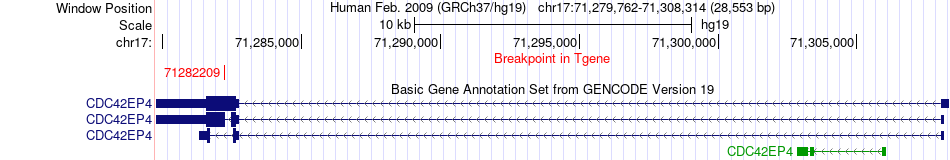
| Most frequent breakpoint | KLK3(51361401)-CDC42EP4(71282209), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | KLK3-CDC42EP4 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | KLK3 | GO:0031639 | plasminogen activation | 89876 |
| Hgene | KLK3 | GO:0051919 | positive regulation of fibrinolysis | 89876 |
| Tgene | CDC42EP4 | GO:0008360 | regulation of cell shape | 11035016 |
| Tgene | CDC42EP4 | GO:0031274 | positive regulation of pseudopodium assembly | 11035016 |
 Fusion gene breakpoints across KLK3 (5'-gene) Fusion gene breakpoints across KLK3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across CDC42EP4 (3'-gene) Fusion gene breakpoints across CDC42EP4 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChiTaRS5.0 | N/A | DA874433 | KLK3 | chr19 | 51361401 | + | CDC42EP4 | chr17 | 71282209 | - |
Top |
Fusion Gene ORF analysis for KLK3-CDC42EP4 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000326003 | ENST00000581014 | KLK3 | chr19 | 51361401 | + | CDC42EP4 | chr17 | 71282209 | - |
| 5CDS-intron | ENST00000326003 | ENST00000582303 | KLK3 | chr19 | 51361401 | + | CDC42EP4 | chr17 | 71282209 | - |
| 5CDS-intron | ENST00000360617 | ENST00000581014 | KLK3 | chr19 | 51361401 | + | CDC42EP4 | chr17 | 71282209 | - |
| 5CDS-intron | ENST00000360617 | ENST00000582303 | KLK3 | chr19 | 51361401 | + | CDC42EP4 | chr17 | 71282209 | - |
| 5CDS-intron | ENST00000593997 | ENST00000581014 | KLK3 | chr19 | 51361401 | + | CDC42EP4 | chr17 | 71282209 | - |
| 5CDS-intron | ENST00000593997 | ENST00000582303 | KLK3 | chr19 | 51361401 | + | CDC42EP4 | chr17 | 71282209 | - |
| Frame-shift | ENST00000326003 | ENST00000335793 | KLK3 | chr19 | 51361401 | + | CDC42EP4 | chr17 | 71282209 | - |
| Frame-shift | ENST00000360617 | ENST00000335793 | KLK3 | chr19 | 51361401 | + | CDC42EP4 | chr17 | 71282209 | - |
| Frame-shift | ENST00000593997 | ENST00000335793 | KLK3 | chr19 | 51361401 | + | CDC42EP4 | chr17 | 71282209 | - |
| In-frame | ENST00000326003 | ENST00000439510 | KLK3 | chr19 | 51361401 | + | CDC42EP4 | chr17 | 71282209 | - |
| In-frame | ENST00000360617 | ENST00000439510 | KLK3 | chr19 | 51361401 | + | CDC42EP4 | chr17 | 71282209 | - |
| In-frame | ENST00000593997 | ENST00000439510 | KLK3 | chr19 | 51361401 | + | CDC42EP4 | chr17 | 71282209 | - |
| intron-3CDS | ENST00000595952 | ENST00000335793 | KLK3 | chr19 | 51361401 | + | CDC42EP4 | chr17 | 71282209 | - |
| intron-3CDS | ENST00000595952 | ENST00000439510 | KLK3 | chr19 | 51361401 | + | CDC42EP4 | chr17 | 71282209 | - |
| intron-3CDS | ENST00000597483 | ENST00000335793 | KLK3 | chr19 | 51361401 | + | CDC42EP4 | chr17 | 71282209 | - |
| intron-3CDS | ENST00000597483 | ENST00000439510 | KLK3 | chr19 | 51361401 | + | CDC42EP4 | chr17 | 71282209 | - |
| intron-intron | ENST00000595952 | ENST00000581014 | KLK3 | chr19 | 51361401 | + | CDC42EP4 | chr17 | 71282209 | - |
| intron-intron | ENST00000595952 | ENST00000582303 | KLK3 | chr19 | 51361401 | + | CDC42EP4 | chr17 | 71282209 | - |
| intron-intron | ENST00000597483 | ENST00000581014 | KLK3 | chr19 | 51361401 | + | CDC42EP4 | chr17 | 71282209 | - |
| intron-intron | ENST00000597483 | ENST00000582303 | KLK3 | chr19 | 51361401 | + | CDC42EP4 | chr17 | 71282209 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000326003 | KLK3 | chr19 | 51361401 | + | ENST00000439510 | CDC42EP4 | chr17 | 71282209 | - | 2863 | 417 | 17 | 1057 | 346 |
| ENST00000593997 | KLK3 | chr19 | 51361401 | + | ENST00000439510 | CDC42EP4 | chr17 | 71282209 | - | 2843 | 397 | 21 | 1037 | 338 |
| ENST00000360617 | KLK3 | chr19 | 51361401 | + | ENST00000439510 | CDC42EP4 | chr17 | 71282209 | - | 2822 | 376 | 0 | 1016 | 338 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000326003 | ENST00000439510 | KLK3 | chr19 | 51361401 | + | CDC42EP4 | chr17 | 71282209 | - | 0.017097333 | 0.98290265 |
| ENST00000593997 | ENST00000439510 | KLK3 | chr19 | 51361401 | + | CDC42EP4 | chr17 | 71282209 | - | 0.01705637 | 0.98294365 |
| ENST00000360617 | ENST00000439510 | KLK3 | chr19 | 51361401 | + | CDC42EP4 | chr17 | 71282209 | - | 0.016405694 | 0.98359436 |
Top |
Fusion Genomic Features for KLK3-CDC42EP4 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
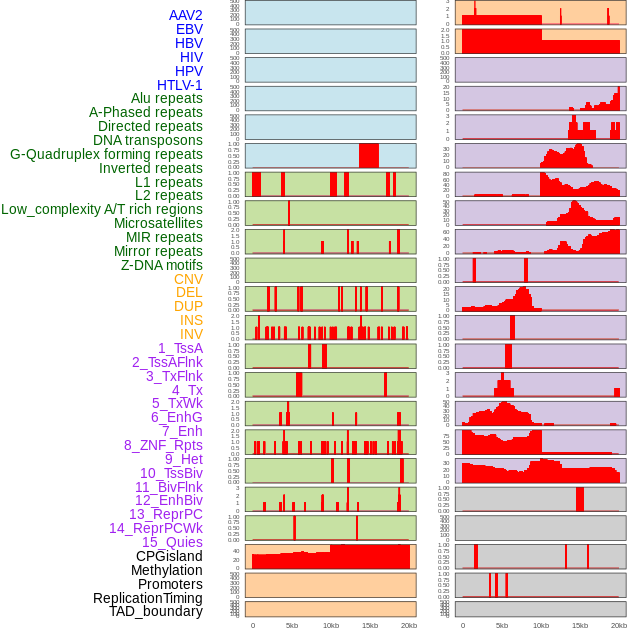
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for KLK3-CDC42EP4 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:51361401/chr17:71282209) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| KLK3 | CDC42EP4 |
| FUNCTION: Hydrolyzes semenogelin-1 thus leading to the liquefaction of the seminal coagulum. | FUNCTION: Probably involved in the organization of the actin cytoskeleton. May act downstream of CDC42 to induce actin filament assembly leading to cell shape changes. Induces pseudopodia formation, when overexpressed in fibroblasts. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | CDC42EP4 | chr19:51361401 | chr17:71282209 | ENST00000335793 | 0 | 2 | 70_74 | 0 | 357.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | CDC42EP4 | chr19:51361401 | chr17:71282209 | ENST00000335793 | 0 | 2 | 27_41 | 0 | 357.0 | Domain | CRIB |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KLK3 | chr19:51361401 | chr17:71282209 | ENST00000326003 | + | 1 | 5 | 25_258 | 0 | 262.0 | Domain | Peptidase S1 |
| Hgene | KLK3 | chr19:51361401 | chr17:71282209 | ENST00000360617 | + | 1 | 5 | 25_258 | 0 | 239.0 | Domain | Peptidase S1 |
| Hgene | KLK3 | chr19:51361401 | chr17:71282209 | ENST00000593997 | + | 1 | 4 | 25_258 | 0 | 228.0 | Domain | Peptidase S1 |
| Hgene | KLK3 | chr19:51361401 | chr17:71282209 | ENST00000595952 | + | 1 | 5 | 25_258 | 0 | 219.0 | Domain | Peptidase S1 |
Top |
Fusion Gene Sequence for KLK3-CDC42EP4 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >43076_43076_1_KLK3-CDC42EP4_KLK3_chr19_51361401_ENST00000326003_CDC42EP4_chr17_71282209_ENST00000439510_length(transcript)=2863nt_BP=417nt AGCCCCAAGCTTACCACCTGCACCCGGAGAGCTGTGTCACCATGTGGGTCCCGGTTGTCTTCCTCACCCTGTCCGTGACGTGGATTGGTG CTGCACCCCTCATCCTGTCTCGGATTGTGGGAGGCTGGGAGTGCGAGAAGCATTCCCAACCCTGGCAGGTGCTTGTGGCCTCTCGTGGCA GGGCAGTCTGCGGCGGTGTTCTGGTGCACCCCCAGTGGGTCCTCACAGCTGCCCACTGCATCAGGAATCGATTCCTCAGGCCAGGTGATG ACTCCAGCCACGACCTCATGCTGCTCCGCCTGTCAGAGCCTGCCGAGCTCACGGATGCTGTGAAGGTCATGGACCTGCCCACCCAGGAGC CAGCACTGGGGACCACCTGCTACGCCTCAGGCTGGGGCAGCATTGAACCAGAGGAGTTGAAGAAGGCCAATGACGGGGAGGGCGGCGATG AGGAGGCGGGCACGGAGGAGGCAGTGCCCCGTCGGAATGGGGCCGCGGGTCCACATTCCCCTGACCCCCTCCTCGATGAGCAGGCCTTTG GGGATCTGACAGATCTGCCTGTCGTGCCCAAGGCCACGTACGGGCTGAAGCATGCGGAGTCCATCATGTCCTTCCACATCGACCTGGGGC CCTCCATGCTGGGTGACGTCCTCAGCATCATGGACAAGGAGGAGTGGGACCCCGAGGAGGGGGAGGGTGGTTACCATGGCGATGAGGGCG CCGCTGGCACCATCACCCAGGCTCCCCCGTACGCCGTGGCGGCCCCTCCCCTGGCAAGGCAGGAAGGCAAGGCTGGCCCAGACTTGCCCT CCCTCCCCTCCCATGCTCTGGAGGATGAGGGGTGGGCAGCAGCGGCCCCCAGCCCCGGCTCAGCCCGCAGCATGGGCAGCCACACCACAC GGGACAGCAGCTCCCTCTCCAGCTGCACCTCAGGCATCCTGGAGGAGCGCAGCCCTGCCTTCCGGGGGCCGGACAGGGCCCGGGCTGCTG TCTCAAGACAGCCAGACAAGGAGTTCTCCTTCATGGATGAGGAGGAGGAGGATGAAATCCGTGTGTGAGGCGGACAGTGGGTGGCCACCG GGAGCTCTTGGCTGCATCTTCTCCCTGCCCCCACCCCACTATGACCTTTGACCCTACGGCGCAGGGGCAGCCAGGACCCTTGATTCAGAC CATGGACCCTGGACCTTGTAGATGAGGGACACTGGCCTGGCCCTCGGGTCTTCGGAGGACGTAGGGGGCTGGCATGGGTGCCGACTGGCT GCCTGACTTCATCATGCTCCCTGCACTTAGGCTGCGTGGGACAAGGGCTGTGTTGTCACAGCAGGAATAGGTTTTCCTCTGTTGGCCTCC CTTTCCTCCACCCTGGCCTCAAATGGATGCCAGATGCCAACCCCAGTTCTGGCCACGTACAGCCAGCGGGTCAGCCCAGAGGCAGCCTCA GCTCCAGGGCTAAGGACTCTCGGCTCCCATTTTCTCTGCTGGCGTTTCTGCTGTGCCCAGCAGTGGCTGCTGGGGAAGCAGCTGCAGCAG GAGGGAGACGGTCTTGCCTCTCAGCCCCTCCCTGCCCCACCCCAGCTCCTGCCCTGGAAATCTGGAGCCCCTTGGAGCTGAGCTGGACGG GGGGCCAGCTGCGAGCATGTGCACTAAACGCAGCCCTTTCCAGGGGAAGAGAACAGGATGGAGAATGGAAGGAAAGCCCCCCAGGCTTCG TGAATTGCAAGAAGGGACCCTTCCAGGATGACACTAGGAACAGGGCTAGGGCACTCGCTCAGTCCCTAGGGGCTTGTTTGTTCTTTATTA TTGTGTTTAAATCCTTATAGAGCAATATCAGGATGGTGTTAATAGGTCTGCCTCAGAATGAGAATCAATCCTTTTAGAAAACCTTTATAC TAAGCCTCCTCTTCAAAATTCACAGTGGCGATTAGCGGACTGGAGTCTGGTGGCGATTAGCGGACTGGAGTCTGGGGACATCCGTGGCAA AGACACCAGCTCAACTTTAGTGCTTCCCAACTTTATTTAGAATGACATGGGGTGGGTGTCTGGTGTGTGTGTTTTCCCTACGCACCTCCC ATAGCTATTAACAACTGAGGAAGGCCAGTGCAGAATATTTTTGGAGAACGATTTTTTTTTTAAATAATATATCATTCCTATGGGGGGAAA GCCTTTTTTTTCTTTTTGGCTGAGTTATTCCCTCCCTCCCCTCAATACCCTCAGTACTGACTACTTCCCTTTCTTTTCTCAGGCCTCCCC CCACCGACTTTTGAGGCCAGGGTTGGCCAGATTTAGCAAAACCAAAACAGAGTGCTGAGTTAAACGCAAATTTCAGGTAAACAAAAGATA ATTTTCTAGCATTAATATGCCCCACGCAATATTTGGAACACTTATGTGAAAAATGATTTGTTTTTCTGAAATTCACGTTTCTCTCTGAGT CCTGTAACTGTCCCCGAGGGGATTGAGCAGAAGCTCGGGTATGAGCCCTGAGGTTGACTGCCGGTTATTTTTCTGTCCTGGGAACAGCCT GACCCACCTCCCTGTCTCCATGTAGCCAGTGAGGGGAGGGGGAGACACAGAACCAACCACAGCCAGGGGCGTCCCCATGGCGACTGTGGC CCGGCCCCTCCTCTCTTGCCTGACTCTCCTCTCTTGCCTGACTCTAGACACTAACTTAGTTCCAGGTTCGGTGCCCTGTTGGTGCTCCTG TTTCCAATAGCTTAGGTCCCATGGTGGGGGAGGAACCTCAGGGGCTATGCAGCCCCCGCCAGCTGCCCTCGAATCCCGTCCAGGCCAATT >43076_43076_1_KLK3-CDC42EP4_KLK3_chr19_51361401_ENST00000326003_CDC42EP4_chr17_71282209_ENST00000439510_length(amino acids)=346AA_BP=134 MHPESCVTMWVPVVFLTLSVTWIGAAPLILSRIVGGWECEKHSQPWQVLVASRGRAVCGGVLVHPQWVLTAAHCIRNRFLRPGDDSSHDL MLLRLSEPAELTDAVKVMDLPTQEPALGTTCYASGWGSIEPEELKKANDGEGGDEEAGTEEAVPRRNGAAGPHSPDPLLDEQAFGDLTDL PVVPKATYGLKHAESIMSFHIDLGPSMLGDVLSIMDKEEWDPEEGEGGYHGDEGAAGTITQAPPYAVAAPPLARQEGKAGPDLPSLPSHA -------------------------------------------------------------- >43076_43076_2_KLK3-CDC42EP4_KLK3_chr19_51361401_ENST00000360617_CDC42EP4_chr17_71282209_ENST00000439510_length(transcript)=2822nt_BP=376nt ATGTGGGTCCCGGTTGTCTTCCTCACCCTGTCCGTGACGTGGATTGGTGCTGCACCCCTCATCCTGTCTCGGATTGTGGGAGGCTGGGAG TGCGAGAAGCATTCCCAACCCTGGCAGGTGCTTGTGGCCTCTCGTGGCAGGGCAGTCTGCGGCGGTGTTCTGGTGCACCCCCAGTGGGTC CTCACAGCTGCCCACTGCATCAGGAATCGATTCCTCAGGCCAGGTGATGACTCCAGCCACGACCTCATGCTGCTCCGCCTGTCAGAGCCT GCCGAGCTCACGGATGCTGTGAAGGTCATGGACCTGCCCACCCAGGAGCCAGCACTGGGGACCACCTGCTACGCCTCAGGCTGGGGCAGC ATTGAACCAGAGGAGTTGAAGAAGGCCAATGACGGGGAGGGCGGCGATGAGGAGGCGGGCACGGAGGAGGCAGTGCCCCGTCGGAATGGG GCCGCGGGTCCACATTCCCCTGACCCCCTCCTCGATGAGCAGGCCTTTGGGGATCTGACAGATCTGCCTGTCGTGCCCAAGGCCACGTAC GGGCTGAAGCATGCGGAGTCCATCATGTCCTTCCACATCGACCTGGGGCCCTCCATGCTGGGTGACGTCCTCAGCATCATGGACAAGGAG GAGTGGGACCCCGAGGAGGGGGAGGGTGGTTACCATGGCGATGAGGGCGCCGCTGGCACCATCACCCAGGCTCCCCCGTACGCCGTGGCG GCCCCTCCCCTGGCAAGGCAGGAAGGCAAGGCTGGCCCAGACTTGCCCTCCCTCCCCTCCCATGCTCTGGAGGATGAGGGGTGGGCAGCA GCGGCCCCCAGCCCCGGCTCAGCCCGCAGCATGGGCAGCCACACCACACGGGACAGCAGCTCCCTCTCCAGCTGCACCTCAGGCATCCTG GAGGAGCGCAGCCCTGCCTTCCGGGGGCCGGACAGGGCCCGGGCTGCTGTCTCAAGACAGCCAGACAAGGAGTTCTCCTTCATGGATGAG GAGGAGGAGGATGAAATCCGTGTGTGAGGCGGACAGTGGGTGGCCACCGGGAGCTCTTGGCTGCATCTTCTCCCTGCCCCCACCCCACTA TGACCTTTGACCCTACGGCGCAGGGGCAGCCAGGACCCTTGATTCAGACCATGGACCCTGGACCTTGTAGATGAGGGACACTGGCCTGGC CCTCGGGTCTTCGGAGGACGTAGGGGGCTGGCATGGGTGCCGACTGGCTGCCTGACTTCATCATGCTCCCTGCACTTAGGCTGCGTGGGA CAAGGGCTGTGTTGTCACAGCAGGAATAGGTTTTCCTCTGTTGGCCTCCCTTTCCTCCACCCTGGCCTCAAATGGATGCCAGATGCCAAC CCCAGTTCTGGCCACGTACAGCCAGCGGGTCAGCCCAGAGGCAGCCTCAGCTCCAGGGCTAAGGACTCTCGGCTCCCATTTTCTCTGCTG GCGTTTCTGCTGTGCCCAGCAGTGGCTGCTGGGGAAGCAGCTGCAGCAGGAGGGAGACGGTCTTGCCTCTCAGCCCCTCCCTGCCCCACC CCAGCTCCTGCCCTGGAAATCTGGAGCCCCTTGGAGCTGAGCTGGACGGGGGGCCAGCTGCGAGCATGTGCACTAAACGCAGCCCTTTCC AGGGGAAGAGAACAGGATGGAGAATGGAAGGAAAGCCCCCCAGGCTTCGTGAATTGCAAGAAGGGACCCTTCCAGGATGACACTAGGAAC AGGGCTAGGGCACTCGCTCAGTCCCTAGGGGCTTGTTTGTTCTTTATTATTGTGTTTAAATCCTTATAGAGCAATATCAGGATGGTGTTA ATAGGTCTGCCTCAGAATGAGAATCAATCCTTTTAGAAAACCTTTATACTAAGCCTCCTCTTCAAAATTCACAGTGGCGATTAGCGGACT GGAGTCTGGTGGCGATTAGCGGACTGGAGTCTGGGGACATCCGTGGCAAAGACACCAGCTCAACTTTAGTGCTTCCCAACTTTATTTAGA ATGACATGGGGTGGGTGTCTGGTGTGTGTGTTTTCCCTACGCACCTCCCATAGCTATTAACAACTGAGGAAGGCCAGTGCAGAATATTTT TGGAGAACGATTTTTTTTTTAAATAATATATCATTCCTATGGGGGGAAAGCCTTTTTTTTCTTTTTGGCTGAGTTATTCCCTCCCTCCCC TCAATACCCTCAGTACTGACTACTTCCCTTTCTTTTCTCAGGCCTCCCCCCACCGACTTTTGAGGCCAGGGTTGGCCAGATTTAGCAAAA CCAAAACAGAGTGCTGAGTTAAACGCAAATTTCAGGTAAACAAAAGATAATTTTCTAGCATTAATATGCCCCACGCAATATTTGGAACAC TTATGTGAAAAATGATTTGTTTTTCTGAAATTCACGTTTCTCTCTGAGTCCTGTAACTGTCCCCGAGGGGATTGAGCAGAAGCTCGGGTA TGAGCCCTGAGGTTGACTGCCGGTTATTTTTCTGTCCTGGGAACAGCCTGACCCACCTCCCTGTCTCCATGTAGCCAGTGAGGGGAGGGG GAGACACAGAACCAACCACAGCCAGGGGCGTCCCCATGGCGACTGTGGCCCGGCCCCTCCTCTCTTGCCTGACTCTCCTCTCTTGCCTGA CTCTAGACACTAACTTAGTTCCAGGTTCGGTGCCCTGTTGGTGCTCCTGTTTCCAATAGCTTAGGTCCCATGGTGGGGGAGGAACCTCAG GGGCTATGCAGCCCCCGCCAGCTGCCCTCGAATCCCGTCCAGGCCAATTCCAGATTCTAAACTGATTTTTTTCATGATATTGTCAAAACA >43076_43076_2_KLK3-CDC42EP4_KLK3_chr19_51361401_ENST00000360617_CDC42EP4_chr17_71282209_ENST00000439510_length(amino acids)=338AA_BP=126 MWVPVVFLTLSVTWIGAAPLILSRIVGGWECEKHSQPWQVLVASRGRAVCGGVLVHPQWVLTAAHCIRNRFLRPGDDSSHDLMLLRLSEP AELTDAVKVMDLPTQEPALGTTCYASGWGSIEPEELKKANDGEGGDEEAGTEEAVPRRNGAAGPHSPDPLLDEQAFGDLTDLPVVPKATY GLKHAESIMSFHIDLGPSMLGDVLSIMDKEEWDPEEGEGGYHGDEGAAGTITQAPPYAVAAPPLARQEGKAGPDLPSLPSHALEDEGWAA -------------------------------------------------------------- >43076_43076_3_KLK3-CDC42EP4_KLK3_chr19_51361401_ENST00000593997_CDC42EP4_chr17_71282209_ENST00000439510_length(transcript)=2843nt_BP=397nt CACCCGGAGAGCTGTGTCACCATGTGGGTCCCGGTTGTCTTCCTCACCCTGTCCGTGACGTGGATTGGTGCTGCACCCCTCATCCTGTCT CGGATTGTGGGAGGCTGGGAGTGCGAGAAGCATTCCCAACCCTGGCAGGTGCTTGTGGCCTCTCGTGGCAGGGCAGTCTGCGGCGGTGTT CTGGTGCACCCCCAGTGGGTCCTCACAGCTGCCCACTGCATCAGGAATCGATTCCTCAGGCCAGGTGATGACTCCAGCCACGACCTCATG CTGCTCCGCCTGTCAGAGCCTGCCGAGCTCACGGATGCTGTGAAGGTCATGGACCTGCCCACCCAGGAGCCAGCACTGGGGACCACCTGC TACGCCTCAGGCTGGGGCAGCATTGAACCAGAGGAGTTGAAGAAGGCCAATGACGGGGAGGGCGGCGATGAGGAGGCGGGCACGGAGGAG GCAGTGCCCCGTCGGAATGGGGCCGCGGGTCCACATTCCCCTGACCCCCTCCTCGATGAGCAGGCCTTTGGGGATCTGACAGATCTGCCT GTCGTGCCCAAGGCCACGTACGGGCTGAAGCATGCGGAGTCCATCATGTCCTTCCACATCGACCTGGGGCCCTCCATGCTGGGTGACGTC CTCAGCATCATGGACAAGGAGGAGTGGGACCCCGAGGAGGGGGAGGGTGGTTACCATGGCGATGAGGGCGCCGCTGGCACCATCACCCAG GCTCCCCCGTACGCCGTGGCGGCCCCTCCCCTGGCAAGGCAGGAAGGCAAGGCTGGCCCAGACTTGCCCTCCCTCCCCTCCCATGCTCTG GAGGATGAGGGGTGGGCAGCAGCGGCCCCCAGCCCCGGCTCAGCCCGCAGCATGGGCAGCCACACCACACGGGACAGCAGCTCCCTCTCC AGCTGCACCTCAGGCATCCTGGAGGAGCGCAGCCCTGCCTTCCGGGGGCCGGACAGGGCCCGGGCTGCTGTCTCAAGACAGCCAGACAAG GAGTTCTCCTTCATGGATGAGGAGGAGGAGGATGAAATCCGTGTGTGAGGCGGACAGTGGGTGGCCACCGGGAGCTCTTGGCTGCATCTT CTCCCTGCCCCCACCCCACTATGACCTTTGACCCTACGGCGCAGGGGCAGCCAGGACCCTTGATTCAGACCATGGACCCTGGACCTTGTA GATGAGGGACACTGGCCTGGCCCTCGGGTCTTCGGAGGACGTAGGGGGCTGGCATGGGTGCCGACTGGCTGCCTGACTTCATCATGCTCC CTGCACTTAGGCTGCGTGGGACAAGGGCTGTGTTGTCACAGCAGGAATAGGTTTTCCTCTGTTGGCCTCCCTTTCCTCCACCCTGGCCTC AAATGGATGCCAGATGCCAACCCCAGTTCTGGCCACGTACAGCCAGCGGGTCAGCCCAGAGGCAGCCTCAGCTCCAGGGCTAAGGACTCT CGGCTCCCATTTTCTCTGCTGGCGTTTCTGCTGTGCCCAGCAGTGGCTGCTGGGGAAGCAGCTGCAGCAGGAGGGAGACGGTCTTGCCTC TCAGCCCCTCCCTGCCCCACCCCAGCTCCTGCCCTGGAAATCTGGAGCCCCTTGGAGCTGAGCTGGACGGGGGGCCAGCTGCGAGCATGT GCACTAAACGCAGCCCTTTCCAGGGGAAGAGAACAGGATGGAGAATGGAAGGAAAGCCCCCCAGGCTTCGTGAATTGCAAGAAGGGACCC TTCCAGGATGACACTAGGAACAGGGCTAGGGCACTCGCTCAGTCCCTAGGGGCTTGTTTGTTCTTTATTATTGTGTTTAAATCCTTATAG AGCAATATCAGGATGGTGTTAATAGGTCTGCCTCAGAATGAGAATCAATCCTTTTAGAAAACCTTTATACTAAGCCTCCTCTTCAAAATT CACAGTGGCGATTAGCGGACTGGAGTCTGGTGGCGATTAGCGGACTGGAGTCTGGGGACATCCGTGGCAAAGACACCAGCTCAACTTTAG TGCTTCCCAACTTTATTTAGAATGACATGGGGTGGGTGTCTGGTGTGTGTGTTTTCCCTACGCACCTCCCATAGCTATTAACAACTGAGG AAGGCCAGTGCAGAATATTTTTGGAGAACGATTTTTTTTTTAAATAATATATCATTCCTATGGGGGGAAAGCCTTTTTTTTCTTTTTGGC TGAGTTATTCCCTCCCTCCCCTCAATACCCTCAGTACTGACTACTTCCCTTTCTTTTCTCAGGCCTCCCCCCACCGACTTTTGAGGCCAG GGTTGGCCAGATTTAGCAAAACCAAAACAGAGTGCTGAGTTAAACGCAAATTTCAGGTAAACAAAAGATAATTTTCTAGCATTAATATGC CCCACGCAATATTTGGAACACTTATGTGAAAAATGATTTGTTTTTCTGAAATTCACGTTTCTCTCTGAGTCCTGTAACTGTCCCCGAGGG GATTGAGCAGAAGCTCGGGTATGAGCCCTGAGGTTGACTGCCGGTTATTTTTCTGTCCTGGGAACAGCCTGACCCACCTCCCTGTCTCCA TGTAGCCAGTGAGGGGAGGGGGAGACACAGAACCAACCACAGCCAGGGGCGTCCCCATGGCGACTGTGGCCCGGCCCCTCCTCTCTTGCC TGACTCTCCTCTCTTGCCTGACTCTAGACACTAACTTAGTTCCAGGTTCGGTGCCCTGTTGGTGCTCCTGTTTCCAATAGCTTAGGTCCC ATGGTGGGGGAGGAACCTCAGGGGCTATGCAGCCCCCGCCAGCTGCCCTCGAATCCCGTCCAGGCCAATTCCAGATTCTAAACTGATTTT >43076_43076_3_KLK3-CDC42EP4_KLK3_chr19_51361401_ENST00000593997_CDC42EP4_chr17_71282209_ENST00000439510_length(amino acids)=338AA_BP=126 MWVPVVFLTLSVTWIGAAPLILSRIVGGWECEKHSQPWQVLVASRGRAVCGGVLVHPQWVLTAAHCIRNRFLRPGDDSSHDLMLLRLSEP AELTDAVKVMDLPTQEPALGTTCYASGWGSIEPEELKKANDGEGGDEEAGTEEAVPRRNGAAGPHSPDPLLDEQAFGDLTDLPVVPKATY GLKHAESIMSFHIDLGPSMLGDVLSIMDKEEWDPEEGEGGYHGDEGAAGTITQAPPYAVAAPPLARQEGKAGPDLPSLPSHALEDEGWAA -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for KLK3-CDC42EP4 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for KLK3-CDC42EP4 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for KLK3-CDC42EP4 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
