
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:KPNA1-AQR (FusionGDB2 ID:43352) |
Fusion Gene Summary for KPNA1-AQR |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: KPNA1-AQR | Fusion gene ID: 43352 | Hgene | Tgene | Gene symbol | KPNA1 | AQR | Gene ID | 3836 | 9716 |
| Gene name | karyopherin subunit alpha 1 | aquarius intron-binding spliceosomal factor | |
| Synonyms | IPOA5|NPI-1|RCH2|SRP1 | IBP160|fSAP164 | |
| Cytomap | 3q21.1 | 15q14 | |
| Type of gene | protein-coding | protein-coding | |
| Description | importin subunit alpha-5RAG cohort protein 2SRP1-betaimportin alpha 5importin subunit alpha-1importin-alpha-S1karyopherin alpha 1 (importin alpha 5)nucleoprotein interactor 1recombination activating gene cohort 2 | RNA helicase aquariusaquarius homologfunctional spliceosome-associated protein 164intron-binding protein aquariusintron-binding protein of 160 kDa | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | P52294 | O60306 | |
| Ensembl transtripts involved in fusion gene | ENST00000344337, ENST00000466923, | ENST00000156471, | |
| Fusion gene scores | * DoF score | 19 X 10 X 13=2470 | 13 X 15 X 5=975 |
| # samples | 26 | 16 | |
| ** MAII score | log2(26/2470*10)=-3.24792751344359 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(16/975*10)=-2.60733031374961 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: KPNA1 [Title/Abstract] AND AQR [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | KPNA1(122215284)-AQR(35219339), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | AQR | GO:0000398 | mRNA splicing, via spliceosome | 28076346 |
 Fusion gene breakpoints across KPNA1 (5'-gene) Fusion gene breakpoints across KPNA1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
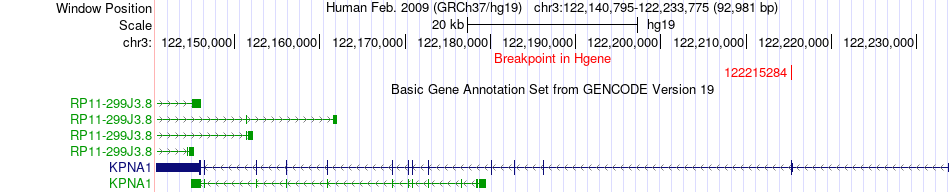 |
 Fusion gene breakpoints across AQR (3'-gene) Fusion gene breakpoints across AQR (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
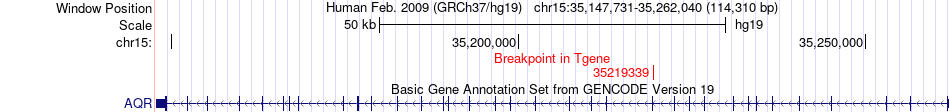 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-HU-A4GN-01A | KPNA1 | chr3 | 122215284 | - | AQR | chr15 | 35219339 | - |
Top |
Fusion Gene ORF analysis for KPNA1-AQR |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000344337 | ENST00000156471 | KPNA1 | chr3 | 122215284 | - | AQR | chr15 | 35219339 | - |
| intron-3CDS | ENST00000466923 | ENST00000156471 | KPNA1 | chr3 | 122215284 | - | AQR | chr15 | 35219339 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000344337 | KPNA1 | chr3 | 122215284 | - | ENST00000156471 | AQR | chr15 | 35219339 | - | 5011 | 306 | 51 | 3749 | 1232 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000344337 | ENST00000156471 | KPNA1 | chr3 | 122215284 | - | AQR | chr15 | 35219339 | - | 0.000331271 | 0.9996687 |
Top |
Fusion Genomic Features for KPNA1-AQR |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
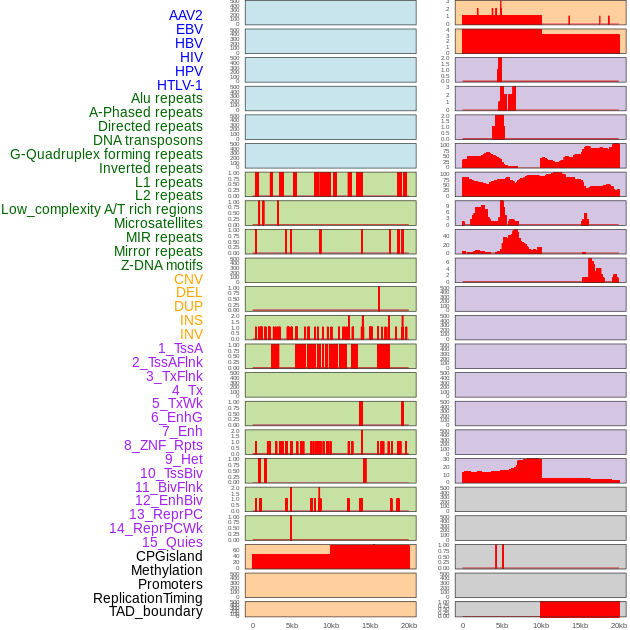 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for KPNA1-AQR |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:122215284/chr15:35219339) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| KPNA1 | AQR |
| FUNCTION: Functions in nuclear protein import as an adapter protein for nuclear receptor KPNB1. Binds specifically and directly to substrates containing either a simple or bipartite NLS motif. Docking of the importin/substrate complex to the nuclear pore complex (NPC) is mediated by KPNB1 through binding to nucleoporin FxFG repeats and the complex is subsequently translocated through the pore by an energy requiring, Ran-dependent mechanism. At the nucleoplasmic side of the NPC, Ran binds to importin-beta and the three components separate and importin-alpha and -beta are re-exported from the nucleus to the cytoplasm where GTP hydrolysis releases Ran from importin. The directionality of nuclear import is thought to be conferred by an asymmetric distribution of the GTP- and GDP-bound forms of Ran between the cytoplasm and nucleus. In vitro, mediates the nuclear import of human cytomegalovirus UL84 by recognizing a non-classical NLS. | FUNCTION: Involved in pre-mRNA splicing as component of the spliceosome (PubMed:11991638, PubMed:25599396, PubMed:28502770, PubMed:28076346). Intron-binding spliceosomal protein required to link pre-mRNA splicing and snoRNP (small nucleolar ribonucleoprotein) biogenesis (PubMed:16949364). Plays a key role in position-dependent assembly of intron-encoded box C/D small snoRNP, splicing being required for snoRNP assembly (PubMed:16949364). May act by helping the folding of the snoRNA sequence. Binds to intron of pre-mRNAs in a sequence-independent manner, contacting the region between snoRNA and the branchpoint of introns (40 nucleotides upstream of the branchpoint) during the late stages of splicing (PubMed:16949364). Has ATP-dependent RNA helicase activity and can unwind double-stranded RNA molecules with a 3' overhang (in vitro) (PubMed:25599396). {ECO:0000269|PubMed:11991638, ECO:0000269|PubMed:16949364, ECO:0000269|PubMed:25599396, ECO:0000269|PubMed:28076346, ECO:0000269|PubMed:28502770}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KPNA1 | chr3:122215284 | chr15:35219339 | ENST00000344337 | - | 2 | 14 | 25_28 | 43 | 539.0 | Compositional bias | Note=Poly-Arg |
| Tgene | AQR | chr3:122215284 | chr15:35219339 | ENST00000156471 | 11 | 35 | 826_831 | 338 | 1486.0 | Nucleotide binding | ATP | |
| Tgene | AQR | chr3:122215284 | chr15:35219339 | ENST00000156471 | 11 | 35 | 417_1485 | 338 | 1486.0 | Region | Required for assembly of the IB complex |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KPNA1 | chr3:122215284 | chr15:35219339 | ENST00000344337 | - | 2 | 14 | 1_57 | 43 | 539.0 | Domain | IBB |
| Hgene | KPNA1 | chr3:122215284 | chr15:35219339 | ENST00000344337 | - | 2 | 14 | 42_51 | 43 | 539.0 | Motif | Nuclear localization signal |
| Hgene | KPNA1 | chr3:122215284 | chr15:35219339 | ENST00000344337 | - | 2 | 14 | 149_241 | 43 | 539.0 | Region | NLS binding site (major) |
| Hgene | KPNA1 | chr3:122215284 | chr15:35219339 | ENST00000344337 | - | 2 | 14 | 245_437 | 43 | 539.0 | Region | Note=Binding to RAG1 |
| Hgene | KPNA1 | chr3:122215284 | chr15:35219339 | ENST00000344337 | - | 2 | 14 | 318_406 | 43 | 539.0 | Region | NLS binding site (minor) |
| Hgene | KPNA1 | chr3:122215284 | chr15:35219339 | ENST00000344337 | - | 2 | 14 | 118_161 | 43 | 539.0 | Repeat | Note=ARM 2 |
| Hgene | KPNA1 | chr3:122215284 | chr15:35219339 | ENST00000344337 | - | 2 | 14 | 162_206 | 43 | 539.0 | Repeat | Note=ARM 3 |
| Hgene | KPNA1 | chr3:122215284 | chr15:35219339 | ENST00000344337 | - | 2 | 14 | 207_245 | 43 | 539.0 | Repeat | Note=ARM 4 |
| Hgene | KPNA1 | chr3:122215284 | chr15:35219339 | ENST00000344337 | - | 2 | 14 | 246_290 | 43 | 539.0 | Repeat | Note=ARM 5 |
| Hgene | KPNA1 | chr3:122215284 | chr15:35219339 | ENST00000344337 | - | 2 | 14 | 291_330 | 43 | 539.0 | Repeat | Note=ARM 6 |
| Hgene | KPNA1 | chr3:122215284 | chr15:35219339 | ENST00000344337 | - | 2 | 14 | 331_372 | 43 | 539.0 | Repeat | Note=ARM 7 |
| Hgene | KPNA1 | chr3:122215284 | chr15:35219339 | ENST00000344337 | - | 2 | 14 | 373_412 | 43 | 539.0 | Repeat | Note=ARM 8 |
| Hgene | KPNA1 | chr3:122215284 | chr15:35219339 | ENST00000344337 | - | 2 | 14 | 413_457 | 43 | 539.0 | Repeat | Note=ARM 9 |
| Hgene | KPNA1 | chr3:122215284 | chr15:35219339 | ENST00000344337 | - | 2 | 14 | 460_504 | 43 | 539.0 | Repeat | Note=ARM 10%3B atypical |
| Hgene | KPNA1 | chr3:122215284 | chr15:35219339 | ENST00000344337 | - | 2 | 14 | 77_117 | 43 | 539.0 | Repeat | Note=ARM 1%3B truncated |
| Tgene | AQR | chr3:122215284 | chr15:35219339 | ENST00000156471 | 11 | 35 | 1_416 | 338 | 1486.0 | Region | Helical region with structural similarity to ARM repeat domains |
Top |
Fusion Gene Sequence for KPNA1-AQR |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >43352_43352_1_KPNA1-AQR_KPNA1_chr3_122215284_ENST00000344337_AQR_chr15_35219339_ENST00000156471_length(transcript)=5011nt_BP=306nt GCCGGCTGAGCTGAGTCTCGCTGCTCGGTGCGAGGCGGCGGAGAGCGAGGCCTGGTGAGCACCGCCGAGGCGCGGGCCAGCTCTTCGAGG TTGTGCGCGGGAGTGGCACGGCGGGCGGGCGAGCGAGGGGCTAACTTCAGCGGTGGCACCGGGATCGGTTGCCTTGAGCCTGAAATCATG ACCACCCCAGGAAAAGAGAACTTTCGCCTGAAAAGTTACAAGAACAAATCTCTGAATCCCGATGAGATGCGCAGGAGGAGGGAGGAAGAA GGACTGCAGTTACGAAAGCAGAAAAGAGAAGAGCAGAGAGCTGCTTTTGCACATTTTCCTGAACTCTATGATTTTGCCCTCTCAAATGTG GCAGAAGTAGATACTCGGGAGTCCTTGGTCAAGTTTTTTGGACCTCTTAGTTCAAACACACTCCACCAGGTGGCATCATACCTCTGCTTG TTGCCAACTCTTCCTAAAAATGAAGACACAACTTTTGATAAAGAATTTCTTCTAGAATTGCTGGTATCTCGTCATGAACGTCGAATTTCT CAGATTCAGCAGTTGAACCAGATGCCTTTGTATCCAACTGAGAAAATTATATGGGATGAAAATATTGTCCCAACTGAGTACTATTCTGGA GAAGGTTGTCTTGCTCTTCCCAAATTGAATTTGCAGTTTTTGACTCTTCATGACTACCTGCTAAGGAACTTTAACCTCTTCCGCTTAGAA TCAACTTATGAAATTCGTCAGGACATTGAAGATAGTGTCAGCAGAATGAAGCCATGGCAATCTGAATATGGCGGTGTAGTGTTTGGTGGT TGGGCGCGAATGGCCCAGCCCATTGTGGCTTTCACTGTCGTTGAAGTGGCCAAACCCAACATAGGTGAAAACTGGCCAACCCGAGTTCGT GCAGATGTTACCATAAATCTCAATGTCAGAGATCACATCAAAGATGAATGGGAAGGTCTTCGTAAGCATGATGTATGCTTTTTAATTACC GTACGTCCCACAAAACCTTATGGCACTAAGTTTGACCGGAGGAGACCTTTTATTGAGCAGGTTGGCCTGGTTTATGTCAGAGGCTGTGAA ATTCAGGGCATGCTGGATGATAAAGGACGTGTCATTGAAGATGGACCTGAACCCAGACCCAATCTTAGAGGAGAATCAAGGACATTTAGA GTGTTTTTGGATCCAAACCAGTATCAACAAGATATGACCAATACTATACAAAATGGAGCAGAGGATGTGTATGAAACTTTTAATATAATA ATGAGGAGAAAACCAAAGGAAAATAACTTTAAGGCTGTGCTGGAGACTATTCGGAACCTGATGAATACTGATTGTGTGGTACCTGACTGG CTGCACGATATCATTTTAGGTTATGGGGACCCAAGTAGTGCACATTATTCGAAAATGCCCAATCAGATTGCCACCCTTGATTTCAATGAT ACATTTCTCTCCATTGAGCATTTAAAAGCCAGCTTCCCTGGTCATAATGTTAAAGTAACTGTAGAAGACCCTGCTCTACAAATACCCCCT TTCAGGATAACTTTTCCAGTAAGAAGTGGAAAAGGGAAGAAAAGGAAAGATGCGGATGTGGAAGATGAAGACACCGAGGAAGCAAAAACC TTAATTGTTGAGCCCCATGTTATTCCTAATAGGGGTCCTTATCCTTATAATCAACCCAAACGTAATACGATTCAGTTCACTCATACACAG ATAGAAGCCATCCGTGCTGGAATGCAGCCTGGGCTGACTATGGTTGTGGGCCCACCTGGTACAGGCAAAACAGATGTGGCAGTTCAGATC ATATCCAACATCTACCACAACTTCCCAGAACAGAGGACTCTAATTGTTACTCATTCCAATCAGGCCCTAAACCAGTTGTTTGAGAAAATC ATGGCATTAGACATTGATGAGCGCCACCTACTGCGTCTTGGTCATGGAGAAGAAGAGCTGGAGACAGAGAAAGATTTCAGCAGGTATGGA AGAGTTAATTATGTTCTGGCTCGAAGAATAGAACTTTTAGAAGAAGTCAAACGATTGCAAAAGAGTCTAGGGGTTCCAGGAGATGCCTCA TATACCTGTGAAACTGCAGGCTATTTCTTCTTATACCAGGTAATGTCTCGCTGGGAAGAGTATATCAGCAAAGTGAAAAATAAAGGTAGT ACATTGCCAGATGTTACGGAAGTCTCCACTTTCTTCCCTTTCCATGAATACTTTGCAAATGCTCCTCAACCCATTTTTAAAGGAAGATCT TATGAAGAAGACATGGAAATTGCTGAAGGATGTTTCAGGCATATTAAGAAAATCTTTACGCAGCTTGAGGAATTCAGAGCCTCTGAATTG CTTCGAAGTGGACTGGACAGATCTAAATACCTTTTAGTGAAAGAAGCCAAAATTATTGCTATGACCTGTACTCATGCTGCCTTAAAACGA CATGACTTGGTCAAGCTAGGTTTCAAGTATGACAACATTTTGATGGAAGAGGCTGCTCAGATTCTGGAGATAGAAACTTTTATCCCTCTT CTTCTACAGAATCCTCAGGATGGATTTAGCCGACTAAAACGATGGATTATGATTGGCGATCATCACCAGTTACCTCCAGTTATTAAGAAC ATGGCCTTTCAAAAGTACTCAAACATGGAGCAGTCTCTCTTCACTCGCTTTGTTCGCGTTGGAGTTCCGACTGTTGACCTTGATGCTCAA GGGAGAGCCAGAGCAAGCTTGTGCAACCTCTACAACTGGCGATACAAGAATCTAGGAAACTTACCCCATGTGCAGCTCTTGCCAGAGTTT AGTACAGCAAATGCTGGCTTACTGTATGACTTCCAGCTCATTAATGTTGAAGATTTTCAAGGAGTGGGAGAATCTGAACCTAATCCTTAC TTCTATCAGAATCTTGGAGAGGCAGAATATGTAGTAGCACTTTTTATGTACATGTGTTTACTTGGTTACCCTGCTGACAAAATCAGTATT CTAACAACATATAATGGCCAAAAGCATCTTATTCGCGACATCATCAATAGACGATGTGGAAACAATCCATTGATTGGAAGACCAAACAAG GTGACAACTGTTGATAGATTTCAAGGTCAACAGAATGACTATATTCTTCTTTCTCTGGTACGAACCAGGGCAGTGGGCCATCTGAGGGAT GTCCGTCGCTTGGTAGTGGCCATGTCTAGAGCCAGACTTGGACTTTATATCTTCGCCAGAGTATCCCTCTTCCAAAACTGTTTTGAACTG ACTCCAGCTTTCAGTCAGCTCACAGCTCGCCCCCTTCATTTGCATATAATTCCAACAGAACCTTTCCCAACTACTAGAAAGAATGGAGAG AGACCATCTCATGAAGTACAAATAATAAAAAATATGCCCCAGATGGCAAACTTTGTATACAACATGTACATGCATTTGATACAGACTACA CATCATTATCATCAGACTTTATTACAACTACCACCTGCTATGGTAGAAGAGGGTGAGGAAGTTCAAAATCAAGAAACAGAATTGGAAACA GAAGAAGAGGCCATGACTGTTCAAGCTGACATCATACCCAGTCCAACAGACACCAGCTGCCGTCAAGAAACTCCAGCCTTTCAAACTGAC ACCACCCCCAGTGAGACAGGAGCCACTTCCACTCCAGAAGCCATCCCTGCTTTATCTGAGACCACCCCTACTGTGGTAGGAGCTGTATCT GCACCGGCAGAAGCTAACACACCTCAGGATGCCACATCTGCCCCGGAAGAGACCAAGTAGCCAAACTGTAGTCCTTCTAAAGGAGGACAT GGCAGTCAAAAAGTCTGAGTAAAGCTGTTTTTTGTATTTTATATTTGCTTCTGCCATTTTACTGTCACTAATTAATGTTTAGTTCTTATA TTTGTTAACTGATTTCGGTGTCTTGAATATATTTTTTTAAATTATGTGTATGAACAATTCTAGTTTCATTTGTTCAATCAGAAGAGCAAA TAACCATTCCTTTCATGTTTTGATCACTGAGTGTGTCTGTAATCATACCTACATTAAAATCATTTTCTATGAATATATAATATATACTTC ACATTTTTAGTGAACTTCTCTAAAGAAGAGGACAGAATATACTGGACTTAACCACGAATACCCTTGAGTGTCCAAATTGGGAAGGAACTT GTTTCTTCTGTTATACTATCAAATGCTTAAATTCTGTTTCCTTTTTTCTTACCTTTGTTTGCTGTCTTTATGTAAAGATATTATTAGCAG TAAGATACAGGTTTCTTCAATCTATCAAAGTGGTTACATGCATTTCCTTCTCTCAAATCTTAGAATGCCTTAGAATCACCTAAGATGTCT GTTTATTCTTGAGGCACACCTCAGACTGAATGAGTCAGAATCTCAGGATTAGAGCCTAGGAATCTATATTTTAGGAGGTTCTCCACACCA CTAATGTTTGGGTTGCTGGCAGAGTGGCATCAGCCAAGGCTGGTTAGGCTGAGCCAGGATTTGTCATCAGCTAAATAGGAGGACTCTGCC AAATCAATCTGGGCAAGAAGCAGGATCCTGAGTCTGGGCAATGAGAGTCAGAGAACTTAACTCCTGTGTGCAGGGTAGAATTTTCCCATG GCAGAAATTCTGAGTTTTCTGCCTTATTTTTCTTGGGATAACCAGCCACAAGGAGACCTTCAGGTAAAATGTGGTTCAAATATGGGAAGG CAGAGGTACCTGGGACCTTGCTACTCAAGTATGATTTGTGAACCAACAGCATCAACATCGTATTGAAGTTTGTTATTTATAGAAATGCAG AAGTTTGGGCTCTGCTCCAAACCTATTAAATCTAAATCTACATTGCAACAAAGTCCCCAGGTGATTCACAGGCACTTTGTAGTTTGAGAA GCACTGTACTAAGATTACTGTGTTGTCTTCACATGAATGAGGCTATAATGTAGAATTACCAGTGAATGTGACTAATGTTTTCTGACAAGG >43352_43352_1_KPNA1-AQR_KPNA1_chr3_122215284_ENST00000344337_AQR_chr15_35219339_ENST00000156471_length(amino acids)=1232AA_BP=85 MVSTAEARASSSRLCAGVARRAGERGANFSGGTGIGCLEPEIMTTPGKENFRLKSYKNKSLNPDEMRRRREEEGLQLRKQKREEQRAAFA HFPELYDFALSNVAEVDTRESLVKFFGPLSSNTLHQVASYLCLLPTLPKNEDTTFDKEFLLELLVSRHERRISQIQQLNQMPLYPTEKII WDENIVPTEYYSGEGCLALPKLNLQFLTLHDYLLRNFNLFRLESTYEIRQDIEDSVSRMKPWQSEYGGVVFGGWARMAQPIVAFTVVEVA KPNIGENWPTRVRADVTINLNVRDHIKDEWEGLRKHDVCFLITVRPTKPYGTKFDRRRPFIEQVGLVYVRGCEIQGMLDDKGRVIEDGPE PRPNLRGESRTFRVFLDPNQYQQDMTNTIQNGAEDVYETFNIIMRRKPKENNFKAVLETIRNLMNTDCVVPDWLHDIILGYGDPSSAHYS KMPNQIATLDFNDTFLSIEHLKASFPGHNVKVTVEDPALQIPPFRITFPVRSGKGKKRKDADVEDEDTEEAKTLIVEPHVIPNRGPYPYN QPKRNTIQFTHTQIEAIRAGMQPGLTMVVGPPGTGKTDVAVQIISNIYHNFPEQRTLIVTHSNQALNQLFEKIMALDIDERHLLRLGHGE EELETEKDFSRYGRVNYVLARRIELLEEVKRLQKSLGVPGDASYTCETAGYFFLYQVMSRWEEYISKVKNKGSTLPDVTEVSTFFPFHEY FANAPQPIFKGRSYEEDMEIAEGCFRHIKKIFTQLEEFRASELLRSGLDRSKYLLVKEAKIIAMTCTHAALKRHDLVKLGFKYDNILMEE AAQILEIETFIPLLLQNPQDGFSRLKRWIMIGDHHQLPPVIKNMAFQKYSNMEQSLFTRFVRVGVPTVDLDAQGRARASLCNLYNWRYKN LGNLPHVQLLPEFSTANAGLLYDFQLINVEDFQGVGESEPNPYFYQNLGEAEYVVALFMYMCLLGYPADKISILTTYNGQKHLIRDIINR RCGNNPLIGRPNKVTTVDRFQGQQNDYILLSLVRTRAVGHLRDVRRLVVAMSRARLGLYIFARVSLFQNCFELTPAFSQLTARPLHLHII PTEPFPTTRKNGERPSHEVQIIKNMPQMANFVYNMYMHLIQTTHHYHQTLLQLPPAMVEEGEEVQNQETELETEEEAMTVQADIIPSPTD -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for KPNA1-AQR |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for KPNA1-AQR |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for KPNA1-AQR |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
