
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:KREMEN1-EMID1 (FusionGDB2 ID:43447) |
Fusion Gene Summary for KREMEN1-EMID1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: KREMEN1-EMID1 | Fusion gene ID: 43447 | Hgene | Tgene | Gene symbol | KREMEN1 | EMID1 | Gene ID | 83999 | 129080 |
| Gene name | kringle containing transmembrane protein 1 | EMI domain containing 1 | |
| Synonyms | ECTD13|KREMEN|KRM1 | EMI5|EMU1 | |
| Cytomap | 22q12.1 | 22q12.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | kremen protein 1dickkopf receptorkringle domain-containing transmembrane protein 1kringle-coding gene marking the eye and the nosekringle-containing protein marking the eye and the nose | EMI domain-containing protein 1emilin and multimerin domain-containing protein 1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q96MU8 | Q96A84 | |
| Ensembl transtripts involved in fusion gene | ENST00000327813, ENST00000400335, ENST00000400338, ENST00000407188, ENST00000479755, | ENST00000484039, ENST00000334018, ENST00000404755, ENST00000404820, | |
| Fusion gene scores | * DoF score | 9 X 8 X 7=504 | 9 X 12 X 7=756 |
| # samples | 11 | 12 | |
| ** MAII score | log2(11/504*10)=-2.19592020997526 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(12/756*10)=-2.65535182861255 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: KREMEN1 [Title/Abstract] AND EMID1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | EMID1(29611619)-KREMEN1(29490246), # samples:2 KREMEN1(29494941)-EMID1(29629368), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across KREMEN1 (5'-gene) Fusion gene breakpoints across KREMEN1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
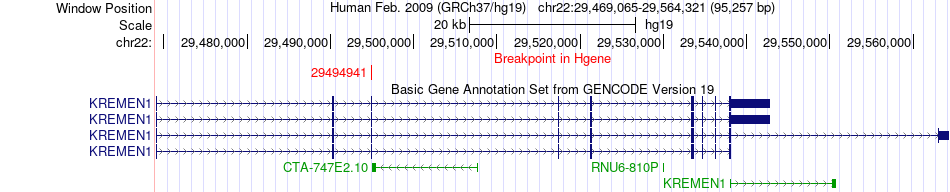 |
 Fusion gene breakpoints across EMID1 (3'-gene) Fusion gene breakpoints across EMID1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
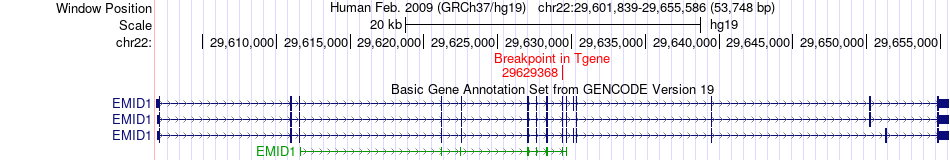 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BLCA | TCGA-FD-A3SJ-01A | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + |
Top |
Fusion Gene ORF analysis for KREMEN1-EMID1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000327813 | ENST00000484039 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + |
| 5CDS-3UTR | ENST00000400335 | ENST00000484039 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + |
| 5CDS-3UTR | ENST00000400338 | ENST00000484039 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + |
| 5CDS-3UTR | ENST00000407188 | ENST00000484039 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + |
| In-frame | ENST00000327813 | ENST00000334018 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + |
| In-frame | ENST00000327813 | ENST00000404755 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + |
| In-frame | ENST00000327813 | ENST00000404820 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + |
| In-frame | ENST00000400335 | ENST00000334018 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + |
| In-frame | ENST00000400335 | ENST00000404755 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + |
| In-frame | ENST00000400335 | ENST00000404820 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + |
| In-frame | ENST00000400338 | ENST00000334018 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + |
| In-frame | ENST00000400338 | ENST00000404755 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + |
| In-frame | ENST00000400338 | ENST00000404820 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + |
| In-frame | ENST00000407188 | ENST00000334018 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + |
| In-frame | ENST00000407188 | ENST00000404755 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + |
| In-frame | ENST00000407188 | ENST00000404820 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + |
| intron-3CDS | ENST00000479755 | ENST00000334018 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + |
| intron-3CDS | ENST00000479755 | ENST00000404755 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + |
| intron-3CDS | ENST00000479755 | ENST00000404820 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + |
| intron-3UTR | ENST00000479755 | ENST00000484039 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000400338 | KREMEN1 | chr22 | 29494941 | + | ENST00000334018 | EMID1 | chr22 | 29629368 | + | 1567 | 405 | 53 | 913 | 286 |
| ENST00000400338 | KREMEN1 | chr22 | 29494941 | + | ENST00000404820 | EMID1 | chr22 | 29629368 | + | 1573 | 405 | 53 | 919 | 288 |
| ENST00000400338 | KREMEN1 | chr22 | 29494941 | + | ENST00000404755 | EMID1 | chr22 | 29629368 | + | 1504 | 405 | 53 | 850 | 265 |
| ENST00000400335 | KREMEN1 | chr22 | 29494941 | + | ENST00000334018 | EMID1 | chr22 | 29629368 | + | 1567 | 405 | 53 | 913 | 286 |
| ENST00000400335 | KREMEN1 | chr22 | 29494941 | + | ENST00000404820 | EMID1 | chr22 | 29629368 | + | 1573 | 405 | 53 | 919 | 288 |
| ENST00000400335 | KREMEN1 | chr22 | 29494941 | + | ENST00000404755 | EMID1 | chr22 | 29629368 | + | 1504 | 405 | 53 | 850 | 265 |
| ENST00000327813 | KREMEN1 | chr22 | 29494941 | + | ENST00000334018 | EMID1 | chr22 | 29629368 | + | 1527 | 365 | 13 | 873 | 286 |
| ENST00000327813 | KREMEN1 | chr22 | 29494941 | + | ENST00000404820 | EMID1 | chr22 | 29629368 | + | 1533 | 365 | 13 | 879 | 288 |
| ENST00000327813 | KREMEN1 | chr22 | 29494941 | + | ENST00000404755 | EMID1 | chr22 | 29629368 | + | 1464 | 365 | 13 | 810 | 265 |
| ENST00000407188 | KREMEN1 | chr22 | 29494941 | + | ENST00000334018 | EMID1 | chr22 | 29629368 | + | 1508 | 346 | 0 | 854 | 284 |
| ENST00000407188 | KREMEN1 | chr22 | 29494941 | + | ENST00000404820 | EMID1 | chr22 | 29629368 | + | 1514 | 346 | 0 | 860 | 286 |
| ENST00000407188 | KREMEN1 | chr22 | 29494941 | + | ENST00000404755 | EMID1 | chr22 | 29629368 | + | 1445 | 346 | 0 | 791 | 263 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000400338 | ENST00000334018 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + | 0.109114654 | 0.8908854 |
| ENST00000400338 | ENST00000404820 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + | 0.17209055 | 0.82790947 |
| ENST00000400338 | ENST00000404755 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + | 0.105327085 | 0.89467293 |
| ENST00000400335 | ENST00000334018 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + | 0.109114654 | 0.8908854 |
| ENST00000400335 | ENST00000404820 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + | 0.17209055 | 0.82790947 |
| ENST00000400335 | ENST00000404755 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + | 0.105327085 | 0.89467293 |
| ENST00000327813 | ENST00000334018 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + | 0.10050351 | 0.8994965 |
| ENST00000327813 | ENST00000404820 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + | 0.15615174 | 0.8438483 |
| ENST00000327813 | ENST00000404755 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + | 0.09907669 | 0.9009233 |
| ENST00000407188 | ENST00000334018 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + | 0.04984928 | 0.9501507 |
| ENST00000407188 | ENST00000404820 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + | 0.100657396 | 0.8993426 |
| ENST00000407188 | ENST00000404755 | KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629368 | + | 0.052433092 | 0.9475669 |
Top |
Fusion Genomic Features for KREMEN1-EMID1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629367 | + | 4.51E-05 | 0.9999548 |
| KREMEN1 | chr22 | 29494941 | + | EMID1 | chr22 | 29629367 | + | 4.51E-05 | 0.9999548 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for KREMEN1-EMID1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr22:29611619/chr22:29490246) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| KREMEN1 | EMID1 |
| FUNCTION: Receptor for Dickkopf proteins. Cooperates with DKK1/2 to inhibit Wnt/beta-catenin signaling by promoting the endocytosis of Wnt receptors LRP5 and LRP6. In the absence of DKK1, potentiates Wnt-beta-catenin signaling by maintaining LRP5 or LRP6 at the cell membrane. Can trigger apoptosis in a Wnt-independent manner and this apoptotic activity is inhibited upon binding of the ligand DKK1. Plays a role in limb development; attenuates Wnt signaling in the developing limb to allow normal limb patterning and can also negatively regulate bone formation. Modulates cell fate decisions in the developing cochlea with an inhibitory role in hair cell fate specification. {ECO:0000250|UniProtKB:Q90Y90, ECO:0000250|UniProtKB:Q99N43}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KREMEN1 | chr22:29494941 | chr22:29629368 | ENST00000327813 | + | 3 | 10 | 31_114 | 117 | 493.0 | Domain | Kringle |
| Hgene | KREMEN1 | chr22:29494941 | chr22:29629368 | ENST00000400335 | + | 3 | 9 | 31_114 | 117 | 459.0 | Domain | Kringle |
| Hgene | KREMEN1 | chr22:29494941 | chr22:29629368 | ENST00000407188 | + | 3 | 9 | 31_114 | 115 | 474.0 | Domain | Kringle |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KREMEN1 | chr22:29494941 | chr22:29629368 | ENST00000327813 | + | 3 | 10 | 116_210 | 117 | 493.0 | Domain | WSC |
| Hgene | KREMEN1 | chr22:29494941 | chr22:29629368 | ENST00000327813 | + | 3 | 10 | 214_321 | 117 | 493.0 | Domain | CUB |
| Hgene | KREMEN1 | chr22:29494941 | chr22:29629368 | ENST00000400335 | + | 3 | 9 | 116_210 | 117 | 459.0 | Domain | WSC |
| Hgene | KREMEN1 | chr22:29494941 | chr22:29629368 | ENST00000400335 | + | 3 | 9 | 214_321 | 117 | 459.0 | Domain | CUB |
| Hgene | KREMEN1 | chr22:29494941 | chr22:29629368 | ENST00000407188 | + | 3 | 9 | 116_210 | 115 | 474.0 | Domain | WSC |
| Hgene | KREMEN1 | chr22:29494941 | chr22:29629368 | ENST00000407188 | + | 3 | 9 | 214_321 | 115 | 474.0 | Domain | CUB |
| Hgene | KREMEN1 | chr22:29494941 | chr22:29629368 | ENST00000327813 | + | 3 | 10 | 414_473 | 117 | 493.0 | Region | Essential for apoptotic activity |
| Hgene | KREMEN1 | chr22:29494941 | chr22:29629368 | ENST00000400335 | + | 3 | 9 | 414_473 | 117 | 459.0 | Region | Essential for apoptotic activity |
| Hgene | KREMEN1 | chr22:29494941 | chr22:29629368 | ENST00000407188 | + | 3 | 9 | 414_473 | 115 | 474.0 | Region | Essential for apoptotic activity |
| Hgene | KREMEN1 | chr22:29494941 | chr22:29629368 | ENST00000327813 | + | 3 | 10 | 21_392 | 117 | 493.0 | Topological domain | Extracellular |
| Hgene | KREMEN1 | chr22:29494941 | chr22:29629368 | ENST00000400335 | + | 3 | 9 | 21_392 | 117 | 459.0 | Topological domain | Extracellular |
| Hgene | KREMEN1 | chr22:29494941 | chr22:29629368 | ENST00000407188 | + | 3 | 9 | 21_392 | 115 | 474.0 | Topological domain | Extracellular |
| Hgene | KREMEN1 | chr22:29494941 | chr22:29629368 | ENST00000327813 | + | 3 | 10 | 393_413 | 117 | 493.0 | Transmembrane | Helical |
| Hgene | KREMEN1 | chr22:29494941 | chr22:29629368 | ENST00000400335 | + | 3 | 9 | 393_413 | 117 | 459.0 | Transmembrane | Helical |
| Hgene | KREMEN1 | chr22:29494941 | chr22:29629368 | ENST00000407188 | + | 3 | 9 | 393_413 | 115 | 474.0 | Transmembrane | Helical |
| Tgene | EMID1 | chr22:29494941 | chr22:29629368 | ENST00000334018 | 7 | 15 | 179_368 | 274 | 444.0 | Domain | Note=Collagen-like | |
| Tgene | EMID1 | chr22:29494941 | chr22:29629368 | ENST00000334018 | 7 | 15 | 33_106 | 274 | 444.0 | Domain | EMI |
Top |
Fusion Gene Sequence for KREMEN1-EMID1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >43447_43447_1_KREMEN1-EMID1_KREMEN1_chr22_29494941_ENST00000327813_EMID1_chr22_29629368_ENST00000334018_length(transcript)=1527nt_BP=365nt GCACTGACGGCCCATGGCGCCGCCAGCCGCCCGCCTCGCCCTGCTCTCCGCCGCGGCGCTCACGCTGGCGGCCCGGCCCGCGCCTAGCCC CGGCCTCGGCCCCGGACCCGAGTGTTTCACAGCCAATGGTGCGGATTATAGGGGAACACAGAACTGGACAGCACTACAAGGCGGGAAGCC ATGTCTGTTTTGGAACGAGACTTTCCAGCATCCATACAACACTCTGAAATACCCCAACGGGGAGGGGGGCCTGGGTGAGCACAACTATTG CAGAAATCCAGATGGAGACGTGAGCCCCTGGTGCTATGTGGCAGAGCACGAGGATGGTGTCTACTGGAAGTACTGTGAGATACCTGCTTG CCAGAGAGACCCATTGCTGTCCAACACCTTCACTGAGACCAACAACCACTGGCCCCAGGGACCCACTGGGCCTCCAGGCCCTCCAGGGCC CATGGGTCCCCCTGGGCCTCCTGGCCCCACAGGTGTCCCTGGGAGTCCTGGTCACATAGGACCCCCAGGCCCCACTGGACCCAAAGGAAT CTCTGGCCACCCAGGAGAGAAGGGCGAGAGAGGACTGCGTGGGGAGCCTGGCCCCCAAGGCTCTGCTGGGCAGCGGGGGGAACCTGGCCC TAAGGGAGACCCTGGTGAGAAGAGCCACTGGGGGGAGGGGTTGCACCAGCTACGCGAGGCTTTGAAGATTTTAGCTGAGAGGGTTTTAAT CTTGGAAACAATGATTGGGCTCTATGAACCAGAGCTGGGGTCTGGGGCGGGCCCTGCCGGCACAGGCACCCCCAGCCTCCTTCGGGGCAA GAGGGGCGGACATGCAACCAACTACCGGATCGTGGCCCCCAGGAGCCGGGACGAGAGAGGCTGAGGGTGGTGGCGGCCCCTGAGGCAGAC CAGGCCAGGCTTCCCCTCCTACCTGGACTCGGCCAGCTGCCTCCAGGGACCGCCCGTCCATATTTATTAATGTCCTCAGGGTCCCTTCTG CCATCTAGGCCTTAGGGGTAAGCAGGTCTCAGTCCTGGCACCATGCACATGTCTGAGGCTGAGCAAGGGCTGAGAGGAGAGGCTTGGGCC TCAGTTTCCCTCTGTGAAGTGGGGGGAGGCAGGCCTTCAAGGAGGGATAGAGGTACAAGGCTTCGTCTCATCTGCTGTCTGAGCATCCAG GCCCAAAGGCACTGAGGGAGTCAGGAGCTGGGGCTCGGCACATGCAGAGATGACAGGGCAGGGGGCAGTCTTCCTCCCCCTCCCCGACCA AACCTCGGGGAGCCCTCCTGTGCCCCTCCCTCCTTGTTGTCCAGTGCTGGGCTCCCCACCCCGAGGTCAGGCTGCCCAATCCTCTGACTG GATCACCGGGGGCTTCTTGCCTCAGTTCTTCCCTCTGAGCCCCCAGGCCCTCCCGCATCTCAGGTTGGGGATGGGGACATGGAGAGGAAG >43447_43447_1_KREMEN1-EMID1_KREMEN1_chr22_29494941_ENST00000327813_EMID1_chr22_29629368_ENST00000334018_length(amino acids)=286AA_BP=135 MAPPAARLALLSAAALTLAARPAPSPGLGPGPECFTANGADYRGTQNWTALQGGKPCLFWNETFQHPYNTLKYPNGEGGLGEHNYCRNPD GDVSPWCYVAEHEDGVYWKYCEIPACQRDPLLSNTFTETNNHWPQGPTGPPGPPGPMGPPGPPGPTGVPGSPGHIGPPGPTGPKGISGHP GEKGERGLRGEPGPQGSAGQRGEPGPKGDPGEKSHWGEGLHQLREALKILAERVLILETMIGLYEPELGSGAGPAGTGTPSLLRGKRGGH -------------------------------------------------------------- >43447_43447_2_KREMEN1-EMID1_KREMEN1_chr22_29494941_ENST00000327813_EMID1_chr22_29629368_ENST00000404755_length(transcript)=1464nt_BP=365nt GCACTGACGGCCCATGGCGCCGCCAGCCGCCCGCCTCGCCCTGCTCTCCGCCGCGGCGCTCACGCTGGCGGCCCGGCCCGCGCCTAGCCC CGGCCTCGGCCCCGGACCCGAGTGTTTCACAGCCAATGGTGCGGATTATAGGGGAACACAGAACTGGACAGCACTACAAGGCGGGAAGCC ATGTCTGTTTTGGAACGAGACTTTCCAGCATCCATACAACACTCTGAAATACCCCAACGGGGAGGGGGGCCTGGGTGAGCACAACTATTG CAGAAATCCAGATGGAGACGTGAGCCCCTGGTGCTATGTGGCAGAGCACGAGGATGGTGTCTACTGGAAGTACTGTGAGATACCTGCTTG CCAGAGAGACCCATTGCTGTCCAACACCTTCACTGAGACCAACAACCACTGGCCCCAGGGACCCACTGGGCCTCCAGGCCCTCCAGGGCC CATGGGTCCCCCTGGGCCTCCTGGCCCCACAGGTGTCCCTGGGAGTCCTGGTCACATAGGACTGCGTGGGGAGCCTGGCCCCCAAGGCTC TGCTGGGCAGCGGGGGGAACCTGGCCCTAAGGGAGACCCTGGTGAGAAGAGCCACTGGGGGGAGGGGTTGCACCAGCTACGCGAGGCTTT GAAGATTTTAGCTGAGAGGGTTTTAATCTTGGAAACAATGATTGGGCTCTATGAACCAGAGCTGGGGTCTGGGGCGGGCCCTGCCGGCAC AGGCACCCCCAGCCTCCTTCGGGGCAAGAGGGGCGGACATGCAACCAACTACCGGATCGTGGCCCCCAGGAGCCGGGACGAGAGAGGCTG AGGGTGGTGGCGGCCCCTGAGGCAGACCAGGCCAGGCTTCCCCTCCTACCTGGACTCGGCCAGCTGCCTCCAGGGACCGCCCGTCCATAT TTATTAATGTCCTCAGGGTCCCTTCTGCCATCTAGGCCTTAGGGGTAAGCAGGTCTCAGTCCTGGCACCATGCACATGTCTGAGGCTGAG CAAGGGCTGAGAGGAGAGGCTTGGGCCTCAGTTTCCCTCTGTGAAGTGGGGGGAGGCAGGCCTTCAAGGAGGGATAGAGGTACAAGGCTT CGTCTCATCTGCTGTCTGAGCATCCAGGCCCAAAGGCACTGAGGGAGTCAGGAGCTGGGGCTCGGCACATGCAGAGATGACAGGGCAGGG GGCAGTCTTCCTCCCCCTCCCCGACCAAACCTCGGGGAGCCCTCCTGTGCCCCTCCCTCCTTGTTGTCCAGTGCTGGGCTCCCCACCCCG AGGTCAGGCTGCCCAATCCTCTGACTGGATCACCGGGGGCTTCTTGCCTCAGTTCTTCCCTCTGAGCCCCCAGGCCCTCCCGCATCTCAG GTTGGGGATGGGGACATGGAGAGGAAGGGGCCGCCTACTCCTGCAAATGCTTGTGACAGATGCCAGGAGGTAGATGTGTGCTGGCCAATA >43447_43447_2_KREMEN1-EMID1_KREMEN1_chr22_29494941_ENST00000327813_EMID1_chr22_29629368_ENST00000404755_length(amino acids)=265AA_BP=135 MAPPAARLALLSAAALTLAARPAPSPGLGPGPECFTANGADYRGTQNWTALQGGKPCLFWNETFQHPYNTLKYPNGEGGLGEHNYCRNPD GDVSPWCYVAEHEDGVYWKYCEIPACQRDPLLSNTFTETNNHWPQGPTGPPGPPGPMGPPGPPGPTGVPGSPGHIGLRGEPGPQGSAGQR -------------------------------------------------------------- >43447_43447_3_KREMEN1-EMID1_KREMEN1_chr22_29494941_ENST00000327813_EMID1_chr22_29629368_ENST00000404820_length(transcript)=1533nt_BP=365nt GCACTGACGGCCCATGGCGCCGCCAGCCGCCCGCCTCGCCCTGCTCTCCGCCGCGGCGCTCACGCTGGCGGCCCGGCCCGCGCCTAGCCC CGGCCTCGGCCCCGGACCCGAGTGTTTCACAGCCAATGGTGCGGATTATAGGGGAACACAGAACTGGACAGCACTACAAGGCGGGAAGCC ATGTCTGTTTTGGAACGAGACTTTCCAGCATCCATACAACACTCTGAAATACCCCAACGGGGAGGGGGGCCTGGGTGAGCACAACTATTG CAGAAATCCAGATGGAGACGTGAGCCCCTGGTGCTATGTGGCAGAGCACGAGGATGGTGTCTACTGGAAGTACTGTGAGATACCTGCTTG CCAGAGAGACCCATTGCTGTCCAACACCTTCACTGAGACCAACAACCACTGGCCCCAGGGACCCACTGGGCCTCCAGGCCCTCCAGGGCC CATGGGTCCCCCTGGGCCTCCTGGCCCCACAGGTGTCCCTGGGAGTCCTGGTCACATAGGACCCCCAGGCCCCACTGGACCCAAAGGAAT CTCTGGCCACCCAGGAGAGAAGGGCGAGAGAGGACTGCGTGGGGAGCCTGGCCCCCAAGGCTCTGCTGGGCAGCGGGGGGAACCTGGCCC TAAGGGAGACCCTGGTGAGAAGAGCCACTGGGCTCCTAGCTTACAGAGCTTCCTGCAGCAGCAGGCTCAGCTGGAGCTCCTGGCCAGACG GGTCACCCTCCTGGAAGCCATCATCTGGCCAGAACCAGAGCTGGGGTCTGGGGCGGGCCCTGCCGGCACAGGCACCCCCAGCCTCCTTCG GGGCAAGAGGGGCGGACATGCAACCAACTACCGGATCGTGGCCCCCAGGAGCCGGGACGAGAGAGGCTGAGGGTGGTGGCGGCCCCTGAG GCAGACCAGGCCAGGCTTCCCCTCCTACCTGGACTCGGCCAGCTGCCTCCAGGGACCGCCCGTCCATATTTATTAATGTCCTCAGGGTCC CTTCTGCCATCTAGGCCTTAGGGGTAAGCAGGTCTCAGTCCTGGCACCATGCACATGTCTGAGGCTGAGCAAGGGCTGAGAGGAGAGGCT TGGGCCTCAGTTTCCCTCTGTGAAGTGGGGGGAGGCAGGCCTTCAAGGAGGGATAGAGGTACAAGGCTTCGTCTCATCTGCTGTCTGAGC ATCCAGGCCCAAAGGCACTGAGGGAGTCAGGAGCTGGGGCTCGGCACATGCAGAGATGACAGGGCAGGGGGCAGTCTTCCTCCCCCTCCC CGACCAAACCTCGGGGAGCCCTCCTGTGCCCCTCCCTCCTTGTTGTCCAGTGCTGGGCTCCCCACCCCGAGGTCAGGCTGCCCAATCCTC TGACTGGATCACCGGGGGCTTCTTGCCTCAGTTCTTCCCTCTGAGCCCCCAGGCCCTCCCGCATCTCAGGTTGGGGATGGGGACATGGAG AGGAAGGGGCCGCCTACTCCTGCAAATGCTTGTGACAGATGCCAGGAGGTAGATGTGTGCTGGCCAATAAAGGCCCCTACCTGATTCCCC >43447_43447_3_KREMEN1-EMID1_KREMEN1_chr22_29494941_ENST00000327813_EMID1_chr22_29629368_ENST00000404820_length(amino acids)=288AA_BP=135 MAPPAARLALLSAAALTLAARPAPSPGLGPGPECFTANGADYRGTQNWTALQGGKPCLFWNETFQHPYNTLKYPNGEGGLGEHNYCRNPD GDVSPWCYVAEHEDGVYWKYCEIPACQRDPLLSNTFTETNNHWPQGPTGPPGPPGPMGPPGPPGPTGVPGSPGHIGPPGPTGPKGISGHP GEKGERGLRGEPGPQGSAGQRGEPGPKGDPGEKSHWAPSLQSFLQQQAQLELLARRVTLLEAIIWPEPELGSGAGPAGTGTPSLLRGKRG -------------------------------------------------------------- >43447_43447_4_KREMEN1-EMID1_KREMEN1_chr22_29494941_ENST00000400335_EMID1_chr22_29629368_ENST00000334018_length(transcript)=1567nt_BP=405nt CGCGCTGCCCCCTTTACCCCGGGCCGCGCCCCGGGGCCCCGCACTGACGGCCCATGGCGCCGCCAGCCGCCCGCCTCGCCCTGCTCTCCG CCGCGGCGCTCACGCTGGCGGCCCGGCCCGCGCCTAGCCCCGGCCTCGGCCCCGGACCCGAGTGTTTCACAGCCAATGGTGCGGATTATA GGGGAACACAGAACTGGACAGCACTACAAGGCGGGAAGCCATGTCTGTTTTGGAACGAGACTTTCCAGCATCCATACAACACTCTGAAAT ACCCCAACGGGGAGGGGGGCCTGGGTGAGCACAACTATTGCAGAAATCCAGATGGAGACGTGAGCCCCTGGTGCTATGTGGCAGAGCACG AGGATGGTGTCTACTGGAAGTACTGTGAGATACCTGCTTGCCAGAGAGACCCATTGCTGTCCAACACCTTCACTGAGACCAACAACCACT GGCCCCAGGGACCCACTGGGCCTCCAGGCCCTCCAGGGCCCATGGGTCCCCCTGGGCCTCCTGGCCCCACAGGTGTCCCTGGGAGTCCTG GTCACATAGGACCCCCAGGCCCCACTGGACCCAAAGGAATCTCTGGCCACCCAGGAGAGAAGGGCGAGAGAGGACTGCGTGGGGAGCCTG GCCCCCAAGGCTCTGCTGGGCAGCGGGGGGAACCTGGCCCTAAGGGAGACCCTGGTGAGAAGAGCCACTGGGGGGAGGGGTTGCACCAGC TACGCGAGGCTTTGAAGATTTTAGCTGAGAGGGTTTTAATCTTGGAAACAATGATTGGGCTCTATGAACCAGAGCTGGGGTCTGGGGCGG GCCCTGCCGGCACAGGCACCCCCAGCCTCCTTCGGGGCAAGAGGGGCGGACATGCAACCAACTACCGGATCGTGGCCCCCAGGAGCCGGG ACGAGAGAGGCTGAGGGTGGTGGCGGCCCCTGAGGCAGACCAGGCCAGGCTTCCCCTCCTACCTGGACTCGGCCAGCTGCCTCCAGGGAC CGCCCGTCCATATTTATTAATGTCCTCAGGGTCCCTTCTGCCATCTAGGCCTTAGGGGTAAGCAGGTCTCAGTCCTGGCACCATGCACAT GTCTGAGGCTGAGCAAGGGCTGAGAGGAGAGGCTTGGGCCTCAGTTTCCCTCTGTGAAGTGGGGGGAGGCAGGCCTTCAAGGAGGGATAG AGGTACAAGGCTTCGTCTCATCTGCTGTCTGAGCATCCAGGCCCAAAGGCACTGAGGGAGTCAGGAGCTGGGGCTCGGCACATGCAGAGA TGACAGGGCAGGGGGCAGTCTTCCTCCCCCTCCCCGACCAAACCTCGGGGAGCCCTCCTGTGCCCCTCCCTCCTTGTTGTCCAGTGCTGG GCTCCCCACCCCGAGGTCAGGCTGCCCAATCCTCTGACTGGATCACCGGGGGCTTCTTGCCTCAGTTCTTCCCTCTGAGCCCCCAGGCCC TCCCGCATCTCAGGTTGGGGATGGGGACATGGAGAGGAAGGGGCCGCCTACTCCTGCAAATGCTTGTGACAGATGCCAGGAGGTAGATGT >43447_43447_4_KREMEN1-EMID1_KREMEN1_chr22_29494941_ENST00000400335_EMID1_chr22_29629368_ENST00000334018_length(amino acids)=286AA_BP=135 MAPPAARLALLSAAALTLAARPAPSPGLGPGPECFTANGADYRGTQNWTALQGGKPCLFWNETFQHPYNTLKYPNGEGGLGEHNYCRNPD GDVSPWCYVAEHEDGVYWKYCEIPACQRDPLLSNTFTETNNHWPQGPTGPPGPPGPMGPPGPPGPTGVPGSPGHIGPPGPTGPKGISGHP GEKGERGLRGEPGPQGSAGQRGEPGPKGDPGEKSHWGEGLHQLREALKILAERVLILETMIGLYEPELGSGAGPAGTGTPSLLRGKRGGH -------------------------------------------------------------- >43447_43447_5_KREMEN1-EMID1_KREMEN1_chr22_29494941_ENST00000400335_EMID1_chr22_29629368_ENST00000404755_length(transcript)=1504nt_BP=405nt CGCGCTGCCCCCTTTACCCCGGGCCGCGCCCCGGGGCCCCGCACTGACGGCCCATGGCGCCGCCAGCCGCCCGCCTCGCCCTGCTCTCCG CCGCGGCGCTCACGCTGGCGGCCCGGCCCGCGCCTAGCCCCGGCCTCGGCCCCGGACCCGAGTGTTTCACAGCCAATGGTGCGGATTATA GGGGAACACAGAACTGGACAGCACTACAAGGCGGGAAGCCATGTCTGTTTTGGAACGAGACTTTCCAGCATCCATACAACACTCTGAAAT ACCCCAACGGGGAGGGGGGCCTGGGTGAGCACAACTATTGCAGAAATCCAGATGGAGACGTGAGCCCCTGGTGCTATGTGGCAGAGCACG AGGATGGTGTCTACTGGAAGTACTGTGAGATACCTGCTTGCCAGAGAGACCCATTGCTGTCCAACACCTTCACTGAGACCAACAACCACT GGCCCCAGGGACCCACTGGGCCTCCAGGCCCTCCAGGGCCCATGGGTCCCCCTGGGCCTCCTGGCCCCACAGGTGTCCCTGGGAGTCCTG GTCACATAGGACTGCGTGGGGAGCCTGGCCCCCAAGGCTCTGCTGGGCAGCGGGGGGAACCTGGCCCTAAGGGAGACCCTGGTGAGAAGA GCCACTGGGGGGAGGGGTTGCACCAGCTACGCGAGGCTTTGAAGATTTTAGCTGAGAGGGTTTTAATCTTGGAAACAATGATTGGGCTCT ATGAACCAGAGCTGGGGTCTGGGGCGGGCCCTGCCGGCACAGGCACCCCCAGCCTCCTTCGGGGCAAGAGGGGCGGACATGCAACCAACT ACCGGATCGTGGCCCCCAGGAGCCGGGACGAGAGAGGCTGAGGGTGGTGGCGGCCCCTGAGGCAGACCAGGCCAGGCTTCCCCTCCTACC TGGACTCGGCCAGCTGCCTCCAGGGACCGCCCGTCCATATTTATTAATGTCCTCAGGGTCCCTTCTGCCATCTAGGCCTTAGGGGTAAGC AGGTCTCAGTCCTGGCACCATGCACATGTCTGAGGCTGAGCAAGGGCTGAGAGGAGAGGCTTGGGCCTCAGTTTCCCTCTGTGAAGTGGG GGGAGGCAGGCCTTCAAGGAGGGATAGAGGTACAAGGCTTCGTCTCATCTGCTGTCTGAGCATCCAGGCCCAAAGGCACTGAGGGAGTCA GGAGCTGGGGCTCGGCACATGCAGAGATGACAGGGCAGGGGGCAGTCTTCCTCCCCCTCCCCGACCAAACCTCGGGGAGCCCTCCTGTGC CCCTCCCTCCTTGTTGTCCAGTGCTGGGCTCCCCACCCCGAGGTCAGGCTGCCCAATCCTCTGACTGGATCACCGGGGGCTTCTTGCCTC AGTTCTTCCCTCTGAGCCCCCAGGCCCTCCCGCATCTCAGGTTGGGGATGGGGACATGGAGAGGAAGGGGCCGCCTACTCCTGCAAATGC >43447_43447_5_KREMEN1-EMID1_KREMEN1_chr22_29494941_ENST00000400335_EMID1_chr22_29629368_ENST00000404755_length(amino acids)=265AA_BP=135 MAPPAARLALLSAAALTLAARPAPSPGLGPGPECFTANGADYRGTQNWTALQGGKPCLFWNETFQHPYNTLKYPNGEGGLGEHNYCRNPD GDVSPWCYVAEHEDGVYWKYCEIPACQRDPLLSNTFTETNNHWPQGPTGPPGPPGPMGPPGPPGPTGVPGSPGHIGLRGEPGPQGSAGQR -------------------------------------------------------------- >43447_43447_6_KREMEN1-EMID1_KREMEN1_chr22_29494941_ENST00000400335_EMID1_chr22_29629368_ENST00000404820_length(transcript)=1573nt_BP=405nt CGCGCTGCCCCCTTTACCCCGGGCCGCGCCCCGGGGCCCCGCACTGACGGCCCATGGCGCCGCCAGCCGCCCGCCTCGCCCTGCTCTCCG CCGCGGCGCTCACGCTGGCGGCCCGGCCCGCGCCTAGCCCCGGCCTCGGCCCCGGACCCGAGTGTTTCACAGCCAATGGTGCGGATTATA GGGGAACACAGAACTGGACAGCACTACAAGGCGGGAAGCCATGTCTGTTTTGGAACGAGACTTTCCAGCATCCATACAACACTCTGAAAT ACCCCAACGGGGAGGGGGGCCTGGGTGAGCACAACTATTGCAGAAATCCAGATGGAGACGTGAGCCCCTGGTGCTATGTGGCAGAGCACG AGGATGGTGTCTACTGGAAGTACTGTGAGATACCTGCTTGCCAGAGAGACCCATTGCTGTCCAACACCTTCACTGAGACCAACAACCACT GGCCCCAGGGACCCACTGGGCCTCCAGGCCCTCCAGGGCCCATGGGTCCCCCTGGGCCTCCTGGCCCCACAGGTGTCCCTGGGAGTCCTG GTCACATAGGACCCCCAGGCCCCACTGGACCCAAAGGAATCTCTGGCCACCCAGGAGAGAAGGGCGAGAGAGGACTGCGTGGGGAGCCTG GCCCCCAAGGCTCTGCTGGGCAGCGGGGGGAACCTGGCCCTAAGGGAGACCCTGGTGAGAAGAGCCACTGGGCTCCTAGCTTACAGAGCT TCCTGCAGCAGCAGGCTCAGCTGGAGCTCCTGGCCAGACGGGTCACCCTCCTGGAAGCCATCATCTGGCCAGAACCAGAGCTGGGGTCTG GGGCGGGCCCTGCCGGCACAGGCACCCCCAGCCTCCTTCGGGGCAAGAGGGGCGGACATGCAACCAACTACCGGATCGTGGCCCCCAGGA GCCGGGACGAGAGAGGCTGAGGGTGGTGGCGGCCCCTGAGGCAGACCAGGCCAGGCTTCCCCTCCTACCTGGACTCGGCCAGCTGCCTCC AGGGACCGCCCGTCCATATTTATTAATGTCCTCAGGGTCCCTTCTGCCATCTAGGCCTTAGGGGTAAGCAGGTCTCAGTCCTGGCACCAT GCACATGTCTGAGGCTGAGCAAGGGCTGAGAGGAGAGGCTTGGGCCTCAGTTTCCCTCTGTGAAGTGGGGGGAGGCAGGCCTTCAAGGAG GGATAGAGGTACAAGGCTTCGTCTCATCTGCTGTCTGAGCATCCAGGCCCAAAGGCACTGAGGGAGTCAGGAGCTGGGGCTCGGCACATG CAGAGATGACAGGGCAGGGGGCAGTCTTCCTCCCCCTCCCCGACCAAACCTCGGGGAGCCCTCCTGTGCCCCTCCCTCCTTGTTGTCCAG TGCTGGGCTCCCCACCCCGAGGTCAGGCTGCCCAATCCTCTGACTGGATCACCGGGGGCTTCTTGCCTCAGTTCTTCCCTCTGAGCCCCC AGGCCCTCCCGCATCTCAGGTTGGGGATGGGGACATGGAGAGGAAGGGGCCGCCTACTCCTGCAAATGCTTGTGACAGATGCCAGGAGGT >43447_43447_6_KREMEN1-EMID1_KREMEN1_chr22_29494941_ENST00000400335_EMID1_chr22_29629368_ENST00000404820_length(amino acids)=288AA_BP=135 MAPPAARLALLSAAALTLAARPAPSPGLGPGPECFTANGADYRGTQNWTALQGGKPCLFWNETFQHPYNTLKYPNGEGGLGEHNYCRNPD GDVSPWCYVAEHEDGVYWKYCEIPACQRDPLLSNTFTETNNHWPQGPTGPPGPPGPMGPPGPPGPTGVPGSPGHIGPPGPTGPKGISGHP GEKGERGLRGEPGPQGSAGQRGEPGPKGDPGEKSHWAPSLQSFLQQQAQLELLARRVTLLEAIIWPEPELGSGAGPAGTGTPSLLRGKRG -------------------------------------------------------------- >43447_43447_7_KREMEN1-EMID1_KREMEN1_chr22_29494941_ENST00000400338_EMID1_chr22_29629368_ENST00000334018_length(transcript)=1567nt_BP=405nt CGCGCTGCCCCCTTTACCCCGGGCCGCGCCCCGGGGCCCCGCACTGACGGCCCATGGCGCCGCCAGCCGCCCGCCTCGCCCTGCTCTCCG CCGCGGCGCTCACGCTGGCGGCCCGGCCCGCGCCTAGCCCCGGCCTCGGCCCCGGACCCGAGTGTTTCACAGCCAATGGTGCGGATTATA GGGGAACACAGAACTGGACAGCACTACAAGGCGGGAAGCCATGTCTGTTTTGGAACGAGACTTTCCAGCATCCATACAACACTCTGAAAT ACCCCAACGGGGAGGGGGGCCTGGGTGAGCACAACTATTGCAGAAATCCAGATGGAGACGTGAGCCCCTGGTGCTATGTGGCAGAGCACG AGGATGGTGTCTACTGGAAGTACTGTGAGATACCTGCTTGCCAGAGAGACCCATTGCTGTCCAACACCTTCACTGAGACCAACAACCACT GGCCCCAGGGACCCACTGGGCCTCCAGGCCCTCCAGGGCCCATGGGTCCCCCTGGGCCTCCTGGCCCCACAGGTGTCCCTGGGAGTCCTG GTCACATAGGACCCCCAGGCCCCACTGGACCCAAAGGAATCTCTGGCCACCCAGGAGAGAAGGGCGAGAGAGGACTGCGTGGGGAGCCTG GCCCCCAAGGCTCTGCTGGGCAGCGGGGGGAACCTGGCCCTAAGGGAGACCCTGGTGAGAAGAGCCACTGGGGGGAGGGGTTGCACCAGC TACGCGAGGCTTTGAAGATTTTAGCTGAGAGGGTTTTAATCTTGGAAACAATGATTGGGCTCTATGAACCAGAGCTGGGGTCTGGGGCGG GCCCTGCCGGCACAGGCACCCCCAGCCTCCTTCGGGGCAAGAGGGGCGGACATGCAACCAACTACCGGATCGTGGCCCCCAGGAGCCGGG ACGAGAGAGGCTGAGGGTGGTGGCGGCCCCTGAGGCAGACCAGGCCAGGCTTCCCCTCCTACCTGGACTCGGCCAGCTGCCTCCAGGGAC CGCCCGTCCATATTTATTAATGTCCTCAGGGTCCCTTCTGCCATCTAGGCCTTAGGGGTAAGCAGGTCTCAGTCCTGGCACCATGCACAT GTCTGAGGCTGAGCAAGGGCTGAGAGGAGAGGCTTGGGCCTCAGTTTCCCTCTGTGAAGTGGGGGGAGGCAGGCCTTCAAGGAGGGATAG AGGTACAAGGCTTCGTCTCATCTGCTGTCTGAGCATCCAGGCCCAAAGGCACTGAGGGAGTCAGGAGCTGGGGCTCGGCACATGCAGAGA TGACAGGGCAGGGGGCAGTCTTCCTCCCCCTCCCCGACCAAACCTCGGGGAGCCCTCCTGTGCCCCTCCCTCCTTGTTGTCCAGTGCTGG GCTCCCCACCCCGAGGTCAGGCTGCCCAATCCTCTGACTGGATCACCGGGGGCTTCTTGCCTCAGTTCTTCCCTCTGAGCCCCCAGGCCC TCCCGCATCTCAGGTTGGGGATGGGGACATGGAGAGGAAGGGGCCGCCTACTCCTGCAAATGCTTGTGACAGATGCCAGGAGGTAGATGT >43447_43447_7_KREMEN1-EMID1_KREMEN1_chr22_29494941_ENST00000400338_EMID1_chr22_29629368_ENST00000334018_length(amino acids)=286AA_BP=135 MAPPAARLALLSAAALTLAARPAPSPGLGPGPECFTANGADYRGTQNWTALQGGKPCLFWNETFQHPYNTLKYPNGEGGLGEHNYCRNPD GDVSPWCYVAEHEDGVYWKYCEIPACQRDPLLSNTFTETNNHWPQGPTGPPGPPGPMGPPGPPGPTGVPGSPGHIGPPGPTGPKGISGHP GEKGERGLRGEPGPQGSAGQRGEPGPKGDPGEKSHWGEGLHQLREALKILAERVLILETMIGLYEPELGSGAGPAGTGTPSLLRGKRGGH -------------------------------------------------------------- >43447_43447_8_KREMEN1-EMID1_KREMEN1_chr22_29494941_ENST00000400338_EMID1_chr22_29629368_ENST00000404755_length(transcript)=1504nt_BP=405nt CGCGCTGCCCCCTTTACCCCGGGCCGCGCCCCGGGGCCCCGCACTGACGGCCCATGGCGCCGCCAGCCGCCCGCCTCGCCCTGCTCTCCG CCGCGGCGCTCACGCTGGCGGCCCGGCCCGCGCCTAGCCCCGGCCTCGGCCCCGGACCCGAGTGTTTCACAGCCAATGGTGCGGATTATA GGGGAACACAGAACTGGACAGCACTACAAGGCGGGAAGCCATGTCTGTTTTGGAACGAGACTTTCCAGCATCCATACAACACTCTGAAAT ACCCCAACGGGGAGGGGGGCCTGGGTGAGCACAACTATTGCAGAAATCCAGATGGAGACGTGAGCCCCTGGTGCTATGTGGCAGAGCACG AGGATGGTGTCTACTGGAAGTACTGTGAGATACCTGCTTGCCAGAGAGACCCATTGCTGTCCAACACCTTCACTGAGACCAACAACCACT GGCCCCAGGGACCCACTGGGCCTCCAGGCCCTCCAGGGCCCATGGGTCCCCCTGGGCCTCCTGGCCCCACAGGTGTCCCTGGGAGTCCTG GTCACATAGGACTGCGTGGGGAGCCTGGCCCCCAAGGCTCTGCTGGGCAGCGGGGGGAACCTGGCCCTAAGGGAGACCCTGGTGAGAAGA GCCACTGGGGGGAGGGGTTGCACCAGCTACGCGAGGCTTTGAAGATTTTAGCTGAGAGGGTTTTAATCTTGGAAACAATGATTGGGCTCT ATGAACCAGAGCTGGGGTCTGGGGCGGGCCCTGCCGGCACAGGCACCCCCAGCCTCCTTCGGGGCAAGAGGGGCGGACATGCAACCAACT ACCGGATCGTGGCCCCCAGGAGCCGGGACGAGAGAGGCTGAGGGTGGTGGCGGCCCCTGAGGCAGACCAGGCCAGGCTTCCCCTCCTACC TGGACTCGGCCAGCTGCCTCCAGGGACCGCCCGTCCATATTTATTAATGTCCTCAGGGTCCCTTCTGCCATCTAGGCCTTAGGGGTAAGC AGGTCTCAGTCCTGGCACCATGCACATGTCTGAGGCTGAGCAAGGGCTGAGAGGAGAGGCTTGGGCCTCAGTTTCCCTCTGTGAAGTGGG GGGAGGCAGGCCTTCAAGGAGGGATAGAGGTACAAGGCTTCGTCTCATCTGCTGTCTGAGCATCCAGGCCCAAAGGCACTGAGGGAGTCA GGAGCTGGGGCTCGGCACATGCAGAGATGACAGGGCAGGGGGCAGTCTTCCTCCCCCTCCCCGACCAAACCTCGGGGAGCCCTCCTGTGC CCCTCCCTCCTTGTTGTCCAGTGCTGGGCTCCCCACCCCGAGGTCAGGCTGCCCAATCCTCTGACTGGATCACCGGGGGCTTCTTGCCTC AGTTCTTCCCTCTGAGCCCCCAGGCCCTCCCGCATCTCAGGTTGGGGATGGGGACATGGAGAGGAAGGGGCCGCCTACTCCTGCAAATGC >43447_43447_8_KREMEN1-EMID1_KREMEN1_chr22_29494941_ENST00000400338_EMID1_chr22_29629368_ENST00000404755_length(amino acids)=265AA_BP=135 MAPPAARLALLSAAALTLAARPAPSPGLGPGPECFTANGADYRGTQNWTALQGGKPCLFWNETFQHPYNTLKYPNGEGGLGEHNYCRNPD GDVSPWCYVAEHEDGVYWKYCEIPACQRDPLLSNTFTETNNHWPQGPTGPPGPPGPMGPPGPPGPTGVPGSPGHIGLRGEPGPQGSAGQR -------------------------------------------------------------- >43447_43447_9_KREMEN1-EMID1_KREMEN1_chr22_29494941_ENST00000400338_EMID1_chr22_29629368_ENST00000404820_length(transcript)=1573nt_BP=405nt CGCGCTGCCCCCTTTACCCCGGGCCGCGCCCCGGGGCCCCGCACTGACGGCCCATGGCGCCGCCAGCCGCCCGCCTCGCCCTGCTCTCCG CCGCGGCGCTCACGCTGGCGGCCCGGCCCGCGCCTAGCCCCGGCCTCGGCCCCGGACCCGAGTGTTTCACAGCCAATGGTGCGGATTATA GGGGAACACAGAACTGGACAGCACTACAAGGCGGGAAGCCATGTCTGTTTTGGAACGAGACTTTCCAGCATCCATACAACACTCTGAAAT ACCCCAACGGGGAGGGGGGCCTGGGTGAGCACAACTATTGCAGAAATCCAGATGGAGACGTGAGCCCCTGGTGCTATGTGGCAGAGCACG AGGATGGTGTCTACTGGAAGTACTGTGAGATACCTGCTTGCCAGAGAGACCCATTGCTGTCCAACACCTTCACTGAGACCAACAACCACT GGCCCCAGGGACCCACTGGGCCTCCAGGCCCTCCAGGGCCCATGGGTCCCCCTGGGCCTCCTGGCCCCACAGGTGTCCCTGGGAGTCCTG GTCACATAGGACCCCCAGGCCCCACTGGACCCAAAGGAATCTCTGGCCACCCAGGAGAGAAGGGCGAGAGAGGACTGCGTGGGGAGCCTG GCCCCCAAGGCTCTGCTGGGCAGCGGGGGGAACCTGGCCCTAAGGGAGACCCTGGTGAGAAGAGCCACTGGGCTCCTAGCTTACAGAGCT TCCTGCAGCAGCAGGCTCAGCTGGAGCTCCTGGCCAGACGGGTCACCCTCCTGGAAGCCATCATCTGGCCAGAACCAGAGCTGGGGTCTG GGGCGGGCCCTGCCGGCACAGGCACCCCCAGCCTCCTTCGGGGCAAGAGGGGCGGACATGCAACCAACTACCGGATCGTGGCCCCCAGGA GCCGGGACGAGAGAGGCTGAGGGTGGTGGCGGCCCCTGAGGCAGACCAGGCCAGGCTTCCCCTCCTACCTGGACTCGGCCAGCTGCCTCC AGGGACCGCCCGTCCATATTTATTAATGTCCTCAGGGTCCCTTCTGCCATCTAGGCCTTAGGGGTAAGCAGGTCTCAGTCCTGGCACCAT GCACATGTCTGAGGCTGAGCAAGGGCTGAGAGGAGAGGCTTGGGCCTCAGTTTCCCTCTGTGAAGTGGGGGGAGGCAGGCCTTCAAGGAG GGATAGAGGTACAAGGCTTCGTCTCATCTGCTGTCTGAGCATCCAGGCCCAAAGGCACTGAGGGAGTCAGGAGCTGGGGCTCGGCACATG CAGAGATGACAGGGCAGGGGGCAGTCTTCCTCCCCCTCCCCGACCAAACCTCGGGGAGCCCTCCTGTGCCCCTCCCTCCTTGTTGTCCAG TGCTGGGCTCCCCACCCCGAGGTCAGGCTGCCCAATCCTCTGACTGGATCACCGGGGGCTTCTTGCCTCAGTTCTTCCCTCTGAGCCCCC AGGCCCTCCCGCATCTCAGGTTGGGGATGGGGACATGGAGAGGAAGGGGCCGCCTACTCCTGCAAATGCTTGTGACAGATGCCAGGAGGT >43447_43447_9_KREMEN1-EMID1_KREMEN1_chr22_29494941_ENST00000400338_EMID1_chr22_29629368_ENST00000404820_length(amino acids)=288AA_BP=135 MAPPAARLALLSAAALTLAARPAPSPGLGPGPECFTANGADYRGTQNWTALQGGKPCLFWNETFQHPYNTLKYPNGEGGLGEHNYCRNPD GDVSPWCYVAEHEDGVYWKYCEIPACQRDPLLSNTFTETNNHWPQGPTGPPGPPGPMGPPGPPGPTGVPGSPGHIGPPGPTGPKGISGHP GEKGERGLRGEPGPQGSAGQRGEPGPKGDPGEKSHWAPSLQSFLQQQAQLELLARRVTLLEAIIWPEPELGSGAGPAGTGTPSLLRGKRG -------------------------------------------------------------- >43447_43447_10_KREMEN1-EMID1_KREMEN1_chr22_29494941_ENST00000407188_EMID1_chr22_29629368_ENST00000334018_length(transcript)=1508nt_BP=346nt ATGGCGCCGCCAGCCGCCCGCCTCGCCCTGCTCTCCGCCGCGGCGCTCACGCTGGCGGCCCGGCCCGCGCCTAGCCCCGGCCTCGGCCCC GAGTGTTTCACAGCCAATGGTGCGGATTATAGGGGAACACAGAACTGGACAGCACTACAAGGCGGGAAGCCATGTCTGTTTTGGAACGAG ACTTTCCAGCATCCATACAACACTCTGAAATACCCCAACGGGGAGGGGGGCCTGGGTGAGCACAACTATTGCAGAAATCCAGATGGAGAC GTGAGCCCCTGGTGCTATGTGGCAGAGCACGAGGATGGTGTCTACTGGAAGTACTGTGAGATACCTGCTTGCCAGAGAGACCCATTGCTG TCCAACACCTTCACTGAGACCAACAACCACTGGCCCCAGGGACCCACTGGGCCTCCAGGCCCTCCAGGGCCCATGGGTCCCCCTGGGCCT CCTGGCCCCACAGGTGTCCCTGGGAGTCCTGGTCACATAGGACCCCCAGGCCCCACTGGACCCAAAGGAATCTCTGGCCACCCAGGAGAG AAGGGCGAGAGAGGACTGCGTGGGGAGCCTGGCCCCCAAGGCTCTGCTGGGCAGCGGGGGGAACCTGGCCCTAAGGGAGACCCTGGTGAG AAGAGCCACTGGGGGGAGGGGTTGCACCAGCTACGCGAGGCTTTGAAGATTTTAGCTGAGAGGGTTTTAATCTTGGAAACAATGATTGGG CTCTATGAACCAGAGCTGGGGTCTGGGGCGGGCCCTGCCGGCACAGGCACCCCCAGCCTCCTTCGGGGCAAGAGGGGCGGACATGCAACC AACTACCGGATCGTGGCCCCCAGGAGCCGGGACGAGAGAGGCTGAGGGTGGTGGCGGCCCCTGAGGCAGACCAGGCCAGGCTTCCCCTCC TACCTGGACTCGGCCAGCTGCCTCCAGGGACCGCCCGTCCATATTTATTAATGTCCTCAGGGTCCCTTCTGCCATCTAGGCCTTAGGGGT AAGCAGGTCTCAGTCCTGGCACCATGCACATGTCTGAGGCTGAGCAAGGGCTGAGAGGAGAGGCTTGGGCCTCAGTTTCCCTCTGTGAAG TGGGGGGAGGCAGGCCTTCAAGGAGGGATAGAGGTACAAGGCTTCGTCTCATCTGCTGTCTGAGCATCCAGGCCCAAAGGCACTGAGGGA GTCAGGAGCTGGGGCTCGGCACATGCAGAGATGACAGGGCAGGGGGCAGTCTTCCTCCCCCTCCCCGACCAAACCTCGGGGAGCCCTCCT GTGCCCCTCCCTCCTTGTTGTCCAGTGCTGGGCTCCCCACCCCGAGGTCAGGCTGCCCAATCCTCTGACTGGATCACCGGGGGCTTCTTG CCTCAGTTCTTCCCTCTGAGCCCCCAGGCCCTCCCGCATCTCAGGTTGGGGATGGGGACATGGAGAGGAAGGGGCCGCCTACTCCTGCAA >43447_43447_10_KREMEN1-EMID1_KREMEN1_chr22_29494941_ENST00000407188_EMID1_chr22_29629368_ENST00000334018_length(amino acids)=284AA_BP=133 MAPPAARLALLSAAALTLAARPAPSPGLGPECFTANGADYRGTQNWTALQGGKPCLFWNETFQHPYNTLKYPNGEGGLGEHNYCRNPDGD VSPWCYVAEHEDGVYWKYCEIPACQRDPLLSNTFTETNNHWPQGPTGPPGPPGPMGPPGPPGPTGVPGSPGHIGPPGPTGPKGISGHPGE KGERGLRGEPGPQGSAGQRGEPGPKGDPGEKSHWGEGLHQLREALKILAERVLILETMIGLYEPELGSGAGPAGTGTPSLLRGKRGGHAT -------------------------------------------------------------- >43447_43447_11_KREMEN1-EMID1_KREMEN1_chr22_29494941_ENST00000407188_EMID1_chr22_29629368_ENST00000404755_length(transcript)=1445nt_BP=346nt ATGGCGCCGCCAGCCGCCCGCCTCGCCCTGCTCTCCGCCGCGGCGCTCACGCTGGCGGCCCGGCCCGCGCCTAGCCCCGGCCTCGGCCCC GAGTGTTTCACAGCCAATGGTGCGGATTATAGGGGAACACAGAACTGGACAGCACTACAAGGCGGGAAGCCATGTCTGTTTTGGAACGAG ACTTTCCAGCATCCATACAACACTCTGAAATACCCCAACGGGGAGGGGGGCCTGGGTGAGCACAACTATTGCAGAAATCCAGATGGAGAC GTGAGCCCCTGGTGCTATGTGGCAGAGCACGAGGATGGTGTCTACTGGAAGTACTGTGAGATACCTGCTTGCCAGAGAGACCCATTGCTG TCCAACACCTTCACTGAGACCAACAACCACTGGCCCCAGGGACCCACTGGGCCTCCAGGCCCTCCAGGGCCCATGGGTCCCCCTGGGCCT CCTGGCCCCACAGGTGTCCCTGGGAGTCCTGGTCACATAGGACTGCGTGGGGAGCCTGGCCCCCAAGGCTCTGCTGGGCAGCGGGGGGAA CCTGGCCCTAAGGGAGACCCTGGTGAGAAGAGCCACTGGGGGGAGGGGTTGCACCAGCTACGCGAGGCTTTGAAGATTTTAGCTGAGAGG GTTTTAATCTTGGAAACAATGATTGGGCTCTATGAACCAGAGCTGGGGTCTGGGGCGGGCCCTGCCGGCACAGGCACCCCCAGCCTCCTT CGGGGCAAGAGGGGCGGACATGCAACCAACTACCGGATCGTGGCCCCCAGGAGCCGGGACGAGAGAGGCTGAGGGTGGTGGCGGCCCCTG AGGCAGACCAGGCCAGGCTTCCCCTCCTACCTGGACTCGGCCAGCTGCCTCCAGGGACCGCCCGTCCATATTTATTAATGTCCTCAGGGT CCCTTCTGCCATCTAGGCCTTAGGGGTAAGCAGGTCTCAGTCCTGGCACCATGCACATGTCTGAGGCTGAGCAAGGGCTGAGAGGAGAGG CTTGGGCCTCAGTTTCCCTCTGTGAAGTGGGGGGAGGCAGGCCTTCAAGGAGGGATAGAGGTACAAGGCTTCGTCTCATCTGCTGTCTGA GCATCCAGGCCCAAAGGCACTGAGGGAGTCAGGAGCTGGGGCTCGGCACATGCAGAGATGACAGGGCAGGGGGCAGTCTTCCTCCCCCTC CCCGACCAAACCTCGGGGAGCCCTCCTGTGCCCCTCCCTCCTTGTTGTCCAGTGCTGGGCTCCCCACCCCGAGGTCAGGCTGCCCAATCC TCTGACTGGATCACCGGGGGCTTCTTGCCTCAGTTCTTCCCTCTGAGCCCCCAGGCCCTCCCGCATCTCAGGTTGGGGATGGGGACATGG AGAGGAAGGGGCCGCCTACTCCTGCAAATGCTTGTGACAGATGCCAGGAGGTAGATGTGTGCTGGCCAATAAAGGCCCCTACCTGATTCC >43447_43447_11_KREMEN1-EMID1_KREMEN1_chr22_29494941_ENST00000407188_EMID1_chr22_29629368_ENST00000404755_length(amino acids)=263AA_BP=133 MAPPAARLALLSAAALTLAARPAPSPGLGPECFTANGADYRGTQNWTALQGGKPCLFWNETFQHPYNTLKYPNGEGGLGEHNYCRNPDGD VSPWCYVAEHEDGVYWKYCEIPACQRDPLLSNTFTETNNHWPQGPTGPPGPPGPMGPPGPPGPTGVPGSPGHIGLRGEPGPQGSAGQRGE -------------------------------------------------------------- >43447_43447_12_KREMEN1-EMID1_KREMEN1_chr22_29494941_ENST00000407188_EMID1_chr22_29629368_ENST00000404820_length(transcript)=1514nt_BP=346nt ATGGCGCCGCCAGCCGCCCGCCTCGCCCTGCTCTCCGCCGCGGCGCTCACGCTGGCGGCCCGGCCCGCGCCTAGCCCCGGCCTCGGCCCC GAGTGTTTCACAGCCAATGGTGCGGATTATAGGGGAACACAGAACTGGACAGCACTACAAGGCGGGAAGCCATGTCTGTTTTGGAACGAG ACTTTCCAGCATCCATACAACACTCTGAAATACCCCAACGGGGAGGGGGGCCTGGGTGAGCACAACTATTGCAGAAATCCAGATGGAGAC GTGAGCCCCTGGTGCTATGTGGCAGAGCACGAGGATGGTGTCTACTGGAAGTACTGTGAGATACCTGCTTGCCAGAGAGACCCATTGCTG TCCAACACCTTCACTGAGACCAACAACCACTGGCCCCAGGGACCCACTGGGCCTCCAGGCCCTCCAGGGCCCATGGGTCCCCCTGGGCCT CCTGGCCCCACAGGTGTCCCTGGGAGTCCTGGTCACATAGGACCCCCAGGCCCCACTGGACCCAAAGGAATCTCTGGCCACCCAGGAGAG AAGGGCGAGAGAGGACTGCGTGGGGAGCCTGGCCCCCAAGGCTCTGCTGGGCAGCGGGGGGAACCTGGCCCTAAGGGAGACCCTGGTGAG AAGAGCCACTGGGCTCCTAGCTTACAGAGCTTCCTGCAGCAGCAGGCTCAGCTGGAGCTCCTGGCCAGACGGGTCACCCTCCTGGAAGCC ATCATCTGGCCAGAACCAGAGCTGGGGTCTGGGGCGGGCCCTGCCGGCACAGGCACCCCCAGCCTCCTTCGGGGCAAGAGGGGCGGACAT GCAACCAACTACCGGATCGTGGCCCCCAGGAGCCGGGACGAGAGAGGCTGAGGGTGGTGGCGGCCCCTGAGGCAGACCAGGCCAGGCTTC CCCTCCTACCTGGACTCGGCCAGCTGCCTCCAGGGACCGCCCGTCCATATTTATTAATGTCCTCAGGGTCCCTTCTGCCATCTAGGCCTT AGGGGTAAGCAGGTCTCAGTCCTGGCACCATGCACATGTCTGAGGCTGAGCAAGGGCTGAGAGGAGAGGCTTGGGCCTCAGTTTCCCTCT GTGAAGTGGGGGGAGGCAGGCCTTCAAGGAGGGATAGAGGTACAAGGCTTCGTCTCATCTGCTGTCTGAGCATCCAGGCCCAAAGGCACT GAGGGAGTCAGGAGCTGGGGCTCGGCACATGCAGAGATGACAGGGCAGGGGGCAGTCTTCCTCCCCCTCCCCGACCAAACCTCGGGGAGC CCTCCTGTGCCCCTCCCTCCTTGTTGTCCAGTGCTGGGCTCCCCACCCCGAGGTCAGGCTGCCCAATCCTCTGACTGGATCACCGGGGGC TTCTTGCCTCAGTTCTTCCCTCTGAGCCCCCAGGCCCTCCCGCATCTCAGGTTGGGGATGGGGACATGGAGAGGAAGGGGCCGCCTACTC >43447_43447_12_KREMEN1-EMID1_KREMEN1_chr22_29494941_ENST00000407188_EMID1_chr22_29629368_ENST00000404820_length(amino acids)=286AA_BP=133 MAPPAARLALLSAAALTLAARPAPSPGLGPECFTANGADYRGTQNWTALQGGKPCLFWNETFQHPYNTLKYPNGEGGLGEHNYCRNPDGD VSPWCYVAEHEDGVYWKYCEIPACQRDPLLSNTFTETNNHWPQGPTGPPGPPGPMGPPGPPGPTGVPGSPGHIGPPGPTGPKGISGHPGE KGERGLRGEPGPQGSAGQRGEPGPKGDPGEKSHWAPSLQSFLQQQAQLELLARRVTLLEAIIWPEPELGSGAGPAGTGTPSLLRGKRGGH -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for KREMEN1-EMID1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for KREMEN1-EMID1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for KREMEN1-EMID1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
