
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:KTN1-CCT5 (FusionGDB2 ID:43742) |
Fusion Gene Summary for KTN1-CCT5 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: KTN1-CCT5 | Fusion gene ID: 43742 | Hgene | Tgene | Gene symbol | KTN1 | CCT5 | Gene ID | 3895 | 22948 |
| Gene name | kinectin 1 | chaperonin containing TCP1 subunit 5 | |
| Synonyms | CG1|KNT|MU-RMS-40.19 | CCT-epsilon|CCTE|HEL-S-69|PNAS-102|TCP-1-epsilon | |
| Cytomap | 14q22.3 | 5p15.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | kinectinCG-1 antigenkinesin receptor | T-complex protein 1 subunit epsilonchaperonin containing TCP1, subunit 5 (epsilon)epididymis secretory protein Li 69 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q86UP2 | P48643 | |
| Ensembl transtripts involved in fusion gene | ENST00000395311, ENST00000395314, ENST00000413890, ENST00000416613, ENST00000438792, ENST00000395308, ENST00000395309, ENST00000554507, ENST00000555573, | ENST00000503026, ENST00000506600, ENST00000515390, ENST00000515676, ENST00000280326, | |
| Fusion gene scores | * DoF score | 17 X 19 X 8=2584 | 58 X 14 X 17=13804 |
| # samples | 20 | 61 | |
| ** MAII score | log2(20/2584*10)=-3.6915341649192 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(61/13804*10)=-4.50013332598527 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: KTN1 [Title/Abstract] AND CCT5 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | KTN1(56151301)-CCT5(10250033), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | KTN1-CCT5 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. KTN1-CCT5 seems lost the major protein functional domain in Tgene partner, which is a cell metabolism gene due to the frame-shifted ORF. KTN1-CCT5 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across KTN1 (5'-gene) Fusion gene breakpoints across KTN1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
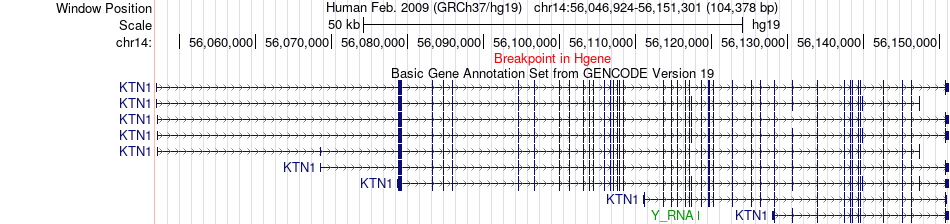 |
 Fusion gene breakpoints across CCT5 (3'-gene) Fusion gene breakpoints across CCT5 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
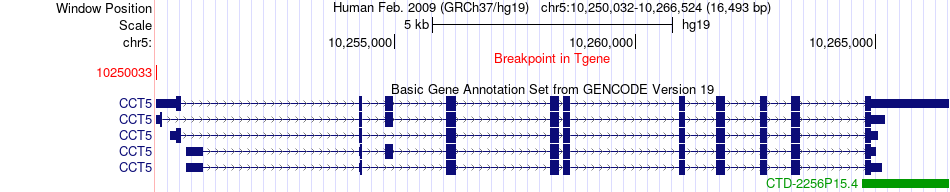 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | ACC | TCGA-OR-A5K5-01A | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
Top |
Fusion Gene ORF analysis for KTN1-CCT5 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000395311 | ENST00000503026 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000395311 | ENST00000506600 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000395311 | ENST00000515390 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000395311 | ENST00000515676 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000395314 | ENST00000503026 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000395314 | ENST00000506600 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000395314 | ENST00000515390 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000395314 | ENST00000515676 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000413890 | ENST00000503026 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000413890 | ENST00000506600 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000413890 | ENST00000515390 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000413890 | ENST00000515676 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000416613 | ENST00000503026 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000416613 | ENST00000506600 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000416613 | ENST00000515390 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000416613 | ENST00000515676 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000438792 | ENST00000503026 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000438792 | ENST00000506600 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000438792 | ENST00000515390 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000438792 | ENST00000515676 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| Frame-shift | ENST00000395311 | ENST00000280326 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| Frame-shift | ENST00000395314 | ENST00000280326 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| Frame-shift | ENST00000416613 | ENST00000280326 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| Frame-shift | ENST00000438792 | ENST00000280326 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| In-frame | ENST00000413890 | ENST00000280326 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| intron-3CDS | ENST00000395308 | ENST00000280326 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| intron-3CDS | ENST00000395309 | ENST00000280326 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| intron-3CDS | ENST00000554507 | ENST00000280326 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| intron-3CDS | ENST00000555573 | ENST00000280326 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000395308 | ENST00000503026 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000395308 | ENST00000506600 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000395308 | ENST00000515390 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000395308 | ENST00000515676 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000395309 | ENST00000503026 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000395309 | ENST00000506600 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000395309 | ENST00000515390 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000395309 | ENST00000515676 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000554507 | ENST00000503026 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000554507 | ENST00000506600 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000554507 | ENST00000515390 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000554507 | ENST00000515676 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000555573 | ENST00000503026 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000555573 | ENST00000506600 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000555573 | ENST00000515390 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000555573 | ENST00000515676 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000413890 | KTN1 | chr14 | 56151301 | - | ENST00000280326 | CCT5 | chr5 | 10250033 | + | 8254 | 4579 | 178 | 4098 | 1306 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000413890 | ENST00000280326 | KTN1 | chr14 | 56151301 | - | CCT5 | chr5 | 10250033 | + | 0.000397499 | 0.9996025 |
Top |
Fusion Genomic Features for KTN1-CCT5 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
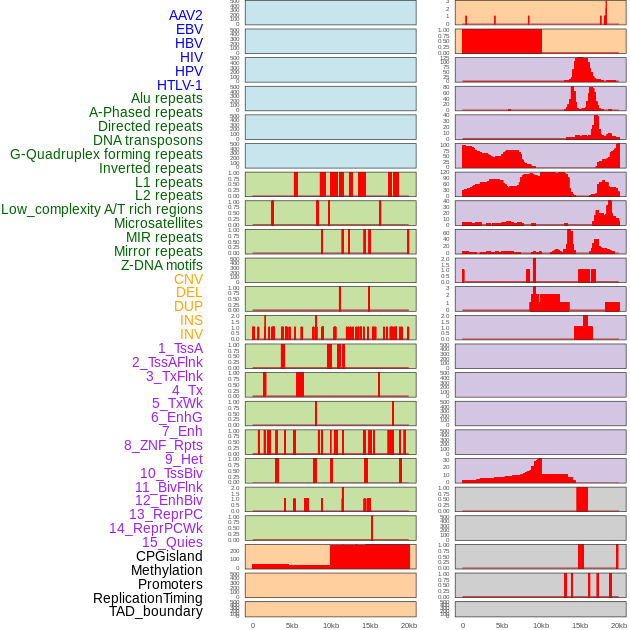 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for KTN1-CCT5 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr14:56151301/chr5:10250033) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| KTN1 | CCT5 |
| FUNCTION: Receptor for kinesin thus involved in kinesin-driven vesicle motility. Accumulates in integrin-based adhesion complexes (IAC) upon integrin aggregation by fibronectin. | FUNCTION: Component of the chaperonin-containing T-complex (TRiC), a molecular chaperone complex that assists the folding of proteins upon ATP hydrolysis (PubMed:25467444). The TRiC complex mediates the folding of WRAP53/TCAB1, thereby regulating telomere maintenance (PubMed:25467444). As part of the TRiC complex may play a role in the assembly of BBSome, a complex involved in ciliogenesis regulating transports vesicles to the cilia (PubMed:20080638). The TRiC complex plays a role in the folding of actin and tubulin (Probable). {ECO:0000269|PubMed:20080638, ECO:0000269|PubMed:25467444, ECO:0000305}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000395311 | - | 42 | 42 | 330_1356 | 1465 | 1307.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000395314 | - | 44 | 44 | 330_1356 | 1516 | 1358.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000413890 | - | 42 | 42 | 330_1356 | 1467 | 1307.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000416613 | - | 44 | 44 | 330_1356 | 1536 | 1321.3333333333333 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000438792 | - | 42 | 42 | 330_1356 | 1459 | 1301.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000395311 | - | 42 | 42 | 1_6 | 1465 | 1307.0 | Topological domain | Cytoplasmic |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000395311 | - | 42 | 42 | 30_1357 | 1465 | 1307.0 | Topological domain | Lumenal |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000395314 | - | 44 | 44 | 1_6 | 1516 | 1358.0 | Topological domain | Cytoplasmic |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000395314 | - | 44 | 44 | 30_1357 | 1516 | 1358.0 | Topological domain | Lumenal |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000413890 | - | 42 | 42 | 1_6 | 1467 | 1307.0 | Topological domain | Cytoplasmic |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000413890 | - | 42 | 42 | 30_1357 | 1467 | 1307.0 | Topological domain | Lumenal |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000416613 | - | 44 | 44 | 1_6 | 1536 | 1321.3333333333333 | Topological domain | Cytoplasmic |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000416613 | - | 44 | 44 | 30_1357 | 1536 | 1321.3333333333333 | Topological domain | Lumenal |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000438792 | - | 42 | 42 | 1_6 | 1459 | 1301.0 | Topological domain | Cytoplasmic |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000438792 | - | 42 | 42 | 30_1357 | 1459 | 1301.0 | Topological domain | Lumenal |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000395311 | - | 42 | 42 | 7_29 | 1465 | 1307.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000395314 | - | 44 | 44 | 7_29 | 1516 | 1358.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000413890 | - | 42 | 42 | 7_29 | 1467 | 1307.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000416613 | - | 44 | 44 | 7_29 | 1536 | 1321.3333333333333 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000438792 | - | 42 | 42 | 7_29 | 1459 | 1301.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000395308 | - | 1 | 43 | 330_1356 | 0 | 1307.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000395309 | - | 1 | 44 | 330_1356 | 0 | 1358.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000395308 | - | 1 | 43 | 1_6 | 0 | 1307.0 | Topological domain | Cytoplasmic |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000395308 | - | 1 | 43 | 30_1357 | 0 | 1307.0 | Topological domain | Lumenal |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000395309 | - | 1 | 44 | 1_6 | 0 | 1358.0 | Topological domain | Cytoplasmic |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000395309 | - | 1 | 44 | 30_1357 | 0 | 1358.0 | Topological domain | Lumenal |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000395308 | - | 1 | 43 | 7_29 | 0 | 1307.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Hgene | KTN1 | chr14:56151301 | chr5:10250033 | ENST00000395309 | - | 1 | 44 | 7_29 | 0 | 1358.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
Top |
Fusion Gene Sequence for KTN1-CCT5 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >43742_43742_1_KTN1-CCT5_KTN1_chr14_56151301_ENST00000413890_CCT5_chr5_10250033_ENST00000280326_length(transcript)=8254nt_BP=4579nt GAAGCCGCCCGTTTCCTGCCGAGCGGCGCGACGGCACCTGAGCGACTGCGGCGGCGGCGGCGGCGGCGGCGGCGCCTCGGAGCGGGCGGC CCGGGCTGTAGTGCCGGCGCCGCCGCGTCTTCCCGGTCTCCTTTCCCGGCCGCACAGGGTTTTATAGGATCACATTGACAAAAGTACCAT GGAGTTTTATGAGTCAGCATATTTTATTGTTCTTATTCCTTCAATAGTTATTACAGTAATTTTCCTCTTCTTCTGGCTTTTCATGAAAGA AACATTATATGATGAAGTTCTTGCAAAACAGAAAAGAGAACAAAAGCTTATTCCTACCAAAACAGATAAAAAGAAAGCAGAAAAGAAAAA GAATAAAAAGAAAGAAATCCAGAATGGAAACCTCCATGAATCCGACTCTGAGAGTGTACCTCGAGACTTTAAATTATCAGATGCTTTGGC AGTAGAAGATGATCAAGTTGCACCTGTTCCATTGAATGTCGTTGAAACTTCAAGTAGTGTTAGGGAAAGAAAAAAGAAGGAAAAGAAACA AAAGCCTGTGCTTGAAGAGCAGGTCATCAAAGAAAGTGACGCATCAAAGATTCCTGGCAAAAAAGTAGAACCTGTCCCAGTTACTAAACA GCCCACCCCTCCCTCTGAAGCAGCTGCCTCGAAGAAGAAACCAGGGCAGAAGAAGTCTAAAAATGGAAGCGATGACCAGGATAAAAAGGT GGAAACTCTCATGGTACCATCAAAAAGGCAAGAAGCATTGCCCCTCCACCAAGAGACTAAACAAGAAAGTGGATCAGGGAAGAAGAAAGC TTCATCAAAGAAACAAAAGACAGAAAATGTCTTCGTAGATGAACCCCTTATTCATGCAACTACTTATATTCCTTTGATGGATAATGCTGA CTCAAGTCCTGTGGTAGATAAGAGAGAGGTTATTGATTTGCTTAAACCTGACCAAGTAGAAGGGATCCAGAAATCTGGGACTAAAAAACT GAAGACCGAAACTGACAAAGAAAATGCTGAAGTGAAGTTTAAAGATTTTCTTCTGTCCTTGAAGACTATGATGTTTTCTGAAGATGAGGC TCTTTGTGTTGTAGACTTGCTAAAGGAGAAGTCTGGTGTAATACAAGATGCTTTAAAGAAGTCAAGTAAGGGAGAATTGACTACGCTTAT ACATCAGCTTCAAGAAAAGGACAAGTTACTCGCTGCTGTGAAGGAAGATGCTGCTGCTACAAAGGATCGGTGTAAGCAGTTAACCCAGGA AATGATGACAGAGAAAGAAAGAAGCAATGTGGTTATAACAAGGATGAAAGATCGAATTGGAACATTAGAAAAGGAACATAATGTATTTCA AAACAAAATACATGTCAGTTATCAAGAGACTCAACAGATGCAGATGAAGTTTCAGCAAGTTCGTGAGCAGATGGAGGCAGAGATAGCTCA CTTGAAGCAGGAAAATGGTATACTGAGAGATGCAGTCAGCAACACTACAAATCAACTGGAAAGCAAGCAGTCTGCAGAACTAAATAAACT ACGCCAGGATTATGCTAGGTTGGTGAATGAGCTGACTGAGAAAACAGGAAAGCTACAGCAAGAGGAAGTCCAAAAGAAGAATGCTGAGCA AGCAGCTACTCAGTTGAAGGTTCAACTACAAGAAGCTGAGAGAAGGTGGGAAGAAGTTCAGAGCTACATCAGGAAGAGAACAGCGGAACA TGAGGCAGCACAGCAAGATTTACAGAGTAAATTTGTGGCCAAAGAAAATGAAGTACAGAGTCTGCATAGTAAGCTTACAGATACCTTGGT ATCAAAACAACAGTTGGAGCAAAGACTAATGCAGTTAATGGAATCAGAGCAGAAAAGGGTGAACAAAGAAGAGTCTCTACAAATGCAGGT TCAGGATATTTTGGAGCAGAATGAGGCTTTGAAAGCTCAAATTCAGCAGTTCCATTCCCAGATAGCAGCCCAGACCTCCGCTTCAGTTCT AGCAGAAGAATTACATAAAGTGATTGCAGAAAAGGATAAGCAGATAAAACAGACTGAAGATTCTTTAGCAAGTGAACGTGATCGTTTAAC AAGTAAAGAAGAGGAACTTAAGGATATACAGAATATGAATTTCTTATTAAAAGCTGAAGTGCAGAAATTACAGGCCCTGGCAAATGAGCA GGCTGCTGCTGCACATGAATTGGAGAAGATGCAACAAAGTGTTTATGTTAAAGATGATAAAATAAGATTGCTGGAAGAGCAACTACAACA TGAAATTTCAAACAAAATGGAAGAATTTAAGATTCTAAATGACCAAAACAAAGCATTAAAATCAGAAGTTCAGAAGCTACAGACTCTTGT TTCTGAACAGCCTAATAAGGATGTTGTGGAACAAATGGAAAAATGCATTCAAGAAAAAGATGAGAAGTTAAAGACTGTGGAAGAATTACT TGAAACTGGACTTATTCAGGTGGCAACTAAAGAAGAGGAGCTGAATGCAATAAGAACAGAAAATTCATCTCTGACAAAAGAAGTTCAAGA CTTAAAAGCTAAGCAAAATGATCAGGTTTCTTTTGCCTCTCTAGTTGAAGAACTTAAGAAAGTGATCCATGAGAAAGATGGAAAGATCAA GTCTGTAGAAGAGCTTCTGGAGGCAGAACTTCTCAAAGTTGCTAACAAGGAGAAAACTGTTCAGTTATCTATCACTTCCAAAGTTCAGGA GCTTCAGAACTTATTAAAAGGAAAAGAGGAACAGATGAATACCATGAAGGCTGTTTTGGAAGAGAAAGAGAAAGACCTAGCCAATACAGG GAAGTGGTTACAGGATCTTCAAGAAGAAAATGAATCTTTAAAAGCACATGTTCAGGAAGTAGCACAACATAACTTGAAAGAGGCCTCTTC TGCATCACAGTTTGAAGAACTTGAGATTGTGTTGAAAGAAAAGGAAAATGAATTGAAGAGGTTAGAAGCCATGCTAAAAGAGAGGGAGAG TGATCTTTCTAGCAAAACACAGCTGTTACAGGATGTACAAGATGAAAACAAATTGTTTAAGTCCCAAATTGAGCAGCTTAAACAACAAAA CTACCAACAGGCATCTTCTTTTCCCCCTCATGAAGAATTATTAAAAGTAATTTCAGAAAGAGAGAAAGAAATAAGTGGTCTCTGGAATGA GTTAGATTCTTTGAAGGATGCAGTTGAACACCAGAGGAAGAAAAACAATGACCTTCGGGAGAAAAACTGGGAAGCAATGGAAGCATTGGC ATCAACTGAAAAAATGCTGCAGGACAAAGTGAACAAGACTTCCAAGGAAAGGCAGCAACAGGTGGAAGCTGTTGAGTTGGAGGCTAAAGA AGTTCTCAAAAAATTATTTCCAAAGGTGTCTGTCCCTTCTAATTTGAGTTATGGTGAATGGTTGCATGGATTTGAAAAAAAGGCAAAAGA ATGTATGGCTGGAACTTCAGGGTCAGAGGAGGTTAAGGTTCTAGAGCACAAGTTGAAAGAAGCTGATGAAATGCACACATTGTTACAGCT AGAGTGTGAAAAATACAAATCCGTCCTTGCAGAAACAGAAGGAATTTTACAGAAGCTACAGAGAAGTGTTGAGCAAGAAGAAAATAAATG GAAAGTTAAGGTCGATGAATCACACAAGACTATTAAACAGATGCAGTCATCATTTACATCTTCAGAACAAGAGCTAGAGCGATTAAGAAG CGAAAATAAGGATATTGAAAATCTGAGAAGAGAACGAGAACATTTGGAAATGGAACTAGAAAAGGCAGAGATGGAACGATCTACCTATGT TACAGAAGTCAGAGAGTTGAAGGCACAGTTAAATGAAACACTCACAAAACTTAGAACTGAACAAAATGAAAGACAGAAGGTAGCTGGTGA TTTGCATAAGGCTCAACAGTCACTGGAGCTTATCCAGTCAAAAATAGTAAAAGCTGCTGGAGACACTACTGTTATTGAAAATAGTGATGT TTCCCCAGAAACGGAGTCTTCTGAGAAGGAGACAATGTCTGTAAGTCTAAATCAGACTGTAACACAGTTACAGCAGTTGCTTCAGGCGGT AAACCAACAGCTCACAAAGGAGAAAGAGCACTACCAGGTGTTAGAGTGAAGTAATTGGGAAACTGTTCATTTGAGGATAAAAAAGGCATT GTATTATATTTTGCCAAATTAAAGCCTTATTTATGTTTTCACCCTTTCTACTTTGTCAGAAACACTGAACAGAGTTTTGTCTTTTCTAAT CCTTGTTAGACTACTGATTTAAAGAAGGAAAAAAAAAAGCCAACTCTGTAGACACCTTCAGAGTTTAGTTTTATAATAAAAACTGTTTGA ATAATTAGACCTTTACATTCCTGAAGATAAACATGTAATCTTTTATCTTATTTTGCTCAATAAAATTGTTCAGAAGATCAAAGTGGTAAA GACAATGTAAAATTTAACATTTTAATACTGATGTTGTACACTGTTTTACTTAACATTTTGGGAAGTAACTGCCTCTGACTTCAACTCAAG AAAACACTTTTTTGTTGCTAATGTAATCGGTTTTTGTAATGGCGTCAGCAAATAAAAGGATGCTTATTATTCAAACTTGAAAAAAAAAAA AAACCGGAAATGGGTCCTACCATCTTCTCGGAGCCGGAGTGCGAAGAAATAAAGAAATAGTGCTTTAAGTCAATGAATTCCTCCTTGGGA CCCACTATCGAGAAACTATCAGTGGTAACGTTTTAAAAAATGACAAATTCAATCTGCTCTTGACTTGTGTGTCCTAAGATTTCCACTAAG TGTCTTCAAACCTCCCCCTCCCCGGCTTCCTGGATAATAGAAGTTCCCGAAGGCCGCCGATTCCAGAAGATACTGTCTGGCGTGAAATTA GTCTCAGTAGAAACATAAGTCCCGCGCGTCTTGTGCTGCGCGTGCGCAAGCTTTTGGGCCCTCCCGAGAAAGGGAAGTGCATTCTCGCTT CCGTAGCGGTCTCCGCCGGTTGGGGGGAAGTAATTCCGGTTGTTGCACCATGGCGTCCATGGGGACCCTCGCCTTCGATGAATATGGGCG CCCTTTCCTCATCATCAAGGATCAGGACCGCAAGTCCCGTCTTATGGGACTTGAGGCCCTCAAGTCTCATATAATGGCAGCAAAGGCTGT AGCAAATACAATGAGAACATCACTTGGACCAAATGGGCTTGATAAGATGATGGTGGATAAGGATGGAGATGTGACTGTAACTAATGATGG GGCCACCATCTTAAGCATGATGGATGTTGATCATCAGATTGCCAAGCTGATGGTGGAACTGTCCAAGTCTCAGGATGATGAAATTGGAGA TGGAACCACAGGAGTGGTTGTCCTGGCTGGTGCCTTGTTAGAAGAAGCGGAGCAATTGCTAGACCGAGGCATTCACCCAATCAGAATAGC CGATGGCTATGAGCAGGCTGCTCGTGTTGCTATTGAACACCTGGACAAGATCAGCGATAGCGTCCTTGTTGACATAAAGGACACCGAACC CCTGATTCAGACAGCAAAAACCACGCTGGGCTCCAAAGTGGTCAACAGTTGTCACCGACAGATGGCTGAGATTGCTGTGAATGCCGTCCT CACTGTAGCAGATATGGAGCGGAGAGACGTTGACTTTGAGCTTATCAAAGTAGAAGGCAAAGTGGGCGGCAGGCTGGAGGACACTAAACT GATTAAGGGCGTGATTGTGGACAAGGATTTCAGTCACCCACAGATGCCAAAAAAAGTGGAAGATGCGAAGATTGCAATTCTCACATGTCC ATTTGAACCACCCAAACCAAAAACAAAGCATAAGCTGGATGTGACCTCTGTCGAAGATTATAAAGCCCTTCAGAAATACGAAAAGGAGAA ATTTGAAGAGATGATTCAACAAATTAAAGAGACTGGTGCTAACCTAGCAATTTGTCAGTGGGGCTTTGATGATGAAGCAAATCACTTACT TCTTCAGAACAACTTGCCTGCGGTTCGCTGGGTAGGAGGACCTGAAATTGAGCTGATTGCCATCGCAACAGGAGGGCGGATCGTCCCCAG GTTCTCAGAGCTCACAGCCGAGAAGCTGGGCTTTGCTGGTCTTGTACAGGAGATCTCATTTGGGACAACTAAGGATAAAATGCTGGTCAT CGAGCAGTGTAAGAACTCCAGAGCTGTAACCATTTTTATTAGAGGAGGAAATAAGATGATCATTGAGGAGGCGAAACGATCCCTTCACGA TGCTTTGTGTGTCATCCGGAACCTCATCCGCGATAATCGTGTGGTGTATGGAGGAGGGGCTGCTGAGATATCCTGTGCCCTGGCAGTTAG CCAAGAGGCGGATAAGTGCCCCACCTTAGAACAGTATGCCATGAGAGCGTTTGCCGACGCACTGGAGGTCATCCCCATGGCCCTCTCTGA AAACAGTGGCATGAATCCCATCCAGACTATGACCGAAGTCCGAGCCAGACAGGTGAAGGAGATGAACCCTGCTCTTGGCATCGACTGTTT GCACAAGGGGACAAATGATATGAAGCAACAGCATGTCATAGAAACCTTGATTGGCAAAAAGCAACAGATATCTCTTGCAACACAAATGGT TAGAATGATTTTGAAGATTGATGACATTCGTAAGCCTGGAGAATCTGAAGAATGAAGACATTGAGAAAACTATGTAGCAAGATCCACTTC TGTGATTAAGTAAATGGATGTCTCGTGATGCATCTACAGTTATTTATTGTTACATCCTTTTCCAGACACTGTAGATGCTATAATAAAAAT AGCTGTTTGGTAACCATAGTTTCACTTGTTCAAAGCTGTGTAATCGTGGGGGTACCATCTCAACTGCTTTTGTATTCATTGTATTAAAAG AATCTGTTTAAACAACCTTTATCTTCTCTTCGGGTTTAAGAAACGTTTATTGTAACAGTAATTAAATGCTGCCTTAATTGAAGGGGTTTG GGTGGATTTTTTTTTCTCAAAATAAGCTGTAGGGACTATTTTAACAGCTTAAACAGGAGCTCTCAAGATGCACTTTCATATTGAGAGGAA TATGGGCTTGATCCTCTTCCTATCTAAATGGGTGGGCCATTTGATTGTAGAGGGTCCACCACAGAATTATGGGATGCCTTAAGTGCTGTT ACTAGGTTGCTCACAGCCTAACCTGGCGTGTTGTTTAGGGCTGATGGAGACCCATGTGAGCCTTTGCTTTCCTCTGGCCCCGGCCCCACC CTGAACACAGCTCATACACAGAATCAGGACCAGCATGTGCAGAGCTGGCCACCAGCACAGGCTTAGGGCAGTTCAGAACCCACTTGTTTC CCTATCAGAGGGACACAGTGAAGTGGAGGTTAAAGTAAATTACAGGAAATAAGGGAGAAATCTTGCAGTTACCATGTTCAGATAGAGTGA CTGAAATTAATTGTACTTACTAAAGTATTAACTAGCTAACAGTGATGGGCCAAGACGCTCCGAGAACTCTACCGGGATTGTCTGTTCTGA CAACCCAGTGAGGCAGATACACTTTCTTACTGCTCACATCTTACAGGTGAGTACTCATAATTGGCCAGCATCTCACCACCAGCAAGTAGT AGGGCCAGGTCAATCCCAGGCAGTCTGACCCCAGAGTGGCCCAGCTCATCCCCTACTCTGTTATTTGCTTGTTAATGATTCTCTAGATTT TCTAAAATAATGTTTCTTAGCATTGTGATGATAAAGCTCATGATGAACTTTATCACTAGTTATGCCACCTTAACTAGTCAGATTTCCTAG AATTAGGAAATGGTGACTCTTGTCTAAATTTGGTTAAGTGATGAATTTGGGTTACCGTCTCATGTGAACCTGGAGATTCACCAGTCTTAA CTTTTGGGTCATTGTGTTTTCTCTACATTCATGCATTGGATGTTTTGCTAAATAACTCCTGTGGATTTAGGAATGTGTGCTAATAGCAAT CTTCCTAATTTTCATGTTTATATGGAACTATGCAGTTGAGTATTGAAAGCTTTAAACTGAGTTTATTTACAAGGACTGAGTCTAGCCTAC AGAGAACATACAGCAGCCTTCTTTGGACCACAGTCTTATCCGAGGGGTCTGTGGTTGTATCAGAAGAGCCACTAAACCAATCCCCCTTTC CAAAATTGAACCTCACAGACGTTCCTGTTTTTTGTGATTGAGAAACTGGTCAATGAACAGGAAGACTTAGAGATGTTTCACAAGCCTTTG >43742_43742_1_KTN1-CCT5_KTN1_chr14_56151301_ENST00000413890_CCT5_chr5_10250033_ENST00000280326_length(amino acids)=1306AA_BP= MEFYESAYFIVLIPSIVITVIFLFFWLFMKETLYDEVLAKQKREQKLIPTKTDKKKAEKKKNKKKEIQNGNLHESDSESVPRDFKLSDAL AVEDDQVAPVPLNVVETSSSVRERKKKEKKQKPVLEEQVIKESDASKIPGKKVEPVPVTKQPTPPSEAAASKKKPGQKKSKNGSDDQDKK VETLMVPSKRQEALPLHQETKQESGSGKKKASSKKQKTENVFVDEPLIHATTYIPLMDNADSSPVVDKREVIDLLKPDQVEGIQKSGTKK LKTETDKENAEVKFKDFLLSLKTMMFSEDEALCVVDLLKEKSGVIQDALKKSSKGELTTLIHQLQEKDKLLAAVKEDAAATKDRCKQLTQ EMMTEKERSNVVITRMKDRIGTLEKEHNVFQNKIHVSYQETQQMQMKFQQVREQMEAEIAHLKQENGILRDAVSNTTNQLESKQSAELNK LRQDYARLVNELTEKTGKLQQEEVQKKNAEQAATQLKVQLQEAERRWEEVQSYIRKRTAEHEAAQQDLQSKFVAKENEVQSLHSKLTDTL VSKQQLEQRLMQLMESEQKRVNKEESLQMQVQDILEQNEALKAQIQQFHSQIAAQTSASVLAEELHKVIAEKDKQIKQTEDSLASERDRL TSKEEELKDIQNMNFLLKAEVQKLQALANEQAAAAHELEKMQQSVYVKDDKIRLLEEQLQHEISNKMEEFKILNDQNKALKSEVQKLQTL VSEQPNKDVVEQMEKCIQEKDEKLKTVEELLETGLIQVATKEEELNAIRTENSSLTKEVQDLKAKQNDQVSFASLVEELKKVIHEKDGKI KSVEELLEAELLKVANKEKTVQLSITSKVQELQNLLKGKEEQMNTMKAVLEEKEKDLANTGKWLQDLQEENESLKAHVQEVAQHNLKEAS SASQFEELEIVLKEKENELKRLEAMLKERESDLSSKTQLLQDVQDENKLFKSQIEQLKQQNYQQASSFPPHEELLKVISEREKEISGLWN ELDSLKDAVEHQRKKNNDLREKNWEAMEALASTEKMLQDKVNKTSKERQQQVEAVELEAKEVLKKLFPKVSVPSNLSYGEWLHGFEKKAK ECMAGTSGSEEVKVLEHKLKEADEMHTLLQLECEKYKSVLAETEGILQKLQRSVEQEENKWKVKVDESHKTIKQMQSSFTSSEQELERLR SENKDIENLRREREHLEMELEKAEMERSTYVTEVRELKAQLNETLTKLRTEQNERQKVAGDLHKAQQSLELIQSKIVKAAGDTTVIENSD -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for KTN1-CCT5 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for KTN1-CCT5 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for KTN1-CCT5 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
