
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:LAMA5-IRF9 (FusionGDB2 ID:43905) |
Fusion Gene Summary for LAMA5-IRF9 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: LAMA5-IRF9 | Fusion gene ID: 43905 | Hgene | Tgene | Gene symbol | LAMA5 | IRF9 | Gene ID | 3911 | 10379 |
| Gene name | laminin subunit alpha 5 | interferon regulatory factor 9 | |
| Synonyms | - | IRF-9|ISGF3|ISGF3G|p48 | |
| Cytomap | 20q13.33 | 14q12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | laminin subunit alpha-5laminin alpha-5 chainlaminin-10 subunit alphalaminin-11 subunit alphalaminin-15 subunit alpha | interferon regulatory factor 9IFN-alpha-responsive transcription factor subunitISGF-3 gammaISGF3 p48 subunitinterferon-stimulated gene factor 3 gammainterferon-stimulated transcription factor 3, gamma (48kD)interferon-stimulated transcription factor | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | O15230 | Q00978 | |
| Ensembl transtripts involved in fusion gene | ENST00000252999, ENST00000370677, ENST00000370692, ENST00000492698, | ENST00000557894, ENST00000396864, | |
| Fusion gene scores | * DoF score | 20 X 20 X 14=5600 | 2 X 2 X 2=8 |
| # samples | 28 | 2 | |
| ** MAII score | log2(28/5600*10)=-4.32192809488736 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(2/8*10)=1.32192809488736 | |
| Context | PubMed: LAMA5 [Title/Abstract] AND IRF9 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | LAMA5(60907427)-IRF9(24632174), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | LAMA5 | GO:0016477 | cell migration | 11821406 |
| Hgene | LAMA5 | GO:0034446 | substrate adhesion-dependent cell spreading | 16236823 |
 Fusion gene breakpoints across LAMA5 (5'-gene) Fusion gene breakpoints across LAMA5 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
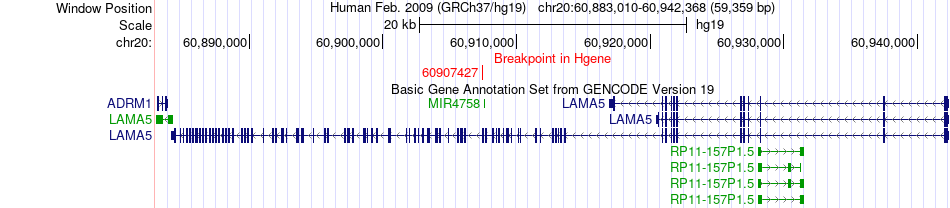 |
 Fusion gene breakpoints across IRF9 (3'-gene) Fusion gene breakpoints across IRF9 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
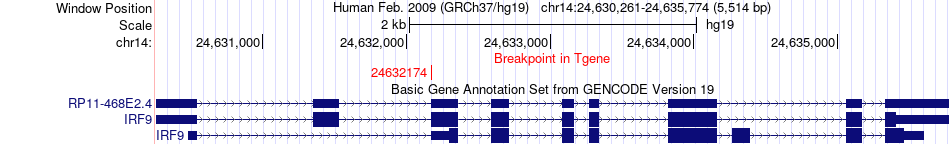 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | ESCA | TCGA-R6-A6Y2 | LAMA5 | chr20 | 60907427 | - | IRF9 | chr14 | 24632174 | + |
Top |
Fusion Gene ORF analysis for LAMA5-IRF9 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000252999 | ENST00000557894 | LAMA5 | chr20 | 60907427 | - | IRF9 | chr14 | 24632174 | + |
| In-frame | ENST00000252999 | ENST00000396864 | LAMA5 | chr20 | 60907427 | - | IRF9 | chr14 | 24632174 | + |
| intron-3CDS | ENST00000370677 | ENST00000396864 | LAMA5 | chr20 | 60907427 | - | IRF9 | chr14 | 24632174 | + |
| intron-3CDS | ENST00000370692 | ENST00000396864 | LAMA5 | chr20 | 60907427 | - | IRF9 | chr14 | 24632174 | + |
| intron-3CDS | ENST00000492698 | ENST00000396864 | LAMA5 | chr20 | 60907427 | - | IRF9 | chr14 | 24632174 | + |
| intron-5UTR | ENST00000370677 | ENST00000557894 | LAMA5 | chr20 | 60907427 | - | IRF9 | chr14 | 24632174 | + |
| intron-5UTR | ENST00000370692 | ENST00000557894 | LAMA5 | chr20 | 60907427 | - | IRF9 | chr14 | 24632174 | + |
| intron-5UTR | ENST00000492698 | ENST00000557894 | LAMA5 | chr20 | 60907427 | - | IRF9 | chr14 | 24632174 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000252999 | LAMA5 | chr20 | 60907427 | - | ENST00000396864 | IRF9 | chr14 | 24632174 | + | 4990 | 3619 | 67 | 4620 | 1517 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000252999 | ENST00000396864 | LAMA5 | chr20 | 60907427 | - | IRF9 | chr14 | 24632174 | + | 0.003689858 | 0.9963102 |
Top |
Fusion Genomic Features for LAMA5-IRF9 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| LAMA5 | chr20 | 60907427 | - | IRF9 | chr14 | 24632174 | + | 0.002227954 | 0.99777204 |
| LAMA5 | chr20 | 60907427 | - | IRF9 | chr14 | 24632174 | + | 0.002227954 | 0.99777204 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
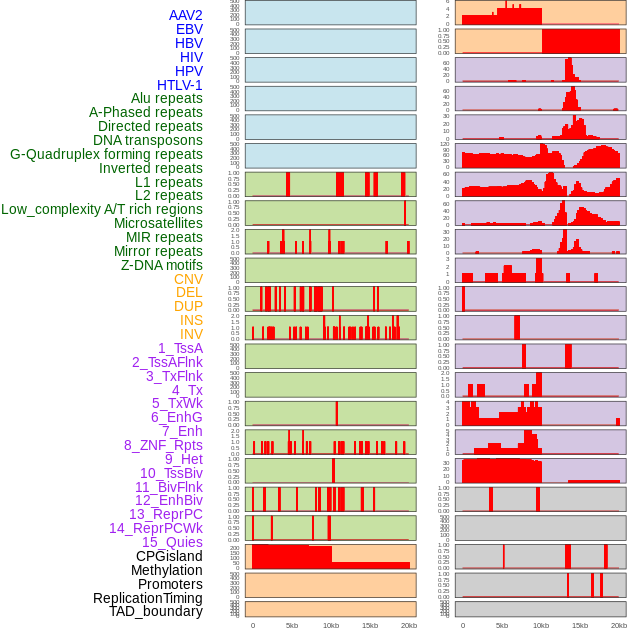 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
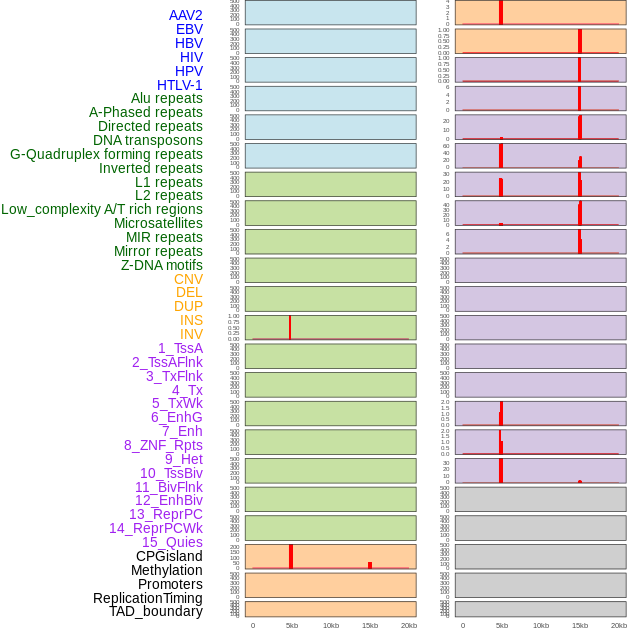 |
Top |
Fusion Protein Features for LAMA5-IRF9 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr20:60907427/chr14:24632174) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| LAMA5 | IRF9 |
| FUNCTION: Binding to cells via a high affinity receptor, laminin is thought to mediate the attachment, migration and organization of cells into tissues during embryonic development by interacting with other extracellular matrix components. | FUNCTION: Transcription factor that plays an essential role in anti-viral immunity. It mediates signaling by type I IFNs (IFN-alpha and IFN-beta). Following type I IFN binding to cell surface receptors, Jak kinases (TYK2 and JAK1) are activated, leading to tyrosine phosphorylation of STAT1 and STAT2. IRF9/ISGF3G associates with the phosphorylated STAT1:STAT2 dimer to form a complex termed ISGF3 transcription factor, that enters the nucleus. ISGF3 binds to the IFN stimulated response element (ISRE) to activate the transcription of interferon stimulated genes, which drive the cell in an antiviral state. {ECO:0000269|PubMed:30143481}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 300_358 | 1184 | 3696.0 | Domain | Laminin EGF-like 1 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 359_428 | 1184 | 3696.0 | Domain | Laminin EGF-like 2 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 41_299 | 1184 | 3696.0 | Domain | Laminin N-terminal |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 429_474 | 1184 | 3696.0 | Domain | Laminin EGF-like 3 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 494_540 | 1184 | 3696.0 | Domain | Laminin EGF-like 4 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 541_586 | 1184 | 3696.0 | Domain | Laminin EGF-like 5 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 587_631 | 1184 | 3696.0 | Domain | Laminin EGF-like 6 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 632_676 | 1184 | 3696.0 | Domain | Laminin EGF-like 7 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 677_722 | 1184 | 3696.0 | Domain | Laminin EGF-like 8 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 723_775 | 1184 | 3696.0 | Domain | Laminin EGF-like 9 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 776_828 | 1184 | 3696.0 | Domain | Laminin EGF-like 10 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 829_850 | 1184 | 3696.0 | Domain | Laminin EGF-like 11%3B truncated |
| Tgene | IRF9 | chr20:60907427 | chr14:24632174 | ENST00000396864 | 1 | 9 | 181_187 | 60 | 394.0 | Compositional bias | Note=Poly-Ser |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 2203_2221 | 1184 | 3696.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 2335_2466 | 1184 | 3696.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 2510_2670 | 1184 | 3696.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 1438_1483 | 1184 | 3696.0 | Domain | Laminin EGF-like 12 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 1484_1527 | 1184 | 3696.0 | Domain | Laminin EGF-like 13 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 1528_1576 | 1184 | 3696.0 | Domain | Laminin EGF-like 14 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 1577_1627 | 1184 | 3696.0 | Domain | Laminin EGF-like 15 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 1628_1637 | 1184 | 3696.0 | Domain | Laminin EGF-like 16%3B first part |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 1641_1830 | 1184 | 3696.0 | Domain | Laminin IV type A |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 1831_1863 | 1184 | 3696.0 | Domain | Laminin EGF-like 16%3B second part |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 1864_1912 | 1184 | 3696.0 | Domain | Laminin EGF-like 17 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 1913_1968 | 1184 | 3696.0 | Domain | Laminin EGF-like 18 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 1969_2022 | 1184 | 3696.0 | Domain | Laminin EGF-like 19 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 2023_2069 | 1184 | 3696.0 | Domain | Laminin EGF-like 20 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 2070_2116 | 1184 | 3696.0 | Domain | Laminin EGF-like 21 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 2117_2166 | 1184 | 3696.0 | Domain | Laminin EGF-like 22 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 2736_2929 | 1184 | 3696.0 | Domain | Laminin G-like 1 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 2941_3115 | 1184 | 3696.0 | Domain | Laminin G-like 2 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 3124_3292 | 1184 | 3696.0 | Domain | Laminin G-like 3 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 3340_3513 | 1184 | 3696.0 | Domain | Laminin G-like 4 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 3520_3692 | 1184 | 3696.0 | Domain | Laminin G-like 5 |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 1722_1724 | 1184 | 3696.0 | Motif | Cell attachment site |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 1838_1840 | 1184 | 3696.0 | Motif | Cell attachment site |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 2167_2735 | 1184 | 3696.0 | Region | Note=Domain II and I |
| Hgene | LAMA5 | chr20:60907427 | chr14:24632174 | ENST00000252999 | - | 28 | 80 | 851_1437 | 1184 | 3696.0 | Region | Note=Domain IV 1 (domain IV B) |
| Tgene | IRF9 | chr20:60907427 | chr14:24632174 | ENST00000396864 | 1 | 9 | 9_116 | 60 | 394.0 | DNA binding | IRF tryptophan pentad repeat |
Top |
Fusion Gene Sequence for LAMA5-IRF9 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >43905_43905_1_LAMA5-IRF9_LAMA5_chr20_60907427_ENST00000252999_IRF9_chr14_24632174_ENST00000396864_length(transcript)=4990nt_BP=3619nt AGACCCGCCGGGCTCCCGCCGCGCGCGCTGTCCCTGGAGCTCGGGGACGCGGCCCGGAGCCGGGAAGATGGCGAAGCGGCTCTGCGCGGG GAGCGCACTGTGTGTTCGCGGCCCCCGGGGCCCCGCGCCGCTGCTGCTGGTCGGGCTGGCGCTGCTGGGCGCGGCGCGGGCGCGGGAGGA GGCGGGCGGCGGCTTCAGCCTGCACCCGCCCTACTTCAACCTGGCCGAGGGCGCCCGCATCGCCGCCTCCGCGACCTGCGGAGAGGAGGC CCCGGCGCGCGGCTCCCCGCGCCCCACCGAGGACCTTTACTGCAAGCTGGTAGGGGGCCCCGTGGCCGGCGGCGACCCCAACCAGACCAT CCGGGGCCAGTACTGTGACATCTGCACGGCTGCCAACAGCAACAAGGCACACCCCGCGAGCAATGCCATCGATGGCACGGAGCGCTGGTG GCAGAGTCCACCGCTGTCCCGCGGCCTGGAGTACAACGAGGTCAACGTCACCCTGGACCTGGGCCAGGTCTTCCACGTGGCCTACGTCCT CATCAAGTTTGCCAACTCACCCCGGCCGGACCTCTGGGTGCTGGAGCGGTCCATGGACTTCGGCCGCACCTACCAGCCCTGGCAGTTCTT TGCCTCCTCCAAGAGGGACTGTCTGGAGCGGTTCGGGCCACAGACGCTGGAGCGCATCACACGGGACGACGCGGCCATCTGCACCACCGA GTACTCACGCATCGTGCCCCTGGAGAACGGAGAGATCGTGGTGTCCCTGGTGAACGGACGTCCGGGCGCCATGAATTTCTCCTACTCGCC GCTGCTACGTGAGTTCACCAAGGCCACCAACGTCCGCCTGCGCTTCCTGCGTACCAACACGCTGCTGGGCCATCTCATGGGGAAGGCGCT GCGGGACCCCACGGTCACCCGCCGGTATTATTACAGCATCAAGGATATCAGCATCGGAGGCCGCTGTGTCTGCCACGGCCACGCGGATGC CTGCGATGCCAAAGACCCCACGGACCCGTTCAGGCTGCAGTGCACCTGCCAGCACAACACCTGCGGGGGCACCTGCGACCGCTGCTGCCC CGGCTTCAATCAGCAGCCGTGGAAGCCTGCGACTGCCAACAGTGCCAACGAGTGCCAGTCCTGTAACTGCTACGGCCATGCCACCGACTG TTACTACGACCCTGAGGTGGACCGGCGCCGCGCCAGCCAGAGCCTGGATGGCACCTATCAGGGTGGGGGTGTCTGTATCGACTGCCAGCA CCACACCACCGGCGTCAACTGTGAGCGCTGCCTGCCCGGCTTCTACCGCTCTCCCAACCACCCTCTCGACTCGCCCCACGTCTGCCGCCG CTGCAACTGCGAGTCCGACTTCACGGATGGCACCTGCGAGGACCTGACGGGTCGATGCTACTGCCGGCCCAACTTCTCTGGGGAGCGGTG TGACGTGTGTGCCGAGGGCTTCACGGGCTTCCCAAGCTGCTACCCGACGCCCTCGTCCTCCAATGACACCAGGGAGCAGGTGCTGCCAGC CGGCCAGATTGTGAATTGTGACTGCAGCGCGGCAGGGACCCAGGGCAACGCCTGCCGGAAGGACCCAAGGGTGGGACGCTGTCTGTGCAA ACCCAACTTCCAAGGCACCCATTGTGAGCTCTGCGCGCCAGGGTTCTACGGCCCCGGCTGCCAGCCCTGCCAGTGTTCCAGCCCTGGAGT GGCCGATGACCGCTGTGACCCTGACACAGGCCAGTGCAGGTGCCGAGTGGGCTTCGAGGGGGCCACATGTGATCGCTGTGCCCCCGGCTA CTTTCACTTCCCTCTCTGCCAGTTGTGTGGCTGCAGCCCTGCAGGAACCTTGCCCGAGGGCTGCGATGAGGCCGGCCGCTGCCTATGCCA GCCTGAGTTTGCTGGACCTCATTGTGACCGGTGCCGCCCTGGCTACCATGGTTTCCCCAACTGCCAAGCATGCACCTGCGACCCTCGGGG AGCCCTGGACCAGCTCTGTGGGGCGGGAGGTTTGTGCCGCTGCCGCCCCGGCTACACAGGCACTGCCTGCCAGGAATGCAGCCCCGGCTT TCACGGCTTCCCCAGCTGTGTCCCCTGCCACTGCTCTGCTGAAGGCTCCCTGCACGCAGCCTGTGACCCCCGGAGTGGGCAGTGCAGCTG CCGGCCCCGTGTGACGGGGCTGCGGTGTGACACATGTGTGCCCGGTGCCTACAACTTCCCCTACTGCGAAGCTGGCTCTTGCCACCCTGC CGGTCTGGCCCCAGTGGATCCTGCCCTTCCTGAGGCACAGGTTCCCTGTATGTGCCGGGCTCACGTGGAGGGGCCGAGCTGTGACCGCTG CAAACCTGGGTTCTGGGGACTGAGCCCCAGCAACCCCGAGGGCTGTACCCGCTGCAGCTGCGACCTCAGGGGCACACTGGGTGGAGTTGC TGAGTGCCAGCCGGGCACCGGCCAGTGCTTCTGCAAGCCCCACGTGTGCGGCCAGGCCTGCGCGTCCTGCAAGGATGGCTTCTTTGGACT GGATCAGGCTGACTATTTTGGCTGCCGCAGCTGCCGGTGTGACATTGGCGGTGCACTGGGCCAGAGCTGTGAACCGAGGACGGGCGTCTG CCGGTGCCGCCCCAACACCCAGGGCCCCACCTGCAGCGAGCCTGCGAGGGACCACTACCTCCCGGACCTGCACCACCTGCGCCTGGAGCT GGAGGAGGCTGCCACACCTGAGGGTCACGCCGTGCGCTTTGGCTTCAACCCCCTCGAGTTCGAGAACTTCAGCTGGAGGGGCTACGCGCA GATGGCACCTGTCCAGCCCAGGATCGTGGCCAGGCTGAACCTGACCTCCCCTGACCTTTTCTGGCTCGTCTTCCGATACGTCAACCGGGG GGCCATGAGTGTGAGCGGGCGGGTCTCTGTGCGAGAGGAGGGCAGGTCGGCCACCTGCGCCAACTGCACAGCACAGAGTCAGCCCGTGGC CTTCCCACCCAGCACGGAGCCTGCCTTCATCACCGTGCCCCAGAGGGGCTTCGGAGAGCCCTTTGTGCTGAACCCTGGCACCTGGGCCCT GCGTGTGGAGGCCGAAGGGGTGCTCCTGGACTACGTGGTTCTGCTGCCTAGCGCATACTACGAGGCGGCGCTCCTGCAGCTGCGGGTGAC TGAGGCCTGCACATACCGTCCCTCTGCCCAGCAGTCTGGCGACAACTGCCTCCTCTACACACACCTCCCCCTGGATGGCTTCCCCTCGGC CGCCGGGCTGGAGGCCCTGTGTCGCCAGGACAACAGCCTGCCCCGGCCCTGCCCCACGGAGCAGCTCAGCCCGTCGCACCCGCCACTGAT CACCTGCACGGGCAGTGATGTGGACGTCCAGCTTCAAGTGGCAGTGCCACAGCCAGGCCGCTATGCCCTAGTGGTGGAGTACGCCAATGA GGATGCCCGCCAGGAGGTGGGCGTGGCCGTGCACACCCCACAGCGGGCCCCCCAGCAGGGGCTGCTCTCCCTGCACCCCTGCCTGTACAG CACCCTGTGCCGGGGCACTGCCCGGGATACCCAGGACCACCTGGCTGTCTTCCACCTGGACTCGGAGGCCAGCGTGAGGCTCACAGCCGA ACAGGCACGCTTCTTCCTGGCCTGGGCAATATTTAAGGGAAAGTATAAGGAGGGGGACACAGGAGGTCCAGCTGTCTGGAAGACTCGCCT GCGCTGTGCACTCAACAAGAGTTCTGAATTTAAGGAGGTTCCTGAGAGGGGCCGCATGGATGTTGCTGAGCCCTACAAGGTGTATCAGTT GCTGCCACCAGGAATCGTCTCTGGCCAGCCAGGGACTCAGAAAGTACCATCAAAGCGACAGCACAGTTCTGTGTCCTCTGAGAGGAAGGA GGAAGAGGATGCCATGCAGAACTGCACACTCAGTCCCTCTGTGCTCCAGGACTCCCTCAATAATGAGGAGGAGGGGGCCAGTGGGGGAGC AGTCCATTCAGACATTGGGAGCAGCAGCAGCAGCAGCAGCCCTGAGCCACAGGAAGTTACAGACACAACTGAGGCCCCCTTTCAAGGGGA TCAGAGGTCCCTGGAGTTTCTGCTTCCTCCAGAGCCAGACTACTCACTGCTGCTCACCTTCATCTACAACGGGCGCGTGGTGGGCGAGGC CCAGGTGCAAAGCCTGGATTGCCGCCTTGTGGCTGAGCCCTCAGGCTCTGAGAGCAGCATGGAGCAGGTGCTGTTCCCCAAGCCTGGCCC ACTGGAGCCCACGCAGCGCCTGCTGAGCCAGCTTGAGAGGGGCATCCTAGTGGCCAGCAACCCCCGAGGCCTCTTCGTGCAGCGCCTTTG CCCCATCCCCATCTCCTGGAATGCACCCCAGGCTCCACCTGGGCCAGGCCCGCATCTGCTGCCCAGCAACGAGTGCGTGGAGCTCTTCAG AACCGCCTACTTCTGCAGAGACTTGGTCAGGTACTTTCAGGGCCTGGGCCCCCCACCGAAGTTCCAGGTAACACTGAATTTCTGGGAAGA GAGCCATGGCTCCAGCCATACTCCACAGAATCTTATCACAGTGAAGATGGAGCAGGCCTTTGCCCGATACTTGCTGGAGCAGACTCCAGA GCAGCAGGCAGCCATTCTGTCCCTGGTGTAGAGCCTGGGGGACCCATCTTCCACCTCACCTCTTTGTTCTTCCTGTCTCCTTTGAAGTAG ACTCATTCTTCACACGATTGACCTGTCCTCTTTGTGATAATTCTCAGTAGTTGTCCGTGATAATCGTGTCCTGAAAATCCTCGCACACAC TGGCTGGTGGAGAACTCAAGGCTAATTTTTTATCCTTTTTTTTTTTTAATTTTGAGATATACGCCCTCTTTCATCTGTAAGGGACTAGGA AATTCCAAATGGTGTGAACCCAGGGGGCCTTTCCCTCTTCCCTGACCTCCCAACTCTAAAGCCAAGCACTTTATATTTTCCTCTTAGATA >43905_43905_1_LAMA5-IRF9_LAMA5_chr20_60907427_ENST00000252999_IRF9_chr14_24632174_ENST00000396864_length(amino acids)=1517AA_BP=1184 MAKRLCAGSALCVRGPRGPAPLLLVGLALLGAARAREEAGGGFSLHPPYFNLAEGARIAASATCGEEAPARGSPRPTEDLYCKLVGGPVA GGDPNQTIRGQYCDICTAANSNKAHPASNAIDGTERWWQSPPLSRGLEYNEVNVTLDLGQVFHVAYVLIKFANSPRPDLWVLERSMDFGR TYQPWQFFASSKRDCLERFGPQTLERITRDDAAICTTEYSRIVPLENGEIVVSLVNGRPGAMNFSYSPLLREFTKATNVRLRFLRTNTLL GHLMGKALRDPTVTRRYYYSIKDISIGGRCVCHGHADACDAKDPTDPFRLQCTCQHNTCGGTCDRCCPGFNQQPWKPATANSANECQSCN CYGHATDCYYDPEVDRRRASQSLDGTYQGGGVCIDCQHHTTGVNCERCLPGFYRSPNHPLDSPHVCRRCNCESDFTDGTCEDLTGRCYCR PNFSGERCDVCAEGFTGFPSCYPTPSSSNDTREQVLPAGQIVNCDCSAAGTQGNACRKDPRVGRCLCKPNFQGTHCELCAPGFYGPGCQP CQCSSPGVADDRCDPDTGQCRCRVGFEGATCDRCAPGYFHFPLCQLCGCSPAGTLPEGCDEAGRCLCQPEFAGPHCDRCRPGYHGFPNCQ ACTCDPRGALDQLCGAGGLCRCRPGYTGTACQECSPGFHGFPSCVPCHCSAEGSLHAACDPRSGQCSCRPRVTGLRCDTCVPGAYNFPYC EAGSCHPAGLAPVDPALPEAQVPCMCRAHVEGPSCDRCKPGFWGLSPSNPEGCTRCSCDLRGTLGGVAECQPGTGQCFCKPHVCGQACAS CKDGFFGLDQADYFGCRSCRCDIGGALGQSCEPRTGVCRCRPNTQGPTCSEPARDHYLPDLHHLRLELEEAATPEGHAVRFGFNPLEFEN FSWRGYAQMAPVQPRIVARLNLTSPDLFWLVFRYVNRGAMSVSGRVSVREEGRSATCANCTAQSQPVAFPPSTEPAFITVPQRGFGEPFV LNPGTWALRVEAEGVLLDYVVLLPSAYYEAALLQLRVTEACTYRPSAQQSGDNCLLYTHLPLDGFPSAAGLEALCRQDNSLPRPCPTEQL SPSHPPLITCTGSDVDVQLQVAVPQPGRYALVVEYANEDARQEVGVAVHTPQRAPQQGLLSLHPCLYSTLCRGTARDTQDHLAVFHLDSE ASVRLTAEQARFFLAWAIFKGKYKEGDTGGPAVWKTRLRCALNKSSEFKEVPERGRMDVAEPYKVYQLLPPGIVSGQPGTQKVPSKRQHS SVSSERKEEEDAMQNCTLSPSVLQDSLNNEEEGASGGAVHSDIGSSSSSSSPEPQEVTDTTEAPFQGDQRSLEFLLPPEPDYSLLLTFIY NGRVVGEAQVQSLDCRLVAEPSGSESSMEQVLFPKPGPLEPTQRLLSQLERGILVASNPRGLFVQRLCPIPISWNAPQAPPGPGPHLLPS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for LAMA5-IRF9 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for LAMA5-IRF9 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for LAMA5-IRF9 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
