
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:LAMTOR1-UVRAG (FusionGDB2 ID:44006) |
Fusion Gene Summary for LAMTOR1-UVRAG |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: LAMTOR1-UVRAG | Fusion gene ID: 44006 | Hgene | Tgene | Gene symbol | LAMTOR1 | UVRAG | Gene ID | 55004 | 7405 |
| Gene name | late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 | UV radiation resistance associated | |
| Synonyms | C11orf59|PDRO|Ragulator1|p18|p27RF-Rho | DHTX|VPS38|p63 | |
| Cytomap | 11q13.4 | 11q13.5 | |
| Type of gene | protein-coding | protein-coding | |
| Description | ragulator complex protein LAMTOR1RhoA activator C11orf59late endosomal/lysosomal adaptor and MAPK and MTOR activator 1lipid raft adaptor protein p18p27Kip1-releasing factor from RhoAp27kip1 releasing factor from RhoAprotein associated with DRMs and | UV radiation resistance-associated gene proteinbeclin 1 binding proteindisrupted in heterotaxy | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | Q6IAA8 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000278671, ENST00000545249, ENST00000539797, ENST00000535107, ENST00000538404, | ENST00000528420, ENST00000525872, ENST00000531818, ENST00000532130, ENST00000533454, ENST00000538870, ENST00000539288, ENST00000356136, | |
| Fusion gene scores | * DoF score | 4 X 4 X 4=64 | 28 X 21 X 11=6468 |
| # samples | 4 | 36 | |
| ** MAII score | log2(4/64*10)=-0.678071905112638 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(36/6468*10)=-4.16725086714399 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: LAMTOR1 [Title/Abstract] AND UVRAG [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | LAMTOR1(71809335)-UVRAG(75562928), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | UVRAG | GO:0071900 | regulation of protein serine/threonine kinase activity | 22542840 |
| Tgene | UVRAG | GO:0097352 | autophagosome maturation | 28306502 |
| Tgene | UVRAG | GO:0097680 | double-strand break repair via classical nonhomologous end joining | 22542840 |
 Fusion gene breakpoints across LAMTOR1 (5'-gene) Fusion gene breakpoints across LAMTOR1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
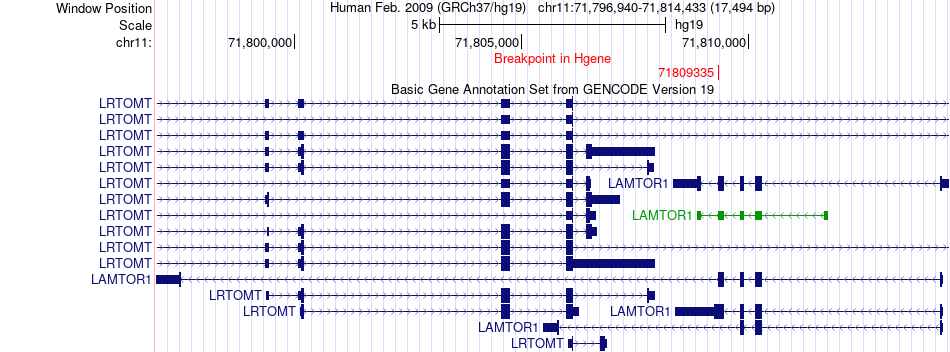 |
 Fusion gene breakpoints across UVRAG (3'-gene) Fusion gene breakpoints across UVRAG (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
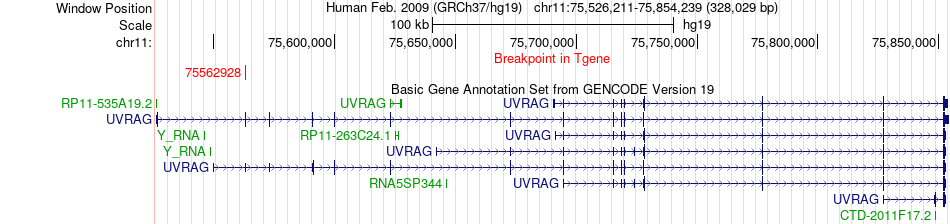 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | HNSC | TCGA-UF-A7JV-01A | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
Top |
Fusion Gene ORF analysis for LAMTOR1-UVRAG |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000278671 | ENST00000528420 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| 5CDS-5UTR | ENST00000545249 | ENST00000528420 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| 5CDS-intron | ENST00000278671 | ENST00000525872 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| 5CDS-intron | ENST00000278671 | ENST00000531818 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| 5CDS-intron | ENST00000278671 | ENST00000532130 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| 5CDS-intron | ENST00000278671 | ENST00000533454 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| 5CDS-intron | ENST00000278671 | ENST00000538870 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| 5CDS-intron | ENST00000278671 | ENST00000539288 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| 5CDS-intron | ENST00000545249 | ENST00000525872 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| 5CDS-intron | ENST00000545249 | ENST00000531818 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| 5CDS-intron | ENST00000545249 | ENST00000532130 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| 5CDS-intron | ENST00000545249 | ENST00000533454 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| 5CDS-intron | ENST00000545249 | ENST00000538870 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| 5CDS-intron | ENST00000545249 | ENST00000539288 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| 5UTR-3CDS | ENST00000539797 | ENST00000356136 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| 5UTR-5UTR | ENST00000539797 | ENST00000528420 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| 5UTR-intron | ENST00000539797 | ENST00000525872 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| 5UTR-intron | ENST00000539797 | ENST00000531818 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| 5UTR-intron | ENST00000539797 | ENST00000532130 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| 5UTR-intron | ENST00000539797 | ENST00000533454 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| 5UTR-intron | ENST00000539797 | ENST00000538870 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| 5UTR-intron | ENST00000539797 | ENST00000539288 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| In-frame | ENST00000278671 | ENST00000356136 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| In-frame | ENST00000545249 | ENST00000356136 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| intron-3CDS | ENST00000535107 | ENST00000356136 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| intron-3CDS | ENST00000538404 | ENST00000356136 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| intron-5UTR | ENST00000535107 | ENST00000528420 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| intron-5UTR | ENST00000538404 | ENST00000528420 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| intron-intron | ENST00000535107 | ENST00000525872 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| intron-intron | ENST00000535107 | ENST00000531818 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| intron-intron | ENST00000535107 | ENST00000532130 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| intron-intron | ENST00000535107 | ENST00000533454 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| intron-intron | ENST00000535107 | ENST00000538870 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| intron-intron | ENST00000535107 | ENST00000539288 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| intron-intron | ENST00000538404 | ENST00000525872 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| intron-intron | ENST00000538404 | ENST00000531818 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| intron-intron | ENST00000538404 | ENST00000532130 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| intron-intron | ENST00000538404 | ENST00000533454 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| intron-intron | ENST00000538404 | ENST00000538870 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
| intron-intron | ENST00000538404 | ENST00000539288 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000545249 | LAMTOR1 | chr11 | 71809335 | - | ENST00000356136 | UVRAG | chr11 | 75562928 | + | 4180 | 415 | 22 | 2397 | 791 |
| ENST00000278671 | LAMTOR1 | chr11 | 71809335 | - | ENST00000356136 | UVRAG | chr11 | 75562928 | + | 4321 | 556 | 163 | 2538 | 791 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000545249 | ENST00000356136 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + | 0.002280787 | 0.9977192 |
| ENST00000278671 | ENST00000356136 | LAMTOR1 | chr11 | 71809335 | - | UVRAG | chr11 | 75562928 | + | 0.002310854 | 0.9976891 |
Top |
Fusion Genomic Features for LAMTOR1-UVRAG |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| LAMTOR1 | chr11 | 71809334 | - | UVRAG | chr11 | 75562927 | + | 2.17E-06 | 0.99999785 |
| LAMTOR1 | chr11 | 71809334 | - | UVRAG | chr11 | 75562927 | + | 2.17E-06 | 0.99999785 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
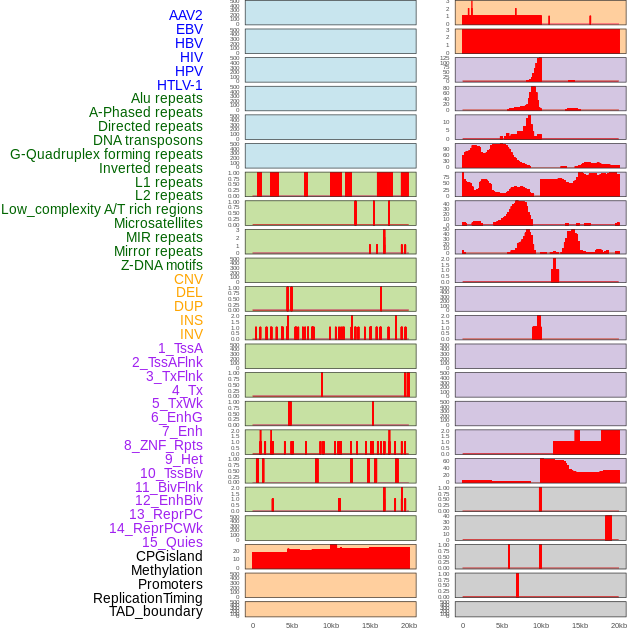 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
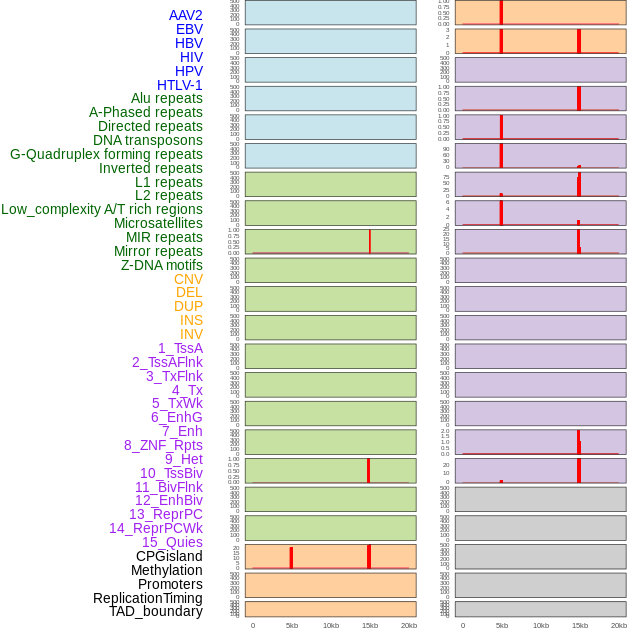 |
Top |
Fusion Protein Features for LAMTOR1-UVRAG |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr11:71809335/chr11:75562928) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| LAMTOR1 | . |
| FUNCTION: As part of the Ragulator complex it is involved in amino acid sensing and activation of mTORC1, a signaling complex promoting cell growth in response to growth factors, energy levels, and amino acids. Activated by amino acids through a mechanism involving the lysosomal V-ATPase, the Ragulator functions as a guanine nucleotide exchange factor activating the small GTPases Rag. Activated Ragulator and Rag GTPases function as a scaffold recruiting mTORC1 to lysosomes where it is in turn activated. LAMTOR1 is directly responsible for anchoring the Ragulator complex to membranes. Also required for late endosomes/lysosomes biogenesis it may regulate both the recycling of receptors through endosomes and the MAPK signaling pathway through recruitment of some of its components to late endosomes. May be involved in cholesterol homeostasis regulating LDL uptake and cholesterol release from late endosomes/lysosomes. May also play a role in RHOA activation. {ECO:0000269|PubMed:19654316, ECO:0000269|PubMed:20381137, ECO:0000269|PubMed:20544018, ECO:0000269|PubMed:22980980}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | UVRAG | chr11:71809335 | chr11:75562928 | ENST00000356136 | 0 | 15 | 224_305 | 39 | 700.0 | Coiled coil | Ontology_term=ECO:0000255 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | UVRAG | chr11:71809335 | chr11:75562928 | ENST00000356136 | 0 | 15 | 23_149 | 39 | 700.0 | Domain | C2 |
Top |
Fusion Gene Sequence for LAMTOR1-UVRAG |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >44006_44006_1_LAMTOR1-UVRAG_LAMTOR1_chr11_71809335_ENST00000278671_UVRAG_chr11_75562928_ENST00000356136_length(transcript)=4321nt_BP=556nt GGGGTATTGACTGAGGCGGCCAAGCGGCTCCGGGACAGGGGGTACGGGGGGTGGGGGCGGGTGGTTGCCTGCGGGAGGCCGCCGCGGGTC ATGTGACCGGAAGGGCTCCTCACGGACGCCGTCCCTCCTCGGCGCGGCCTGAGCGCCCGGCCCGACCCCGGCCATGGGGTGCTGCTACAG CAGCGAGAACGAGGACTCGGACCAGGACCGAGAGGAGCGGAAGCTGCTGCTGGACCCTAGCAGCCCCCCTACCAAAGCTCTCAATGGAGC CGAGCCCAACTACCACAGCCTGCCTTCCGCTCGCACTGATGAGCAGGCCCTGCTCTCTTCCATCCTTGCCAAGACAGCCAGCAACATCAT TGATGTGTCTGCTGCAGACTCACAGGGCATGGAGCAGCATGAGTACATGGACCGTGCCAGGCAGTACAGCACCCGCTTGGCTGTGCTGAG CAGCAGCCTGACCCATTGGAAGAAGCTGCCACCGCTGCCGTCTCTTACCAGCCAGCCCCACCAAGTGCTGGCCAGTGAGCCCATCCCGTT CTCTGATTTGCAGCAGCGGCGTCTTCGACATCTTCGGAACATTGCTGCCCGGAACATTGTTAATAGAAATGGCCATCAGCTCCTTGATAC CTACTTTACACTTCACTTGTGTAGTACTGAAAAGATATATAAAGAATTTTATAGAAGTGAAGTGATTAAGAATTCCTTGAATCCCACGTG GCGAAGTCTCGATTTTGGAATTATGCCAGACCGTCTTGATACATCTGTGTCTTGTTTCGTGGTGAAGATATGGGGTGGAAAGGAGAACAT CTACCAGCTGTTGATTGAATGGAAAGTCTGTTTGGATGGGCTGAAATACTTGGGTCAGCAGATTCATGCCCGAAACCAAAATGAAATAAT TTTTGGGCTGAATGATGGATACTATGGTGCTCCATTTGAACATAAGGGTTATTCAAATGCTCAGAAGACTATTCTTCTGCAGGTGGATCA GAACTGTGTTCGCAATTCTTACGATGTCTTCTCTTTGCTACGGCTTCATAGAGCCCAGTGTGCAATTAAACAGACTCAGGTAACTGTTCA GAAAATTGGAAAGGAAATTGAAGAAAAACTAAGACTCACATCTACAAGCAATGAACTGAAAAAAAAAAGTGAATGCCTGCAGTTAAAAAT TTTGGTGCTTCAGAATGAACTGGAACGGCAGAAGAAAGCTTTGGGACGGGAGGTGGCATTACTGCATAAGCAACAAATTGCATTACAAGA CAAAGGAAGTGCATTTTCAGCTGAGCACCTCAAACTTCAACTCCAGAAGGAATCCCTAAATGAGCTGAGGAAGGAGTGCACTGCAAAAAG AGAACTCTTCTTGAAGACTAATGCTCAGTTGACAATTCGTTGCAGGCAGTTACTCTCTGAGCTTTCCTACATTTACCCTATTGATTTGAA TGAACATAAGGATTACTTTGTATGCGGTGTCAAGTTGCCTAATTCTGAGGACTTCCAAGCAAAAGATGATGGAAGCATTGCTGTTGCCCT TGGTTATACTGCACATCTGGTCTCCATGATTTCCTTTTTCCTACAAGTGCCCCTCAGATATCCTATAATTCATAAGGGGTCTAGATCAAC AATCAAAGACAATATCAATGACAAACTGACGGAAAAGGAGAGAGAGTTTCCACTGTATCCAAAAGGAGGGGAGAAGTTGCAGTTTGATTA TGGTGTCTATCTTCTGAACAAAAATATAGCACAGCTAAGATATCAACATGGACTAGGGACTCCAGACTTGCGGCAAACCCTTCCCAACCT GAAAAACTTCATGGAGCATGGACTAATGGTCAGGTGTGACAGACATCACACCTCCAGTGCAATCCCTGTTCCTAAGAGACAAAGCTCCAT ATTTGGGGGTGCAGATGTAGGCTTCTCTGGGGGGATCCCTTCACCAGACAAAGGACATCGAAAACGGGCCAGCTCTGAGAATGAGAGACT TCAGTACAAAACCCCTCCTCCCAGTTACAACTCAGCATTAGCCCAGCCTGTGACCACCGTCCCCTCCATGGGAGAGACCGAGAGAAAGAT AACATCTCTATCCTCCTCCTTGGATACCTCCTTGGACTTCTCCAAAGAAAACAAGAAAAAAGGAGAGGATCTAGTTGGCAGCTTAAACGG AGGCCACGCGAATGTGCACCCTAGCCAAGAACAAGGAGAAGCCCTCTCCGGGCACCGGGCCACAGTCAATGGCACTCTCCTACCCAGCGA GCAGGCCGGGTCCGCCAGTGTCCAGCTTCCAGGCGAGTTCCACCCAGTCTCAGAAGCTGAGCTCTGCTGTACTGTGGAGCAAGCAGAAGA AATCATCGGGCTGGAAGCCACAGGTTTCGCCTCAGGTGATCAGCTAGAAGCATTTAACTGCATCCCAGTGGACAGTGCTGTGGCAGTAGA GTGTGACGAACAAGTTCTGGGAGAATTTGAAGAGTTCTCCCGAAGGATCTATGCACTGAATGAAAACGTATCCAGCTTCCGCCGGCCGCG CAGGAGTTCCGATAAGTGAAGTGAGCAGGTCAACAGTAGGACTGGGGCAGAAGCTCTGCCTAAAATGAAGTGAAAGCTGCACTTAACCCT TTGTGATAATGATGACACAAAATGAATATTAATGGAGGATATTCCTCGGAAAAACAGACTTTGGGAATGAAGGAGGGACTCAGGATCATT GTTATCAGTGGGCCAAAGTTAGATTTTGCTTTCAAGATTTGCTTTTCGGGCCTGATGATTTTAAAGCAAAAATCACCCTCTAGTTGAAAG AGCTTACAGCTCGAGTCACCTTTTAGCTATTTGTCTGCTTTTTATTTACCCTTGTATGTTATCCTCAGAGGGAAGATGATAATATATAAA TAATATAATGAACACACCCTTAGTTTCTCATAAGCATTTGCCCTCACCATGGTTTATAAAACTTTGGGAAAACGGAATATTCAGAAATAG GTTTCCGCCATGTACTGAAAGGTCTGTGGCCATCTGTGAGGTAGATGAAGAAGCAGCATAGTGGTCTCCTTACATCTAGGCCTAACTGTC CCTCTTCCTGCCCCCGGGTACCACAGTCCACCTTTAGACCCTACTGTCGCCCCATCTTCTCCGTGGATGGGCCATGCGTCCTGAAAACAG GACATCAGATTCACTGGTTCTGTAACCCAGTAGCTGTGACGTTCCATCTCTTCTAACCAGCCATGGCCTTCCCCTCCTCTGCCATACCCT TAATGCGGCCCTCAGATTAGATGAAAAACTTGCTCCTGGTGGATCCCAAGGGACCCTCAAGGACCTCGAGGTTACTGCAGTCAGATGCCA TCTCATCCCCTGTGGGGGCCAAAGTTTTTATGTGGGCAGATGCTGTGGTCAGGAACTAGGCATGCTTTCTGGCAATGCACTCACCAGACA AAAATCCCTTGATGTAAATCCCATGTTAATTTATTAAATTTCAGTCAGAAGGTCAGCATTTACATGACAGAATGTATGTAGAGAGTTGGG GTGTCTGGTAGGCAAACTGCAAGGCAGTTGAGATAGTTGGATTAAGAGGCTAGACGAGACATAGAATACTATTGGTATGTGTGCAATTTC ATGAATATTAAATTATGTTTCGAAGTCCAGTTGTCATTCCCGCATTCAGATTTCATTTGCTGTTGCTTTATACGTTACGTACCCAAGGAC ATTGCCTCAGGGTTGCAAACTCTTTAAAGGAAAATTTATCCATATATCCATGTATTATATAGAAGAATAAAAATTGAGTTTACTTCACCC GACCTGATTTTTTTTTACCAGCTTGGAAATTCCTCTTGAACCTACGTCTCCATATGTCACATCATGACAGGACTAGCCTGAACAAAAGCC ATGTCTATCTAAGCGGAGGCTGTTGACTTCATTCAGTTTGCATATTGTATATAGCAACACAAACACTTGACAGGTATATACTCCAGTCGC CACATTTGTCCTGCATAACAGCTTCACTCACAGGCCTCACCGTCACTTTATTTTGTGTCCAAGCATTCCTGGGCTCAAGTTTAATGTATA GCTACATTGTTGTTTTCCATGTAGAGAACTCACAGGATGACTACATCACAAATAAACCCAACTCTCAGGCAGTCGAAAGCTTTAACTGGA TCTGCAGAAGCCCATCTTCCTCCACATGAAGAAGTGGGGCTCCTTCACCTAGAGCAGGAGCTTCTCTGCAGTGTACTGTCTGTCGCTCAT TTTGGAGTCAGTGTGGGTGGAGACAGCATCGTGGATATGCACGTGCTGCGGGGCCCTCCAGGGAAGTAACATTTACCAAAAAAAAAGAAA >44006_44006_1_LAMTOR1-UVRAG_LAMTOR1_chr11_71809335_ENST00000278671_UVRAG_chr11_75562928_ENST00000356136_length(amino acids)=791AA_BP=130 MGCCYSSENEDSDQDREERKLLLDPSSPPTKALNGAEPNYHSLPSARTDEQALLSSILAKTASNIIDVSAADSQGMEQHEYMDRARQYST RLAVLSSSLTHWKKLPPLPSLTSQPHQVLASEPIPFSDLQQRRLRHLRNIAARNIVNRNGHQLLDTYFTLHLCSTEKIYKEFYRSEVIKN SLNPTWRSLDFGIMPDRLDTSVSCFVVKIWGGKENIYQLLIEWKVCLDGLKYLGQQIHARNQNEIIFGLNDGYYGAPFEHKGYSNAQKTI LLQVDQNCVRNSYDVFSLLRLHRAQCAIKQTQVTVQKIGKEIEEKLRLTSTSNELKKKSECLQLKILVLQNELERQKKALGREVALLHKQ QIALQDKGSAFSAEHLKLQLQKESLNELRKECTAKRELFLKTNAQLTIRCRQLLSELSYIYPIDLNEHKDYFVCGVKLPNSEDFQAKDDG SIAVALGYTAHLVSMISFFLQVPLRYPIIHKGSRSTIKDNINDKLTEKEREFPLYPKGGEKLQFDYGVYLLNKNIAQLRYQHGLGTPDLR QTLPNLKNFMEHGLMVRCDRHHTSSAIPVPKRQSSIFGGADVGFSGGIPSPDKGHRKRASSENERLQYKTPPPSYNSALAQPVTTVPSMG ETERKITSLSSSLDTSLDFSKENKKKGEDLVGSLNGGHANVHPSQEQGEALSGHRATVNGTLLPSEQAGSASVQLPGEFHPVSEAELCCT -------------------------------------------------------------- >44006_44006_2_LAMTOR1-UVRAG_LAMTOR1_chr11_71809335_ENST00000545249_UVRAG_chr11_75562928_ENST00000356136_length(transcript)=4180nt_BP=415nt AGCGCCCGGCCCGACCCCGGCCATGGGGTGCTGCTACAGCAGCGAGAACGAGGACTCGGACCAGGACCGAGAGGAGCGGAAGCTGCTGCT GGACCCTAGCAGCCCCCCTACCAAAGCTCTCAATGGAGCCGAGCCCAACTACCACAGCCTGCCTTCCGCTCGCACTGATGAGCAGGCCCT GCTCTCTTCCATCCTTGCCAAGACAGCCAGCAACATCATTGATGTGTCTGCTGCAGACTCACAGGGCATGGAGCAGCATGAGTACATGGA CCGTGCCAGGCAGTACAGCACCCGCTTGGCTGTGCTGAGCAGCAGCCTGACCCATTGGAAGAAGCTGCCACCGCTGCCGTCTCTTACCAG CCAGCCCCACCAAGTGCTGGCCAGTGAGCCCATCCCGTTCTCTGATTTGCAGCAGCGGCGTCTTCGACATCTTCGGAACATTGCTGCCCG GAACATTGTTAATAGAAATGGCCATCAGCTCCTTGATACCTACTTTACACTTCACTTGTGTAGTACTGAAAAGATATATAAAGAATTTTA TAGAAGTGAAGTGATTAAGAATTCCTTGAATCCCACGTGGCGAAGTCTCGATTTTGGAATTATGCCAGACCGTCTTGATACATCTGTGTC TTGTTTCGTGGTGAAGATATGGGGTGGAAAGGAGAACATCTACCAGCTGTTGATTGAATGGAAAGTCTGTTTGGATGGGCTGAAATACTT GGGTCAGCAGATTCATGCCCGAAACCAAAATGAAATAATTTTTGGGCTGAATGATGGATACTATGGTGCTCCATTTGAACATAAGGGTTA TTCAAATGCTCAGAAGACTATTCTTCTGCAGGTGGATCAGAACTGTGTTCGCAATTCTTACGATGTCTTCTCTTTGCTACGGCTTCATAG AGCCCAGTGTGCAATTAAACAGACTCAGGTAACTGTTCAGAAAATTGGAAAGGAAATTGAAGAAAAACTAAGACTCACATCTACAAGCAA TGAACTGAAAAAAAAAAGTGAATGCCTGCAGTTAAAAATTTTGGTGCTTCAGAATGAACTGGAACGGCAGAAGAAAGCTTTGGGACGGGA GGTGGCATTACTGCATAAGCAACAAATTGCATTACAAGACAAAGGAAGTGCATTTTCAGCTGAGCACCTCAAACTTCAACTCCAGAAGGA ATCCCTAAATGAGCTGAGGAAGGAGTGCACTGCAAAAAGAGAACTCTTCTTGAAGACTAATGCTCAGTTGACAATTCGTTGCAGGCAGTT ACTCTCTGAGCTTTCCTACATTTACCCTATTGATTTGAATGAACATAAGGATTACTTTGTATGCGGTGTCAAGTTGCCTAATTCTGAGGA CTTCCAAGCAAAAGATGATGGAAGCATTGCTGTTGCCCTTGGTTATACTGCACATCTGGTCTCCATGATTTCCTTTTTCCTACAAGTGCC CCTCAGATATCCTATAATTCATAAGGGGTCTAGATCAACAATCAAAGACAATATCAATGACAAACTGACGGAAAAGGAGAGAGAGTTTCC ACTGTATCCAAAAGGAGGGGAGAAGTTGCAGTTTGATTATGGTGTCTATCTTCTGAACAAAAATATAGCACAGCTAAGATATCAACATGG ACTAGGGACTCCAGACTTGCGGCAAACCCTTCCCAACCTGAAAAACTTCATGGAGCATGGACTAATGGTCAGGTGTGACAGACATCACAC CTCCAGTGCAATCCCTGTTCCTAAGAGACAAAGCTCCATATTTGGGGGTGCAGATGTAGGCTTCTCTGGGGGGATCCCTTCACCAGACAA AGGACATCGAAAACGGGCCAGCTCTGAGAATGAGAGACTTCAGTACAAAACCCCTCCTCCCAGTTACAACTCAGCATTAGCCCAGCCTGT GACCACCGTCCCCTCCATGGGAGAGACCGAGAGAAAGATAACATCTCTATCCTCCTCCTTGGATACCTCCTTGGACTTCTCCAAAGAAAA CAAGAAAAAAGGAGAGGATCTAGTTGGCAGCTTAAACGGAGGCCACGCGAATGTGCACCCTAGCCAAGAACAAGGAGAAGCCCTCTCCGG GCACCGGGCCACAGTCAATGGCACTCTCCTACCCAGCGAGCAGGCCGGGTCCGCCAGTGTCCAGCTTCCAGGCGAGTTCCACCCAGTCTC AGAAGCTGAGCTCTGCTGTACTGTGGAGCAAGCAGAAGAAATCATCGGGCTGGAAGCCACAGGTTTCGCCTCAGGTGATCAGCTAGAAGC ATTTAACTGCATCCCAGTGGACAGTGCTGTGGCAGTAGAGTGTGACGAACAAGTTCTGGGAGAATTTGAAGAGTTCTCCCGAAGGATCTA TGCACTGAATGAAAACGTATCCAGCTTCCGCCGGCCGCGCAGGAGTTCCGATAAGTGAAGTGAGCAGGTCAACAGTAGGACTGGGGCAGA AGCTCTGCCTAAAATGAAGTGAAAGCTGCACTTAACCCTTTGTGATAATGATGACACAAAATGAATATTAATGGAGGATATTCCTCGGAA AAACAGACTTTGGGAATGAAGGAGGGACTCAGGATCATTGTTATCAGTGGGCCAAAGTTAGATTTTGCTTTCAAGATTTGCTTTTCGGGC CTGATGATTTTAAAGCAAAAATCACCCTCTAGTTGAAAGAGCTTACAGCTCGAGTCACCTTTTAGCTATTTGTCTGCTTTTTATTTACCC TTGTATGTTATCCTCAGAGGGAAGATGATAATATATAAATAATATAATGAACACACCCTTAGTTTCTCATAAGCATTTGCCCTCACCATG GTTTATAAAACTTTGGGAAAACGGAATATTCAGAAATAGGTTTCCGCCATGTACTGAAAGGTCTGTGGCCATCTGTGAGGTAGATGAAGA AGCAGCATAGTGGTCTCCTTACATCTAGGCCTAACTGTCCCTCTTCCTGCCCCCGGGTACCACAGTCCACCTTTAGACCCTACTGTCGCC CCATCTTCTCCGTGGATGGGCCATGCGTCCTGAAAACAGGACATCAGATTCACTGGTTCTGTAACCCAGTAGCTGTGACGTTCCATCTCT TCTAACCAGCCATGGCCTTCCCCTCCTCTGCCATACCCTTAATGCGGCCCTCAGATTAGATGAAAAACTTGCTCCTGGTGGATCCCAAGG GACCCTCAAGGACCTCGAGGTTACTGCAGTCAGATGCCATCTCATCCCCTGTGGGGGCCAAAGTTTTTATGTGGGCAGATGCTGTGGTCA GGAACTAGGCATGCTTTCTGGCAATGCACTCACCAGACAAAAATCCCTTGATGTAAATCCCATGTTAATTTATTAAATTTCAGTCAGAAG GTCAGCATTTACATGACAGAATGTATGTAGAGAGTTGGGGTGTCTGGTAGGCAAACTGCAAGGCAGTTGAGATAGTTGGATTAAGAGGCT AGACGAGACATAGAATACTATTGGTATGTGTGCAATTTCATGAATATTAAATTATGTTTCGAAGTCCAGTTGTCATTCCCGCATTCAGAT TTCATTTGCTGTTGCTTTATACGTTACGTACCCAAGGACATTGCCTCAGGGTTGCAAACTCTTTAAAGGAAAATTTATCCATATATCCAT GTATTATATAGAAGAATAAAAATTGAGTTTACTTCACCCGACCTGATTTTTTTTTACCAGCTTGGAAATTCCTCTTGAACCTACGTCTCC ATATGTCACATCATGACAGGACTAGCCTGAACAAAAGCCATGTCTATCTAAGCGGAGGCTGTTGACTTCATTCAGTTTGCATATTGTATA TAGCAACACAAACACTTGACAGGTATATACTCCAGTCGCCACATTTGTCCTGCATAACAGCTTCACTCACAGGCCTCACCGTCACTTTAT TTTGTGTCCAAGCATTCCTGGGCTCAAGTTTAATGTATAGCTACATTGTTGTTTTCCATGTAGAGAACTCACAGGATGACTACATCACAA ATAAACCCAACTCTCAGGCAGTCGAAAGCTTTAACTGGATCTGCAGAAGCCCATCTTCCTCCACATGAAGAAGTGGGGCTCCTTCACCTA GAGCAGGAGCTTCTCTGCAGTGTACTGTCTGTCGCTCATTTTGGAGTCAGTGTGGGTGGAGACAGCATCGTGGATATGCACGTGCTGCGG >44006_44006_2_LAMTOR1-UVRAG_LAMTOR1_chr11_71809335_ENST00000545249_UVRAG_chr11_75562928_ENST00000356136_length(amino acids)=791AA_BP=130 MGCCYSSENEDSDQDREERKLLLDPSSPPTKALNGAEPNYHSLPSARTDEQALLSSILAKTASNIIDVSAADSQGMEQHEYMDRARQYST RLAVLSSSLTHWKKLPPLPSLTSQPHQVLASEPIPFSDLQQRRLRHLRNIAARNIVNRNGHQLLDTYFTLHLCSTEKIYKEFYRSEVIKN SLNPTWRSLDFGIMPDRLDTSVSCFVVKIWGGKENIYQLLIEWKVCLDGLKYLGQQIHARNQNEIIFGLNDGYYGAPFEHKGYSNAQKTI LLQVDQNCVRNSYDVFSLLRLHRAQCAIKQTQVTVQKIGKEIEEKLRLTSTSNELKKKSECLQLKILVLQNELERQKKALGREVALLHKQ QIALQDKGSAFSAEHLKLQLQKESLNELRKECTAKRELFLKTNAQLTIRCRQLLSELSYIYPIDLNEHKDYFVCGVKLPNSEDFQAKDDG SIAVALGYTAHLVSMISFFLQVPLRYPIIHKGSRSTIKDNINDKLTEKEREFPLYPKGGEKLQFDYGVYLLNKNIAQLRYQHGLGTPDLR QTLPNLKNFMEHGLMVRCDRHHTSSAIPVPKRQSSIFGGADVGFSGGIPSPDKGHRKRASSENERLQYKTPPPSYNSALAQPVTTVPSMG ETERKITSLSSSLDTSLDFSKENKKKGEDLVGSLNGGHANVHPSQEQGEALSGHRATVNGTLLPSEQAGSASVQLPGEFHPVSEAELCCT -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for LAMTOR1-UVRAG |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | LAMTOR1 | chr11:71809335 | chr11:75562928 | ENST00000278671 | - | 4 | 5 | 121_161 | 131.0 | 162.0 | LAMTOR2 and LAMTOR3 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for LAMTOR1-UVRAG |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for LAMTOR1-UVRAG |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
