
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:LDLR-ZGLP1 (FusionGDB2 ID:44420) |
Fusion Gene Summary for LDLR-ZGLP1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: LDLR-ZGLP1 | Fusion gene ID: 44420 | Hgene | Tgene | Gene symbol | LDLR | ZGLP1 | Gene ID | 3949 | 100125288 |
| Gene name | low density lipoprotein receptor | zinc finger GATA like protein 1 | |
| Synonyms | FH|FHC|FHCL1|LDLCQ2 | GATAD3|GLP-1|GLP1 | |
| Cytomap | 19p13.2 | 19p13.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | low-density lipoprotein receptorLDL receptorlow-density lipoprotein receptor class A domain-containing protein 3 | GATA-type zinc finger protein 1GATA zinc finger domain containing 3GATA-like protein 1 | |
| Modification date | 20200322 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000455727, ENST00000535915, ENST00000545707, ENST00000557933, ENST00000558013, ENST00000558518, ENST00000560628, | ENST00000403352, ENST00000403903, | |
| Fusion gene scores | * DoF score | 23 X 17 X 15=5865 | 4 X 9 X 4=144 |
| # samples | 33 | 5 | |
| ** MAII score | log2(33/5865*10)=-4.15159317867005 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/144*10)=-1.52606881166759 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: LDLR [Title/Abstract] AND ZGLP1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | LDLR(11200291)-ZGLP1(10419067), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | LDLR-ZGLP1 seems lost the major protein functional domain in Hgene partner, which is a cell metabolism gene due to the frame-shifted ORF. LDLR-ZGLP1 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. LDLR-ZGLP1 seems lost the major protein functional domain in Tgene partner, which is a transcription factor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across LDLR (5'-gene) Fusion gene breakpoints across LDLR (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
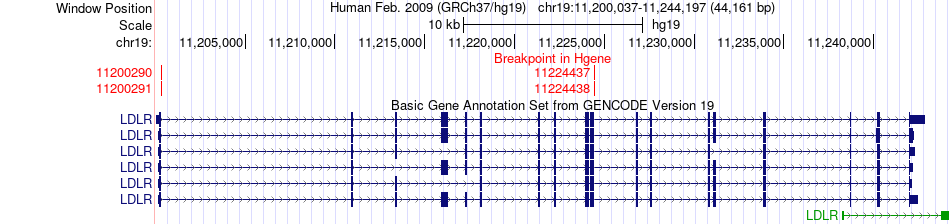 |
 Fusion gene breakpoints across ZGLP1 (3'-gene) Fusion gene breakpoints across ZGLP1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
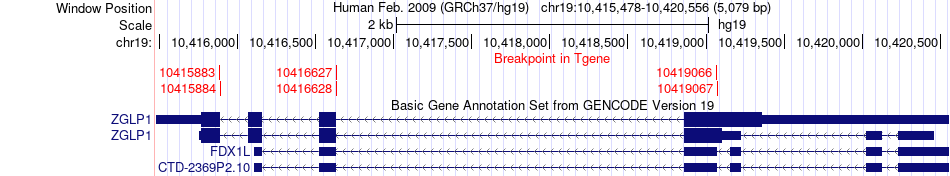 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | ACC | TCGA-OR-A5JP-01A | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10416627 | - |
| ChimerDB4 | ACC | TCGA-OR-A5JP-01A | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10419066 | - |
| ChimerDB4 | ACC | TCGA-OR-A5JP-01A | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10416628 | - |
| ChimerDB4 | ACC | TCGA-OR-A5JP-01A | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10419067 | - |
| ChimerDB4 | ACC | TCGA-OR-A5JP-01A | LDLR | chr19 | 11224437 | + | ZGLP1 | chr19 | 10415883 | - |
| ChimerDB4 | ACC | TCGA-OR-A5JP | LDLR | chr19 | 11224438 | + | ZGLP1 | chr19 | 10415884 | - |
Top |
Fusion Gene ORF analysis for LDLR-ZGLP1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| Frame-shift | ENST00000455727 | ENST00000403352 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10416627 | - |
| Frame-shift | ENST00000455727 | ENST00000403352 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10419066 | - |
| Frame-shift | ENST00000455727 | ENST00000403352 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10416628 | - |
| Frame-shift | ENST00000455727 | ENST00000403352 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10419067 | - |
| Frame-shift | ENST00000455727 | ENST00000403903 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10416627 | - |
| Frame-shift | ENST00000455727 | ENST00000403903 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10419066 | - |
| Frame-shift | ENST00000455727 | ENST00000403903 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10416628 | - |
| Frame-shift | ENST00000455727 | ENST00000403903 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10419067 | - |
| Frame-shift | ENST00000455727 | ENST00000403903 | LDLR | chr19 | 11224437 | + | ZGLP1 | chr19 | 10415883 | - |
| Frame-shift | ENST00000455727 | ENST00000403903 | LDLR | chr19 | 11224438 | + | ZGLP1 | chr19 | 10415884 | - |
| Frame-shift | ENST00000535915 | ENST00000403352 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10416627 | - |
| Frame-shift | ENST00000535915 | ENST00000403352 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10419066 | - |
| Frame-shift | ENST00000535915 | ENST00000403352 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10416628 | - |
| Frame-shift | ENST00000535915 | ENST00000403352 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10419067 | - |
| Frame-shift | ENST00000535915 | ENST00000403903 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10416627 | - |
| Frame-shift | ENST00000535915 | ENST00000403903 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10419066 | - |
| Frame-shift | ENST00000535915 | ENST00000403903 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10416628 | - |
| Frame-shift | ENST00000535915 | ENST00000403903 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10419067 | - |
| Frame-shift | ENST00000535915 | ENST00000403903 | LDLR | chr19 | 11224437 | + | ZGLP1 | chr19 | 10415883 | - |
| Frame-shift | ENST00000535915 | ENST00000403903 | LDLR | chr19 | 11224438 | + | ZGLP1 | chr19 | 10415884 | - |
| Frame-shift | ENST00000545707 | ENST00000403352 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10416627 | - |
| Frame-shift | ENST00000545707 | ENST00000403352 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10419066 | - |
| Frame-shift | ENST00000545707 | ENST00000403352 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10416628 | - |
| Frame-shift | ENST00000545707 | ENST00000403352 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10419067 | - |
| Frame-shift | ENST00000545707 | ENST00000403903 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10416627 | - |
| Frame-shift | ENST00000545707 | ENST00000403903 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10419066 | - |
| Frame-shift | ENST00000545707 | ENST00000403903 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10416628 | - |
| Frame-shift | ENST00000545707 | ENST00000403903 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10419067 | - |
| Frame-shift | ENST00000545707 | ENST00000403903 | LDLR | chr19 | 11224437 | + | ZGLP1 | chr19 | 10415883 | - |
| Frame-shift | ENST00000545707 | ENST00000403903 | LDLR | chr19 | 11224438 | + | ZGLP1 | chr19 | 10415884 | - |
| Frame-shift | ENST00000557933 | ENST00000403352 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10416627 | - |
| Frame-shift | ENST00000557933 | ENST00000403352 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10419066 | - |
| Frame-shift | ENST00000557933 | ENST00000403352 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10416628 | - |
| Frame-shift | ENST00000557933 | ENST00000403352 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10419067 | - |
| Frame-shift | ENST00000557933 | ENST00000403903 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10416627 | - |
| Frame-shift | ENST00000557933 | ENST00000403903 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10419066 | - |
| Frame-shift | ENST00000557933 | ENST00000403903 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10416628 | - |
| Frame-shift | ENST00000557933 | ENST00000403903 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10419067 | - |
| Frame-shift | ENST00000557933 | ENST00000403903 | LDLR | chr19 | 11224437 | + | ZGLP1 | chr19 | 10415883 | - |
| Frame-shift | ENST00000557933 | ENST00000403903 | LDLR | chr19 | 11224438 | + | ZGLP1 | chr19 | 10415884 | - |
| Frame-shift | ENST00000558013 | ENST00000403352 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10416627 | - |
| Frame-shift | ENST00000558013 | ENST00000403352 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10419066 | - |
| Frame-shift | ENST00000558013 | ENST00000403352 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10416628 | - |
| Frame-shift | ENST00000558013 | ENST00000403352 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10419067 | - |
| Frame-shift | ENST00000558013 | ENST00000403903 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10416627 | - |
| Frame-shift | ENST00000558013 | ENST00000403903 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10419066 | - |
| Frame-shift | ENST00000558013 | ENST00000403903 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10416628 | - |
| Frame-shift | ENST00000558013 | ENST00000403903 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10419067 | - |
| Frame-shift | ENST00000558013 | ENST00000403903 | LDLR | chr19 | 11224437 | + | ZGLP1 | chr19 | 10415883 | - |
| Frame-shift | ENST00000558013 | ENST00000403903 | LDLR | chr19 | 11224438 | + | ZGLP1 | chr19 | 10415884 | - |
| Frame-shift | ENST00000558518 | ENST00000403352 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10416627 | - |
| Frame-shift | ENST00000558518 | ENST00000403352 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10419066 | - |
| Frame-shift | ENST00000558518 | ENST00000403352 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10416628 | - |
| Frame-shift | ENST00000558518 | ENST00000403352 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10419067 | - |
| Frame-shift | ENST00000558518 | ENST00000403903 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10416627 | - |
| Frame-shift | ENST00000558518 | ENST00000403903 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10419066 | - |
| Frame-shift | ENST00000558518 | ENST00000403903 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10416628 | - |
| Frame-shift | ENST00000558518 | ENST00000403903 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10419067 | - |
| Frame-shift | ENST00000558518 | ENST00000403903 | LDLR | chr19 | 11224437 | + | ZGLP1 | chr19 | 10415883 | - |
| Frame-shift | ENST00000558518 | ENST00000403903 | LDLR | chr19 | 11224438 | + | ZGLP1 | chr19 | 10415884 | - |
| In-frame | ENST00000455727 | ENST00000403352 | LDLR | chr19 | 11224437 | + | ZGLP1 | chr19 | 10415883 | - |
| In-frame | ENST00000455727 | ENST00000403352 | LDLR | chr19 | 11224438 | + | ZGLP1 | chr19 | 10415884 | - |
| In-frame | ENST00000535915 | ENST00000403352 | LDLR | chr19 | 11224437 | + | ZGLP1 | chr19 | 10415883 | - |
| In-frame | ENST00000535915 | ENST00000403352 | LDLR | chr19 | 11224438 | + | ZGLP1 | chr19 | 10415884 | - |
| In-frame | ENST00000545707 | ENST00000403352 | LDLR | chr19 | 11224437 | + | ZGLP1 | chr19 | 10415883 | - |
| In-frame | ENST00000545707 | ENST00000403352 | LDLR | chr19 | 11224438 | + | ZGLP1 | chr19 | 10415884 | - |
| In-frame | ENST00000557933 | ENST00000403352 | LDLR | chr19 | 11224437 | + | ZGLP1 | chr19 | 10415883 | - |
| In-frame | ENST00000557933 | ENST00000403352 | LDLR | chr19 | 11224438 | + | ZGLP1 | chr19 | 10415884 | - |
| In-frame | ENST00000558013 | ENST00000403352 | LDLR | chr19 | 11224437 | + | ZGLP1 | chr19 | 10415883 | - |
| In-frame | ENST00000558013 | ENST00000403352 | LDLR | chr19 | 11224438 | + | ZGLP1 | chr19 | 10415884 | - |
| In-frame | ENST00000558518 | ENST00000403352 | LDLR | chr19 | 11224437 | + | ZGLP1 | chr19 | 10415883 | - |
| In-frame | ENST00000558518 | ENST00000403352 | LDLR | chr19 | 11224438 | + | ZGLP1 | chr19 | 10415884 | - |
| intron-3CDS | ENST00000560628 | ENST00000403352 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10416627 | - |
| intron-3CDS | ENST00000560628 | ENST00000403352 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10419066 | - |
| intron-3CDS | ENST00000560628 | ENST00000403352 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10416628 | - |
| intron-3CDS | ENST00000560628 | ENST00000403352 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10419067 | - |
| intron-3CDS | ENST00000560628 | ENST00000403352 | LDLR | chr19 | 11224437 | + | ZGLP1 | chr19 | 10415883 | - |
| intron-3CDS | ENST00000560628 | ENST00000403352 | LDLR | chr19 | 11224438 | + | ZGLP1 | chr19 | 10415884 | - |
| intron-3CDS | ENST00000560628 | ENST00000403903 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10416627 | - |
| intron-3CDS | ENST00000560628 | ENST00000403903 | LDLR | chr19 | 11200290 | + | ZGLP1 | chr19 | 10419066 | - |
| intron-3CDS | ENST00000560628 | ENST00000403903 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10416628 | - |
| intron-3CDS | ENST00000560628 | ENST00000403903 | LDLR | chr19 | 11200291 | + | ZGLP1 | chr19 | 10419067 | - |
| intron-3CDS | ENST00000560628 | ENST00000403903 | LDLR | chr19 | 11224437 | + | ZGLP1 | chr19 | 10415883 | - |
| intron-3CDS | ENST00000560628 | ENST00000403903 | LDLR | chr19 | 11224438 | + | ZGLP1 | chr19 | 10415884 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000558518 | LDLR | chr19 | 11224438 | + | ENST00000403352 | ZGLP1 | chr19 | 10415884 | - | 1907 | 1773 | 169 | 1890 | 573 |
| ENST00000557933 | LDLR | chr19 | 11224438 | + | ENST00000403352 | ZGLP1 | chr19 | 10415884 | - | 1807 | 1673 | 69 | 1790 | 573 |
| ENST00000455727 | LDLR | chr19 | 11224438 | + | ENST00000403352 | ZGLP1 | chr19 | 10415884 | - | 1302 | 1168 | 68 | 1285 | 405 |
| ENST00000535915 | LDLR | chr19 | 11224438 | + | ENST00000403352 | ZGLP1 | chr19 | 10415884 | - | 1683 | 1549 | 68 | 1666 | 532 |
| ENST00000545707 | LDLR | chr19 | 11224438 | + | ENST00000403352 | ZGLP1 | chr19 | 10415884 | - | 1425 | 1291 | 68 | 1408 | 446 |
| ENST00000558013 | LDLR | chr19 | 11224438 | + | ENST00000403352 | ZGLP1 | chr19 | 10415884 | - | 1791 | 1657 | 53 | 1774 | 573 |
| ENST00000558518 | LDLR | chr19 | 11224437 | + | ENST00000403352 | ZGLP1 | chr19 | 10415883 | - | 1907 | 1773 | 169 | 1890 | 573 |
| ENST00000557933 | LDLR | chr19 | 11224437 | + | ENST00000403352 | ZGLP1 | chr19 | 10415883 | - | 1807 | 1673 | 69 | 1790 | 573 |
| ENST00000455727 | LDLR | chr19 | 11224437 | + | ENST00000403352 | ZGLP1 | chr19 | 10415883 | - | 1302 | 1168 | 68 | 1285 | 405 |
| ENST00000535915 | LDLR | chr19 | 11224437 | + | ENST00000403352 | ZGLP1 | chr19 | 10415883 | - | 1683 | 1549 | 68 | 1666 | 532 |
| ENST00000545707 | LDLR | chr19 | 11224437 | + | ENST00000403352 | ZGLP1 | chr19 | 10415883 | - | 1425 | 1291 | 68 | 1408 | 446 |
| ENST00000558013 | LDLR | chr19 | 11224437 | + | ENST00000403352 | ZGLP1 | chr19 | 10415883 | - | 1791 | 1657 | 53 | 1774 | 573 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000558518 | ENST00000403352 | LDLR | chr19 | 11224438 | + | ZGLP1 | chr19 | 10415884 | - | 0.016103107 | 0.98389685 |
| ENST00000557933 | ENST00000403352 | LDLR | chr19 | 11224438 | + | ZGLP1 | chr19 | 10415884 | - | 0.016469566 | 0.98353046 |
| ENST00000455727 | ENST00000403352 | LDLR | chr19 | 11224438 | + | ZGLP1 | chr19 | 10415884 | - | 0.017386675 | 0.98261327 |
| ENST00000535915 | ENST00000403352 | LDLR | chr19 | 11224438 | + | ZGLP1 | chr19 | 10415884 | - | 0.015940597 | 0.98405933 |
| ENST00000545707 | ENST00000403352 | LDLR | chr19 | 11224438 | + | ZGLP1 | chr19 | 10415884 | - | 0.020687504 | 0.97931254 |
| ENST00000558013 | ENST00000403352 | LDLR | chr19 | 11224438 | + | ZGLP1 | chr19 | 10415884 | - | 0.016846169 | 0.9831539 |
| ENST00000558518 | ENST00000403352 | LDLR | chr19 | 11224437 | + | ZGLP1 | chr19 | 10415883 | - | 0.016103107 | 0.98389685 |
| ENST00000557933 | ENST00000403352 | LDLR | chr19 | 11224437 | + | ZGLP1 | chr19 | 10415883 | - | 0.016469566 | 0.98353046 |
| ENST00000455727 | ENST00000403352 | LDLR | chr19 | 11224437 | + | ZGLP1 | chr19 | 10415883 | - | 0.017386675 | 0.98261327 |
| ENST00000535915 | ENST00000403352 | LDLR | chr19 | 11224437 | + | ZGLP1 | chr19 | 10415883 | - | 0.015940597 | 0.98405933 |
| ENST00000545707 | ENST00000403352 | LDLR | chr19 | 11224437 | + | ZGLP1 | chr19 | 10415883 | - | 0.020687504 | 0.97931254 |
| ENST00000558013 | ENST00000403352 | LDLR | chr19 | 11224437 | + | ZGLP1 | chr19 | 10415883 | - | 0.016846169 | 0.9831539 |
Top |
Fusion Genomic Features for LDLR-ZGLP1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
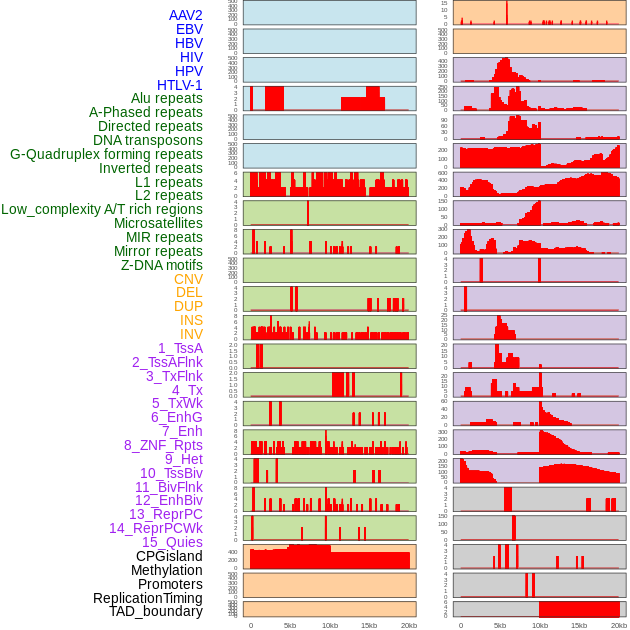 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for LDLR-ZGLP1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:11200291/chr19:10419067) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000455727 | + | 8 | 16 | 107_145 | 360 | 693.0 | Domain | LDL-receptor class A 3 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000455727 | + | 8 | 16 | 146_186 | 360 | 693.0 | Domain | LDL-receptor class A 4 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000455727 | + | 8 | 16 | 195_233 | 360 | 693.0 | Domain | LDL-receptor class A 5 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000455727 | + | 8 | 16 | 234_272 | 360 | 693.0 | Domain | LDL-receptor class A 6 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000455727 | + | 8 | 16 | 25_65 | 360 | 693.0 | Domain | LDL-receptor class A 1 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000455727 | + | 8 | 16 | 274_313 | 360 | 693.0 | Domain | LDL-receptor class A 7 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000455727 | + | 8 | 16 | 314_353 | 360 | 693.0 | Domain | EGF-like 1 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000455727 | + | 8 | 16 | 66_106 | 360 | 693.0 | Domain | LDL-receptor class A 2 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000535915 | + | 9 | 17 | 107_145 | 487 | 820.0 | Domain | LDL-receptor class A 3 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000535915 | + | 9 | 17 | 146_186 | 487 | 820.0 | Domain | LDL-receptor class A 4 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000535915 | + | 9 | 17 | 195_233 | 487 | 820.0 | Domain | LDL-receptor class A 5 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000535915 | + | 9 | 17 | 234_272 | 487 | 820.0 | Domain | LDL-receptor class A 6 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000535915 | + | 9 | 17 | 25_65 | 487 | 820.0 | Domain | LDL-receptor class A 1 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000535915 | + | 9 | 17 | 274_313 | 487 | 820.0 | Domain | LDL-receptor class A 7 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000535915 | + | 9 | 17 | 314_353 | 487 | 820.0 | Domain | EGF-like 1 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000535915 | + | 9 | 17 | 354_393 | 487 | 820.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000535915 | + | 9 | 17 | 66_106 | 487 | 820.0 | Domain | LDL-receptor class A 2 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000545707 | + | 9 | 16 | 107_145 | 401 | 683.0 | Domain | LDL-receptor class A 3 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000545707 | + | 9 | 16 | 146_186 | 401 | 683.0 | Domain | LDL-receptor class A 4 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000545707 | + | 9 | 16 | 195_233 | 401 | 683.0 | Domain | LDL-receptor class A 5 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000545707 | + | 9 | 16 | 234_272 | 401 | 683.0 | Domain | LDL-receptor class A 6 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000545707 | + | 9 | 16 | 25_65 | 401 | 683.0 | Domain | LDL-receptor class A 1 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000545707 | + | 9 | 16 | 274_313 | 401 | 683.0 | Domain | LDL-receptor class A 7 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000545707 | + | 9 | 16 | 314_353 | 401 | 683.0 | Domain | EGF-like 1 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000545707 | + | 9 | 16 | 354_393 | 401 | 683.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000545707 | + | 9 | 16 | 66_106 | 401 | 683.0 | Domain | LDL-receptor class A 2 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558013 | + | 10 | 18 | 107_145 | 528 | 859.0 | Domain | LDL-receptor class A 3 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558013 | + | 10 | 18 | 146_186 | 528 | 859.0 | Domain | LDL-receptor class A 4 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558013 | + | 10 | 18 | 195_233 | 528 | 859.0 | Domain | LDL-receptor class A 5 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558013 | + | 10 | 18 | 234_272 | 528 | 859.0 | Domain | LDL-receptor class A 6 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558013 | + | 10 | 18 | 25_65 | 528 | 859.0 | Domain | LDL-receptor class A 1 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558013 | + | 10 | 18 | 274_313 | 528 | 859.0 | Domain | LDL-receptor class A 7 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558013 | + | 10 | 18 | 314_353 | 528 | 859.0 | Domain | EGF-like 1 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558013 | + | 10 | 18 | 354_393 | 528 | 859.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558013 | + | 10 | 18 | 66_106 | 528 | 859.0 | Domain | LDL-receptor class A 2 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558518 | + | 10 | 18 | 107_145 | 528 | 861.0 | Domain | LDL-receptor class A 3 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558518 | + | 10 | 18 | 146_186 | 528 | 861.0 | Domain | LDL-receptor class A 4 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558518 | + | 10 | 18 | 195_233 | 528 | 861.0 | Domain | LDL-receptor class A 5 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558518 | + | 10 | 18 | 234_272 | 528 | 861.0 | Domain | LDL-receptor class A 6 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558518 | + | 10 | 18 | 25_65 | 528 | 861.0 | Domain | LDL-receptor class A 1 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558518 | + | 10 | 18 | 274_313 | 528 | 861.0 | Domain | LDL-receptor class A 7 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558518 | + | 10 | 18 | 314_353 | 528 | 861.0 | Domain | EGF-like 1 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558518 | + | 10 | 18 | 354_393 | 528 | 861.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558518 | + | 10 | 18 | 66_106 | 528 | 861.0 | Domain | LDL-receptor class A 2 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000455727 | + | 8 | 16 | 107_145 | 360 | 693.0 | Domain | LDL-receptor class A 3 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000455727 | + | 8 | 16 | 146_186 | 360 | 693.0 | Domain | LDL-receptor class A 4 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000455727 | + | 8 | 16 | 195_233 | 360 | 693.0 | Domain | LDL-receptor class A 5 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000455727 | + | 8 | 16 | 234_272 | 360 | 693.0 | Domain | LDL-receptor class A 6 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000455727 | + | 8 | 16 | 25_65 | 360 | 693.0 | Domain | LDL-receptor class A 1 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000455727 | + | 8 | 16 | 274_313 | 360 | 693.0 | Domain | LDL-receptor class A 7 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000455727 | + | 8 | 16 | 314_353 | 360 | 693.0 | Domain | EGF-like 1 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000455727 | + | 8 | 16 | 66_106 | 360 | 693.0 | Domain | LDL-receptor class A 2 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000535915 | + | 9 | 17 | 107_145 | 487 | 820.0 | Domain | LDL-receptor class A 3 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000535915 | + | 9 | 17 | 146_186 | 487 | 820.0 | Domain | LDL-receptor class A 4 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000535915 | + | 9 | 17 | 195_233 | 487 | 820.0 | Domain | LDL-receptor class A 5 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000535915 | + | 9 | 17 | 234_272 | 487 | 820.0 | Domain | LDL-receptor class A 6 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000535915 | + | 9 | 17 | 25_65 | 487 | 820.0 | Domain | LDL-receptor class A 1 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000535915 | + | 9 | 17 | 274_313 | 487 | 820.0 | Domain | LDL-receptor class A 7 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000535915 | + | 9 | 17 | 314_353 | 487 | 820.0 | Domain | EGF-like 1 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000535915 | + | 9 | 17 | 354_393 | 487 | 820.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000535915 | + | 9 | 17 | 66_106 | 487 | 820.0 | Domain | LDL-receptor class A 2 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000545707 | + | 9 | 16 | 107_145 | 401 | 683.0 | Domain | LDL-receptor class A 3 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000545707 | + | 9 | 16 | 146_186 | 401 | 683.0 | Domain | LDL-receptor class A 4 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000545707 | + | 9 | 16 | 195_233 | 401 | 683.0 | Domain | LDL-receptor class A 5 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000545707 | + | 9 | 16 | 234_272 | 401 | 683.0 | Domain | LDL-receptor class A 6 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000545707 | + | 9 | 16 | 25_65 | 401 | 683.0 | Domain | LDL-receptor class A 1 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000545707 | + | 9 | 16 | 274_313 | 401 | 683.0 | Domain | LDL-receptor class A 7 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000545707 | + | 9 | 16 | 314_353 | 401 | 683.0 | Domain | EGF-like 1 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000545707 | + | 9 | 16 | 354_393 | 401 | 683.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000545707 | + | 9 | 16 | 66_106 | 401 | 683.0 | Domain | LDL-receptor class A 2 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558013 | + | 10 | 18 | 107_145 | 528 | 859.0 | Domain | LDL-receptor class A 3 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558013 | + | 10 | 18 | 146_186 | 528 | 859.0 | Domain | LDL-receptor class A 4 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558013 | + | 10 | 18 | 195_233 | 528 | 859.0 | Domain | LDL-receptor class A 5 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558013 | + | 10 | 18 | 234_272 | 528 | 859.0 | Domain | LDL-receptor class A 6 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558013 | + | 10 | 18 | 25_65 | 528 | 859.0 | Domain | LDL-receptor class A 1 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558013 | + | 10 | 18 | 274_313 | 528 | 859.0 | Domain | LDL-receptor class A 7 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558013 | + | 10 | 18 | 314_353 | 528 | 859.0 | Domain | EGF-like 1 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558013 | + | 10 | 18 | 354_393 | 528 | 859.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558013 | + | 10 | 18 | 66_106 | 528 | 859.0 | Domain | LDL-receptor class A 2 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558518 | + | 10 | 18 | 107_145 | 528 | 861.0 | Domain | LDL-receptor class A 3 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558518 | + | 10 | 18 | 146_186 | 528 | 861.0 | Domain | LDL-receptor class A 4 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558518 | + | 10 | 18 | 195_233 | 528 | 861.0 | Domain | LDL-receptor class A 5 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558518 | + | 10 | 18 | 234_272 | 528 | 861.0 | Domain | LDL-receptor class A 6 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558518 | + | 10 | 18 | 25_65 | 528 | 861.0 | Domain | LDL-receptor class A 1 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558518 | + | 10 | 18 | 274_313 | 528 | 861.0 | Domain | LDL-receptor class A 7 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558518 | + | 10 | 18 | 314_353 | 528 | 861.0 | Domain | EGF-like 1 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558518 | + | 10 | 18 | 354_393 | 528 | 861.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558518 | + | 10 | 18 | 66_106 | 528 | 861.0 | Domain | LDL-receptor class A 2 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000535915 | + | 9 | 17 | 397_438 | 487 | 820.0 | Repeat | Note=LDL-receptor class B 1 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000535915 | + | 9 | 17 | 439_485 | 487 | 820.0 | Repeat | Note=LDL-receptor class B 2 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558013 | + | 10 | 18 | 397_438 | 528 | 859.0 | Repeat | Note=LDL-receptor class B 1 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558013 | + | 10 | 18 | 439_485 | 528 | 859.0 | Repeat | Note=LDL-receptor class B 2 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558013 | + | 10 | 18 | 486_528 | 528 | 859.0 | Repeat | Note=LDL-receptor class B 3 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558518 | + | 10 | 18 | 397_438 | 528 | 861.0 | Repeat | Note=LDL-receptor class B 1 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558518 | + | 10 | 18 | 439_485 | 528 | 861.0 | Repeat | Note=LDL-receptor class B 2 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558518 | + | 10 | 18 | 486_528 | 528 | 861.0 | Repeat | Note=LDL-receptor class B 3 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000535915 | + | 9 | 17 | 397_438 | 487 | 820.0 | Repeat | Note=LDL-receptor class B 1 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000535915 | + | 9 | 17 | 439_485 | 487 | 820.0 | Repeat | Note=LDL-receptor class B 2 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558013 | + | 10 | 18 | 397_438 | 528 | 859.0 | Repeat | Note=LDL-receptor class B 1 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558013 | + | 10 | 18 | 439_485 | 528 | 859.0 | Repeat | Note=LDL-receptor class B 2 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558013 | + | 10 | 18 | 486_528 | 528 | 859.0 | Repeat | Note=LDL-receptor class B 3 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558518 | + | 10 | 18 | 397_438 | 528 | 861.0 | Repeat | Note=LDL-receptor class B 1 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558518 | + | 10 | 18 | 439_485 | 528 | 861.0 | Repeat | Note=LDL-receptor class B 2 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558518 | + | 10 | 18 | 486_528 | 528 | 861.0 | Repeat | Note=LDL-receptor class B 3 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000455727 | + | 8 | 16 | 354_393 | 360 | 693.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000455727 | + | 8 | 16 | 663_712 | 360 | 693.0 | Domain | EGF-like 3 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000535915 | + | 9 | 17 | 663_712 | 487 | 820.0 | Domain | EGF-like 3 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000545707 | + | 9 | 16 | 663_712 | 401 | 683.0 | Domain | EGF-like 3 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558013 | + | 10 | 18 | 663_712 | 528 | 859.0 | Domain | EGF-like 3 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558518 | + | 10 | 18 | 663_712 | 528 | 861.0 | Domain | EGF-like 3 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000455727 | + | 8 | 16 | 354_393 | 360 | 693.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000455727 | + | 8 | 16 | 663_712 | 360 | 693.0 | Domain | EGF-like 3 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000535915 | + | 9 | 17 | 663_712 | 487 | 820.0 | Domain | EGF-like 3 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000545707 | + | 9 | 16 | 663_712 | 401 | 683.0 | Domain | EGF-like 3 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558013 | + | 10 | 18 | 663_712 | 528 | 859.0 | Domain | EGF-like 3 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558518 | + | 10 | 18 | 663_712 | 528 | 861.0 | Domain | EGF-like 3 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000455727 | + | 8 | 16 | 823_828 | 360 | 693.0 | Motif | NPXY motif |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000535915 | + | 9 | 17 | 823_828 | 487 | 820.0 | Motif | NPXY motif |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000545707 | + | 9 | 16 | 823_828 | 401 | 683.0 | Motif | NPXY motif |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558013 | + | 10 | 18 | 823_828 | 528 | 859.0 | Motif | NPXY motif |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558518 | + | 10 | 18 | 823_828 | 528 | 861.0 | Motif | NPXY motif |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000455727 | + | 8 | 16 | 823_828 | 360 | 693.0 | Motif | NPXY motif |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000535915 | + | 9 | 17 | 823_828 | 487 | 820.0 | Motif | NPXY motif |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000545707 | + | 9 | 16 | 823_828 | 401 | 683.0 | Motif | NPXY motif |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558013 | + | 10 | 18 | 823_828 | 528 | 859.0 | Motif | NPXY motif |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558518 | + | 10 | 18 | 823_828 | 528 | 861.0 | Motif | NPXY motif |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000455727 | + | 8 | 16 | 721_768 | 360 | 693.0 | Region | Note=Clustered O-linked oligosaccharides |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000455727 | + | 8 | 16 | 811_860 | 360 | 693.0 | Region | Required for MYLIP-triggered down-regulation of LDLR |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000535915 | + | 9 | 17 | 721_768 | 487 | 820.0 | Region | Note=Clustered O-linked oligosaccharides |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000535915 | + | 9 | 17 | 811_860 | 487 | 820.0 | Region | Required for MYLIP-triggered down-regulation of LDLR |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000545707 | + | 9 | 16 | 721_768 | 401 | 683.0 | Region | Note=Clustered O-linked oligosaccharides |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000545707 | + | 9 | 16 | 811_860 | 401 | 683.0 | Region | Required for MYLIP-triggered down-regulation of LDLR |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558013 | + | 10 | 18 | 721_768 | 528 | 859.0 | Region | Note=Clustered O-linked oligosaccharides |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558013 | + | 10 | 18 | 811_860 | 528 | 859.0 | Region | Required for MYLIP-triggered down-regulation of LDLR |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558518 | + | 10 | 18 | 721_768 | 528 | 861.0 | Region | Note=Clustered O-linked oligosaccharides |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558518 | + | 10 | 18 | 811_860 | 528 | 861.0 | Region | Required for MYLIP-triggered down-regulation of LDLR |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000455727 | + | 8 | 16 | 721_768 | 360 | 693.0 | Region | Note=Clustered O-linked oligosaccharides |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000455727 | + | 8 | 16 | 811_860 | 360 | 693.0 | Region | Required for MYLIP-triggered down-regulation of LDLR |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000535915 | + | 9 | 17 | 721_768 | 487 | 820.0 | Region | Note=Clustered O-linked oligosaccharides |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000535915 | + | 9 | 17 | 811_860 | 487 | 820.0 | Region | Required for MYLIP-triggered down-regulation of LDLR |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000545707 | + | 9 | 16 | 721_768 | 401 | 683.0 | Region | Note=Clustered O-linked oligosaccharides |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000545707 | + | 9 | 16 | 811_860 | 401 | 683.0 | Region | Required for MYLIP-triggered down-regulation of LDLR |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558013 | + | 10 | 18 | 721_768 | 528 | 859.0 | Region | Note=Clustered O-linked oligosaccharides |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558013 | + | 10 | 18 | 811_860 | 528 | 859.0 | Region | Required for MYLIP-triggered down-regulation of LDLR |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558518 | + | 10 | 18 | 721_768 | 528 | 861.0 | Region | Note=Clustered O-linked oligosaccharides |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558518 | + | 10 | 18 | 811_860 | 528 | 861.0 | Region | Required for MYLIP-triggered down-regulation of LDLR |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000455727 | + | 8 | 16 | 397_438 | 360 | 693.0 | Repeat | Note=LDL-receptor class B 1 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000455727 | + | 8 | 16 | 439_485 | 360 | 693.0 | Repeat | Note=LDL-receptor class B 2 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000455727 | + | 8 | 16 | 486_528 | 360 | 693.0 | Repeat | Note=LDL-receptor class B 3 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000455727 | + | 8 | 16 | 529_572 | 360 | 693.0 | Repeat | Note=LDL-receptor class B 4 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000455727 | + | 8 | 16 | 573_615 | 360 | 693.0 | Repeat | Note=LDL-receptor class B 5 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000455727 | + | 8 | 16 | 616_658 | 360 | 693.0 | Repeat | Note=LDL-receptor class B 6 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000535915 | + | 9 | 17 | 486_528 | 487 | 820.0 | Repeat | Note=LDL-receptor class B 3 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000535915 | + | 9 | 17 | 529_572 | 487 | 820.0 | Repeat | Note=LDL-receptor class B 4 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000535915 | + | 9 | 17 | 573_615 | 487 | 820.0 | Repeat | Note=LDL-receptor class B 5 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000535915 | + | 9 | 17 | 616_658 | 487 | 820.0 | Repeat | Note=LDL-receptor class B 6 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000545707 | + | 9 | 16 | 397_438 | 401 | 683.0 | Repeat | Note=LDL-receptor class B 1 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000545707 | + | 9 | 16 | 439_485 | 401 | 683.0 | Repeat | Note=LDL-receptor class B 2 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000545707 | + | 9 | 16 | 486_528 | 401 | 683.0 | Repeat | Note=LDL-receptor class B 3 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000545707 | + | 9 | 16 | 529_572 | 401 | 683.0 | Repeat | Note=LDL-receptor class B 4 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000545707 | + | 9 | 16 | 573_615 | 401 | 683.0 | Repeat | Note=LDL-receptor class B 5 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000545707 | + | 9 | 16 | 616_658 | 401 | 683.0 | Repeat | Note=LDL-receptor class B 6 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558013 | + | 10 | 18 | 529_572 | 528 | 859.0 | Repeat | Note=LDL-receptor class B 4 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558013 | + | 10 | 18 | 573_615 | 528 | 859.0 | Repeat | Note=LDL-receptor class B 5 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558013 | + | 10 | 18 | 616_658 | 528 | 859.0 | Repeat | Note=LDL-receptor class B 6 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558518 | + | 10 | 18 | 529_572 | 528 | 861.0 | Repeat | Note=LDL-receptor class B 4 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558518 | + | 10 | 18 | 573_615 | 528 | 861.0 | Repeat | Note=LDL-receptor class B 5 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558518 | + | 10 | 18 | 616_658 | 528 | 861.0 | Repeat | Note=LDL-receptor class B 6 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000455727 | + | 8 | 16 | 397_438 | 360 | 693.0 | Repeat | Note=LDL-receptor class B 1 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000455727 | + | 8 | 16 | 439_485 | 360 | 693.0 | Repeat | Note=LDL-receptor class B 2 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000455727 | + | 8 | 16 | 486_528 | 360 | 693.0 | Repeat | Note=LDL-receptor class B 3 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000455727 | + | 8 | 16 | 529_572 | 360 | 693.0 | Repeat | Note=LDL-receptor class B 4 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000455727 | + | 8 | 16 | 573_615 | 360 | 693.0 | Repeat | Note=LDL-receptor class B 5 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000455727 | + | 8 | 16 | 616_658 | 360 | 693.0 | Repeat | Note=LDL-receptor class B 6 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000535915 | + | 9 | 17 | 486_528 | 487 | 820.0 | Repeat | Note=LDL-receptor class B 3 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000535915 | + | 9 | 17 | 529_572 | 487 | 820.0 | Repeat | Note=LDL-receptor class B 4 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000535915 | + | 9 | 17 | 573_615 | 487 | 820.0 | Repeat | Note=LDL-receptor class B 5 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000535915 | + | 9 | 17 | 616_658 | 487 | 820.0 | Repeat | Note=LDL-receptor class B 6 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000545707 | + | 9 | 16 | 397_438 | 401 | 683.0 | Repeat | Note=LDL-receptor class B 1 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000545707 | + | 9 | 16 | 439_485 | 401 | 683.0 | Repeat | Note=LDL-receptor class B 2 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000545707 | + | 9 | 16 | 486_528 | 401 | 683.0 | Repeat | Note=LDL-receptor class B 3 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000545707 | + | 9 | 16 | 529_572 | 401 | 683.0 | Repeat | Note=LDL-receptor class B 4 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000545707 | + | 9 | 16 | 573_615 | 401 | 683.0 | Repeat | Note=LDL-receptor class B 5 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000545707 | + | 9 | 16 | 616_658 | 401 | 683.0 | Repeat | Note=LDL-receptor class B 6 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558013 | + | 10 | 18 | 529_572 | 528 | 859.0 | Repeat | Note=LDL-receptor class B 4 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558013 | + | 10 | 18 | 573_615 | 528 | 859.0 | Repeat | Note=LDL-receptor class B 5 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558013 | + | 10 | 18 | 616_658 | 528 | 859.0 | Repeat | Note=LDL-receptor class B 6 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558518 | + | 10 | 18 | 529_572 | 528 | 861.0 | Repeat | Note=LDL-receptor class B 4 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558518 | + | 10 | 18 | 573_615 | 528 | 861.0 | Repeat | Note=LDL-receptor class B 5 |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558518 | + | 10 | 18 | 616_658 | 528 | 861.0 | Repeat | Note=LDL-receptor class B 6 |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000455727 | + | 8 | 16 | 22_788 | 360 | 693.0 | Topological domain | Extracellular |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000535915 | + | 9 | 17 | 22_788 | 487 | 820.0 | Topological domain | Extracellular |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000545707 | + | 9 | 16 | 22_788 | 401 | 683.0 | Topological domain | Extracellular |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558013 | + | 10 | 18 | 22_788 | 528 | 859.0 | Topological domain | Extracellular |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558518 | + | 10 | 18 | 22_788 | 528 | 861.0 | Topological domain | Extracellular |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000455727 | + | 8 | 16 | 22_788 | 360 | 693.0 | Topological domain | Extracellular |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000535915 | + | 9 | 17 | 22_788 | 487 | 820.0 | Topological domain | Extracellular |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000545707 | + | 9 | 16 | 22_788 | 401 | 683.0 | Topological domain | Extracellular |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558013 | + | 10 | 18 | 22_788 | 528 | 859.0 | Topological domain | Extracellular |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558518 | + | 10 | 18 | 22_788 | 528 | 861.0 | Topological domain | Extracellular |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000455727 | + | 8 | 16 | 789_810 | 360 | 693.0 | Transmembrane | Helical |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000535915 | + | 9 | 17 | 789_810 | 487 | 820.0 | Transmembrane | Helical |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000545707 | + | 9 | 16 | 789_810 | 401 | 683.0 | Transmembrane | Helical |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558013 | + | 10 | 18 | 789_810 | 528 | 859.0 | Transmembrane | Helical |
| Hgene | LDLR | chr19:11224437 | chr19:10415883 | ENST00000558518 | + | 10 | 18 | 789_810 | 528 | 861.0 | Transmembrane | Helical |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000455727 | + | 8 | 16 | 789_810 | 360 | 693.0 | Transmembrane | Helical |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000535915 | + | 9 | 17 | 789_810 | 487 | 820.0 | Transmembrane | Helical |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000545707 | + | 9 | 16 | 789_810 | 401 | 683.0 | Transmembrane | Helical |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558013 | + | 10 | 18 | 789_810 | 528 | 859.0 | Transmembrane | Helical |
| Hgene | LDLR | chr19:11224438 | chr19:10415884 | ENST00000558518 | + | 10 | 18 | 789_810 | 528 | 861.0 | Transmembrane | Helical |
| Tgene | ZGLP1 | chr19:11224437 | chr19:10415883 | ENST00000403903 | 2 | 4 | 206_230 | 232 | 272.0 | Zinc finger | GATA-type | |
| Tgene | ZGLP1 | chr19:11224438 | chr19:10415884 | ENST00000403903 | 2 | 4 | 206_230 | 232 | 272.0 | Zinc finger | GATA-type |
Top |
Fusion Gene Sequence for LDLR-ZGLP1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >44420_44420_1_LDLR-ZGLP1_LDLR_chr19_11224437_ENST00000455727_ZGLP1_chr19_10415883_ENST00000403352_length(transcript)=1302nt_BP=1168nt GTGCAATCGCGGGAAGCCAGGGTTTCCAGCTAGGACACAGCAGGTCGTGATCCGGGTCGGGACACTGCCTGGCAGAGGCTGCGAGCATGG GGCCCTGGGGCTGGAAATTGCGCTGGACCGTCGCCTTGCTCCTCGCCGCGGCGGGGACTGCAGTGGGCGACAGATGCGAAAGAAACGAGT TCCAGTGCCAAGACGGGAAATGCATCTCCTACAAGTGGGTCTGCGATGGCAGCGCTGAGTGCCAGGATGGCTCTGATGAGTCCCAGGAGA CGTGCTTGTCTGTCACCTGCAAATCCGGGGACTTCAGCTGTGGGGGCCGTGTCAACCGCTGCATTCCTCAGTTCTGGAGGTGCGATGGCC AAGTGGACTGCGACAACGGCTCAGACGAGCAAGGCTGTCTGACACTCTGCGAGGGACCCAACAAGTTCAAGTGTCACAGCGGCGAATGCA TCACCCTGGACAAAGTCTGCAACATGGCTAGAGACTGCCGGGACTGGTCAGATGAACCCATCAAAGAGTGCGGGACCAACGAATGCTTGG ACAACAACGGCGGCTGTTCCCACGTCTGCAATGACCTTAAGATCGGCTACGAGTGCCTGTGCCCCGACGGCTTCCAGCTGGTGGCCCAGC GAAGATGCGAAGATATCGATGAGTGTCAGGATCCCGACACCTGCAGCCAGCTCTGCGTGAACCTGGAGGGTGGCTACAAGTGCCAGTGTG AGGAAGGCTTCCAGCTGGACCCCCACACGAAGGCCTGCAAGGCTGTGGGCTCCATCGCCTACCTCTTCTTCACCAACCGGCACGAGGTCA GGAAGATGACGCTGGACCGGAGCGAGTACACCAGCCTCATCCCCAACCTGAGGAACGTGGTCGCTCTGGACACGGAGGTGGCCAGCAATA GAATCTACTGGTCTGACCTGTCCCAGAGAATGATCTGCAGCACCCAGCTTGACAGAGCCCACGGCGTCTCTTCCTATGACACCGTCATCA GCAGAGACATCCAGGCCCCCGACGGGCTGGCTGTGGACTGGATCCACAGCAACATCTACTGGACCGACTCTGTCCTGGGCACTGTCTCTG TTGCGGATACCAAGGGCGTGAAGAGGAAAACGTTATTCAGGGAGAACGGCTCCAAGCCAAGGGCCATCGTGGTGGATCCTGTTCATGGGT ACAAGAAATACGGCACTCGCTGCTCCAGCTGCTGGCTGGTGCCCAGGAAAAATGTCCAGCCCAAGAGGCTATGTGGCAGATGTGGAGTGT >44420_44420_1_LDLR-ZGLP1_LDLR_chr19_11224437_ENST00000455727_ZGLP1_chr19_10415883_ENST00000403352_length(amino acids)=405AA_BP=367 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCLTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPD GFQLVAQRRCEDIDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGSIAYLFFTNRHEVRKMTLDRSEYTSLIPNLRNVVAL DTEVASNRIYWSDLSQRMICSTQLDRAHGVSSYDTVISRDIQAPDGLAVDWIHSNIYWTDSVLGTVSVADTKGVKRKTLFRENGSKPRAI -------------------------------------------------------------- >44420_44420_2_LDLR-ZGLP1_LDLR_chr19_11224437_ENST00000535915_ZGLP1_chr19_10415883_ENST00000403352_length(transcript)=1683nt_BP=1549nt GTGCAATCGCGGGAAGCCAGGGTTTCCAGCTAGGACACAGCAGGTCGTGATCCGGGTCGGGACACTGCCTGGCAGAGGCTGCGAGCATGG GGCCCTGGGGCTGGAAATTGCGCTGGACCGTCGCCTTGCTCCTCGCCGCGGCGGGGACTGCAGTGGGCGACAGATGCGAAAGAAACGAGT TCCAGTGCCAAGACGGGAAATGCATCTCCTACAAGTGGGTCTGCGATGGCAGCGCTGAGTGCCAGGATGGCTCTGATGAGTCCCAGGAGA CGTGCTCCCCCAAGACGTGCTCCCAGGACGAGTTTCGCTGCCACGATGGGAAGTGCATCTCTCGGCAGTTCGTCTGTGACTCAGACCGGG ACTGCTTGGACGGCTCAGACGAGGCCTCCTGCCCGGTGCTCACCTGTGGTCCCGCCAGCTTCCAGTGCAACAGCTCCACCTGCATCCCCC AGCTGTGGGCCTGCGACAACGACCCCGACTGCGAAGATGGCTCGGATGAGTGGCCGCAGCGCTGTAGGGGTCTTTACGTGTTCCAAGGGG ACAGTAGCCCCTGCTCGGCCTTCGAGTTCCACTGCCTAAGTGGCGAGTGCATCCACTCCAGCTGGCGCTGTGATGGTGGCCCCGACTGCA AGGACAAATCTGACGAGGAAAACTGCGCTGTGGCCACCTGTCGCCCTGACGAATTCCAGTGCTCTGATGGAAACTGCATCCATGGCAGCC GGCAGTGTGACCGGGAATATGACTGCAAGGACATGAGCGATGAAGTTGGCTGCGTTAATGTGACACTCTGCGAGGGACCCAACAAGTTCA AGTGTCACAGCGGCGAATGCATCACCCTGGACAAAGTCTGCAACATGGCTAGAGACTGCCGGGACTGGTCAGATGAACCCATCAAAGAGT GCGGGACCAACGAATGCTTGGACAACAACGGCGGCTGTTCCCACGTCTGCAATGACCTTAAGATCGGCTACGAGTGCCTGTGCCCCGACG GCTTCCAGCTGGTGGCCCAGCGAAGATGCGAAGATATCGATGAGTGTCAGGATCCCGACACCTGCAGCCAGCTCTGCGTGAACCTGGAGG GTGGCTACAAGTGCCAGTGTGAGGAAGGCTTCCAGCTGGACCCCCACACGAAGGCCTGCAAGGCTGTGGGCTCCATCGCCTACCTCTTCT TCACCAACCGGCACGAGGTCAGGAAGATGACGCTGGACCGGAGCGAGTACACCAGCCTCATCCCCAACCTGAGGAACGTGGTCGCTCTGG ACACGGAGGTGGCCAGCAATAGAATCTACTGGTCTGACCTGTCCCAGAGAATGATCTGCAGCACCCAGCTTGACAGAGCCCACGGCGTCT CTTCCTATGACACCGTCATCAGCAGAGACATCCAGGCCCCCGACGGGCTGGCTGTGGACTGGATCCACAGCAACATCTACTGGACCGACT CTGTCCTGGGCACTGTCTCTGTTGCGGATACCAAGGGCGTGAAGAGGAAAACGTTATTCAGGGAGAACGGCTCCAAGCCAAGGGCCATCG TGGTGGATCCTGTTCATGGGTACAAGAAATACGGCACTCGCTGCTCCAGCTGCTGGCTGGTGCCCAGGAAAAATGTCCAGCCCAAGAGGC >44420_44420_2_LDLR-ZGLP1_LDLR_chr19_11224437_ENST00000535915_ZGLP1_chr19_10415883_ENST00000403352_length(amino acids)=532AA_BP=494 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCSPKTCSQDEFRCHDGKCISRQ FVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCEDGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWR CDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDMSDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDW SDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCEDIDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAV GSIAYLFFTNRHEVRKMTLDRSEYTSLIPNLRNVVALDTEVASNRIYWSDLSQRMICSTQLDRAHGVSSYDTVISRDIQAPDGLAVDWIH -------------------------------------------------------------- >44420_44420_3_LDLR-ZGLP1_LDLR_chr19_11224437_ENST00000545707_ZGLP1_chr19_10415883_ENST00000403352_length(transcript)=1425nt_BP=1291nt GTGCAATCGCGGGAAGCCAGGGTTTCCAGCTAGGACACAGCAGGTCGTGATCCGGGTCGGGACACTGCCTGGCAGAGGCTGCGAGCATGG GGCCCTGGGGCTGGAAATTGCGCTGGACCGTCGCCTTGCTCCTCGCCGCGGCGGGGACTGCAGTGGGCGACAGATGCGAAAGAAACGAGT TCCAGTGCCAAGACGGGAAATGCATCTCCTACAAGTGGGTCTGCGATGGCAGCGCTGAGTGCCAGGATGGCTCTGATGAGTCCCAGGAGA CGTGCTTGTCTGTCACCTGCAAATCCGGGGACTTCAGCTGTGGGGGCCGTGTCAACCGCTGCATTCCTCAGTTCTGGAGGTGCGATGGCC AAGTGGACTGCGACAACGGCTCAGACGAGCAAGGCTGTCCTGTGGCCACCTGTCGCCCTGACGAATTCCAGTGCTCTGATGGAAACTGCA TCCATGGCAGCCGGCAGTGTGACCGGGAATATGACTGCAAGGACATGAGCGATGAAGTTGGCTGCGTTAATGTGACACTCTGCGAGGGAC CCAACAAGTTCAAGTGTCACAGCGGCGAATGCATCACCCTGGACAAAGTCTGCAACATGGCTAGAGACTGCCGGGACTGGTCAGATGAAC CCATCAAAGAGTGCGGGACCAACGAATGCTTGGACAACAACGGCGGCTGTTCCCACGTCTGCAATGACCTTAAGATCGGCTACGAGTGCC TGTGCCCCGACGGCTTCCAGCTGGTGGCCCAGCGAAGATGCGAAGATATCGATGAGTGTCAGGATCCCGACACCTGCAGCCAGCTCTGCG TGAACCTGGAGGGTGGCTACAAGTGCCAGTGTGAGGAAGGCTTCCAGCTGGACCCCCACACGAAGGCCTGCAAGGCTGTGGGCTCCATCG CCTACCTCTTCTTCACCAACCGGCACGAGGTCAGGAAGATGACGCTGGACCGGAGCGAGTACACCAGCCTCATCCCCAACCTGAGGAACG TGGTCGCTCTGGACACGGAGGTGGCCAGCAATAGAATCTACTGGTCTGACCTGTCCCAGAGAATGATCTGCAGCACCCAGCTTGACAGAG CCCACGGCGTCTCTTCCTATGACACCGTCATCAGCAGAGACATCCAGGCCCCCGACGGGCTGGCTGTGGACTGGATCCACAGCAACATCT ACTGGACCGACTCTGTCCTGGGCACTGTCTCTGTTGCGGATACCAAGGGCGTGAAGAGGAAAACGTTATTCAGGGAGAACGGCTCCAAGC CAAGGGCCATCGTGGTGGATCCTGTTCATGGGTACAAGAAATACGGCACTCGCTGCTCCAGCTGCTGGCTGGTGCCCAGGAAAAATGTCC >44420_44420_3_LDLR-ZGLP1_LDLR_chr19_11224437_ENST00000545707_ZGLP1_chr19_10415883_ENST00000403352_length(amino acids)=446AA_BP=408 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPVATCRPDEFQCSDGNCIHGSRQCDREYDCKDMSDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARD CRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCEDIDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKA CKAVGSIAYLFFTNRHEVRKMTLDRSEYTSLIPNLRNVVALDTEVASNRIYWSDLSQRMICSTQLDRAHGVSSYDTVISRDIQAPDGLAV -------------------------------------------------------------- >44420_44420_4_LDLR-ZGLP1_LDLR_chr19_11224437_ENST00000557933_ZGLP1_chr19_10415883_ENST00000403352_length(transcript)=1807nt_BP=1673nt AGTGCAATCGCGGGAAGCCAGGGTTTCCAGCTAGGACACAGCAGGTCGTGATCCGGGTCGGGACACTGCCTGGCAGAGGCTGCGAGCATG GGGCCCTGGGGCTGGAAATTGCGCTGGACCGTCGCCTTGCTCCTCGCCGCGGCGGGGACTGCAGTGGGCGACAGATGCGAAAGAAACGAG TTCCAGTGCCAAGACGGGAAATGCATCTCCTACAAGTGGGTCTGCGATGGCAGCGCTGAGTGCCAGGATGGCTCTGATGAGTCCCAGGAG ACGTGCTTGTCTGTCACCTGCAAATCCGGGGACTTCAGCTGTGGGGGCCGTGTCAACCGCTGCATTCCTCAGTTCTGGAGGTGCGATGGC CAAGTGGACTGCGACAACGGCTCAGACGAGCAAGGCTGTCCCCCCAAGACGTGCTCCCAGGACGAGTTTCGCTGCCACGATGGGAAGTGC ATCTCTCGGCAGTTCGTCTGTGACTCAGACCGGGACTGCTTGGACGGCTCAGACGAGGCCTCCTGCCCGGTGCTCACCTGTGGTCCCGCC AGCTTCCAGTGCAACAGCTCCACCTGCATCCCCCAGCTGTGGGCCTGCGACAACGACCCCGACTGCGAAGATGGCTCGGATGAGTGGCCG CAGCGCTGTAGGGGTCTTTACGTGTTCCAAGGGGACAGTAGCCCCTGCTCGGCCTTCGAGTTCCACTGCCTAAGTGGCGAGTGCATCCAC TCCAGCTGGCGCTGTGATGGTGGCCCCGACTGCAAGGACAAATCTGACGAGGAAAACTGCGCTGTGGCCACCTGTCGCCCTGACGAATTC CAGTGCTCTGATGGAAACTGCATCCATGGCAGCCGGCAGTGTGACCGGGAATATGACTGCAAGGACATGAGCGATGAAGTTGGCTGCGTT AATGTGACACTCTGCGAGGGACCCAACAAGTTCAAGTGTCACAGCGGCGAATGCATCACCCTGGACAAAGTCTGCAACATGGCTAGAGAC TGCCGGGACTGGTCAGATGAACCCATCAAAGAGTGCGGGACCAACGAATGCTTGGACAACAACGGCGGCTGTTCCCACGTCTGCAATGAC CTTAAGATCGGCTACGAGTGCCTGTGCCCCGACGGCTTCCAGCTGGTGGCCCAGCGAAGATGCGAAGATATCGATGAGTGTCAGGATCCC GACACCTGCAGCCAGCTCTGCGTGAACCTGGAGGGTGGCTACAAGTGCCAGTGTGAGGAAGGCTTCCAGCTGGACCCCCACACGAAGGCC TGCAAGGCTGTGGGCTCCATCGCCTACCTCTTCTTCACCAACCGGCACGAGGTCAGGAAGATGACGCTGGACCGGAGCGAGTACACCAGC CTCATCCCCAACCTGAGGAACGTGGTCGCTCTGGACACGGAGGTGGCCAGCAATAGAATCTACTGGTCTGACCTGTCCCAGAGAATGATC TGCAGCACCCAGCTTGACAGAGCCCACGGCGTCTCTTCCTATGACACCGTCATCAGCAGAGACATCCAGGCCCCCGACGGGCTGGCTGTG GACTGGATCCACAGCAACATCTACTGGACCGACTCTGTCCTGGGCACTGTCTCTGTTGCGGATACCAAGGGCGTGAAGAGGAAAACGTTA TTCAGGGAGAACGGCTCCAAGCCAAGGGCCATCGTGGTGGATCCTGTTCATGGGTACAAGAAATACGGCACTCGCTGCTCCAGCTGCTGG CTGGTGCCCAGGAAAAATGTCCAGCCCAAGAGGCTATGTGGCAGATGTGGAGTGTCCCTGGACCCCATTCAGGAAGGTTAAACCCAGCTT >44420_44420_4_LDLR-ZGLP1_LDLR_chr19_11224437_ENST00000557933_ZGLP1_chr19_10415883_ENST00000403352_length(amino acids)=573AA_BP=535 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPPKTCSQDEFRCHDGKCISRQFVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCE DGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWRCDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDM SDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCED IDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGSIAYLFFTNRHEVRKMTLDRSEYTSLIPNLRNVVALDTEVASNRIYWS DLSQRMICSTQLDRAHGVSSYDTVISRDIQAPDGLAVDWIHSNIYWTDSVLGTVSVADTKGVKRKTLFRENGSKPRAIVVDPVHGYKKYG -------------------------------------------------------------- >44420_44420_5_LDLR-ZGLP1_LDLR_chr19_11224437_ENST00000558013_ZGLP1_chr19_10415883_ENST00000403352_length(transcript)=1791nt_BP=1657nt GCCAGGGTTTCCAGCTAGGACACAGCAGGTCGTGATCCGGGTCGGGACACTGCCTGGCAGAGGCTGCGAGCATGGGGCCCTGGGGCTGGA AATTGCGCTGGACCGTCGCCTTGCTCCTCGCCGCGGCGGGGACTGCAGTGGGCGACAGATGCGAAAGAAACGAGTTCCAGTGCCAAGACG GGAAATGCATCTCCTACAAGTGGGTCTGCGATGGCAGCGCTGAGTGCCAGGATGGCTCTGATGAGTCCCAGGAGACGTGCTTGTCTGTCA CCTGCAAATCCGGGGACTTCAGCTGTGGGGGCCGTGTCAACCGCTGCATTCCTCAGTTCTGGAGGTGCGATGGCCAAGTGGACTGCGACA ACGGCTCAGACGAGCAAGGCTGTCCCCCCAAGACGTGCTCCCAGGACGAGTTTCGCTGCCACGATGGGAAGTGCATCTCTCGGCAGTTCG TCTGTGACTCAGACCGGGACTGCTTGGACGGCTCAGACGAGGCCTCCTGCCCGGTGCTCACCTGTGGTCCCGCCAGCTTCCAGTGCAACA GCTCCACCTGCATCCCCCAGCTGTGGGCCTGCGACAACGACCCCGACTGCGAAGATGGCTCGGATGAGTGGCCGCAGCGCTGTAGGGGTC TTTACGTGTTCCAAGGGGACAGTAGCCCCTGCTCGGCCTTCGAGTTCCACTGCCTAAGTGGCGAGTGCATCCACTCCAGCTGGCGCTGTG ATGGTGGCCCCGACTGCAAGGACAAATCTGACGAGGAAAACTGCGCTGTGGCCACCTGTCGCCCTGACGAATTCCAGTGCTCTGATGGAA ACTGCATCCATGGCAGCCGGCAGTGTGACCGGGAATATGACTGCAAGGACATGAGCGATGAAGTTGGCTGCGTTAATGTGACACTCTGCG AGGGACCCAACAAGTTCAAGTGTCACAGCGGCGAATGCATCACCCTGGACAAAGTCTGCAACATGGCTAGAGACTGCCGGGACTGGTCAG ATGAACCCATCAAAGAGTGCGGGACCAACGAATGCTTGGACAACAACGGCGGCTGTTCCCACGTCTGCAATGACCTTAAGATCGGCTACG AGTGCCTGTGCCCCGACGGCTTCCAGCTGGTGGCCCAGCGAAGATGCGAAGATATCGATGAGTGTCAGGATCCCGACACCTGCAGCCAGC TCTGCGTGAACCTGGAGGGTGGCTACAAGTGCCAGTGTGAGGAAGGCTTCCAGCTGGACCCCCACACGAAGGCCTGCAAGGCTGTGGGCT CCATCGCCTACCTCTTCTTCACCAACCGGCACGAGGTCAGGAAGATGACGCTGGACCGGAGCGAGTACACCAGCCTCATCCCCAACCTGA GGAACGTGGTCGCTCTGGACACGGAGGTGGCCAGCAATAGAATCTACTGGTCTGACCTGTCCCAGAGAATGATCTGCAGCACCCAGCTTG ACAGAGCCCACGGCGTCTCTTCCTATGACACCGTCATCAGCAGAGACATCCAGGCCCCCGACGGGCTGGCTGTGGACTGGATCCACAGCA ACATCTACTGGACCGACTCTGTCCTGGGCACTGTCTCTGTTGCGGATACCAAGGGCGTGAAGAGGAAAACGTTATTCAGGGAGAACGGCT CCAAGCCAAGGGCCATCGTGGTGGATCCTGTTCATGGGTACAAGAAATACGGCACTCGCTGCTCCAGCTGCTGGCTGGTGCCCAGGAAAA >44420_44420_5_LDLR-ZGLP1_LDLR_chr19_11224437_ENST00000558013_ZGLP1_chr19_10415883_ENST00000403352_length(amino acids)=573AA_BP=535 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPPKTCSQDEFRCHDGKCISRQFVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCE DGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWRCDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDM SDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCED IDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGSIAYLFFTNRHEVRKMTLDRSEYTSLIPNLRNVVALDTEVASNRIYWS DLSQRMICSTQLDRAHGVSSYDTVISRDIQAPDGLAVDWIHSNIYWTDSVLGTVSVADTKGVKRKTLFRENGSKPRAIVVDPVHGYKKYG -------------------------------------------------------------- >44420_44420_6_LDLR-ZGLP1_LDLR_chr19_11224437_ENST00000558518_ZGLP1_chr19_10415883_ENST00000403352_length(transcript)=1907nt_BP=1773nt CTCTTGCAGTGAGGTGAAGACATTTGAAAATCACCCCACTGCAAACTCCTCCCCCTGCTAGAAACCTCACATTGAAATGCTGTAAATGAC GTGGGCCCCGAGTGCAATCGCGGGAAGCCAGGGTTTCCAGCTAGGACACAGCAGGTCGTGATCCGGGTCGGGACACTGCCTGGCAGAGGC TGCGAGCATGGGGCCCTGGGGCTGGAAATTGCGCTGGACCGTCGCCTTGCTCCTCGCCGCGGCGGGGACTGCAGTGGGCGACAGATGCGA AAGAAACGAGTTCCAGTGCCAAGACGGGAAATGCATCTCCTACAAGTGGGTCTGCGATGGCAGCGCTGAGTGCCAGGATGGCTCTGATGA GTCCCAGGAGACGTGCTTGTCTGTCACCTGCAAATCCGGGGACTTCAGCTGTGGGGGCCGTGTCAACCGCTGCATTCCTCAGTTCTGGAG GTGCGATGGCCAAGTGGACTGCGACAACGGCTCAGACGAGCAAGGCTGTCCCCCCAAGACGTGCTCCCAGGACGAGTTTCGCTGCCACGA TGGGAAGTGCATCTCTCGGCAGTTCGTCTGTGACTCAGACCGGGACTGCTTGGACGGCTCAGACGAGGCCTCCTGCCCGGTGCTCACCTG TGGTCCCGCCAGCTTCCAGTGCAACAGCTCCACCTGCATCCCCCAGCTGTGGGCCTGCGACAACGACCCCGACTGCGAAGATGGCTCGGA TGAGTGGCCGCAGCGCTGTAGGGGTCTTTACGTGTTCCAAGGGGACAGTAGCCCCTGCTCGGCCTTCGAGTTCCACTGCCTAAGTGGCGA GTGCATCCACTCCAGCTGGCGCTGTGATGGTGGCCCCGACTGCAAGGACAAATCTGACGAGGAAAACTGCGCTGTGGCCACCTGTCGCCC TGACGAATTCCAGTGCTCTGATGGAAACTGCATCCATGGCAGCCGGCAGTGTGACCGGGAATATGACTGCAAGGACATGAGCGATGAAGT TGGCTGCGTTAATGTGACACTCTGCGAGGGACCCAACAAGTTCAAGTGTCACAGCGGCGAATGCATCACCCTGGACAAAGTCTGCAACAT GGCTAGAGACTGCCGGGACTGGTCAGATGAACCCATCAAAGAGTGCGGGACCAACGAATGCTTGGACAACAACGGCGGCTGTTCCCACGT CTGCAATGACCTTAAGATCGGCTACGAGTGCCTGTGCCCCGACGGCTTCCAGCTGGTGGCCCAGCGAAGATGCGAAGATATCGATGAGTG TCAGGATCCCGACACCTGCAGCCAGCTCTGCGTGAACCTGGAGGGTGGCTACAAGTGCCAGTGTGAGGAAGGCTTCCAGCTGGACCCCCA CACGAAGGCCTGCAAGGCTGTGGGCTCCATCGCCTACCTCTTCTTCACCAACCGGCACGAGGTCAGGAAGATGACGCTGGACCGGAGCGA GTACACCAGCCTCATCCCCAACCTGAGGAACGTGGTCGCTCTGGACACGGAGGTGGCCAGCAATAGAATCTACTGGTCTGACCTGTCCCA GAGAATGATCTGCAGCACCCAGCTTGACAGAGCCCACGGCGTCTCTTCCTATGACACCGTCATCAGCAGAGACATCCAGGCCCCCGACGG GCTGGCTGTGGACTGGATCCACAGCAACATCTACTGGACCGACTCTGTCCTGGGCACTGTCTCTGTTGCGGATACCAAGGGCGTGAAGAG GAAAACGTTATTCAGGGAGAACGGCTCCAAGCCAAGGGCCATCGTGGTGGATCCTGTTCATGGGTACAAGAAATACGGCACTCGCTGCTC CAGCTGCTGGCTGGTGCCCAGGAAAAATGTCCAGCCCAAGAGGCTATGTGGCAGATGTGGAGTGTCCCTGGACCCCATTCAGGAAGGTTA >44420_44420_6_LDLR-ZGLP1_LDLR_chr19_11224437_ENST00000558518_ZGLP1_chr19_10415883_ENST00000403352_length(amino acids)=573AA_BP=535 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPPKTCSQDEFRCHDGKCISRQFVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCE DGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWRCDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDM SDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCED IDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGSIAYLFFTNRHEVRKMTLDRSEYTSLIPNLRNVVALDTEVASNRIYWS DLSQRMICSTQLDRAHGVSSYDTVISRDIQAPDGLAVDWIHSNIYWTDSVLGTVSVADTKGVKRKTLFRENGSKPRAIVVDPVHGYKKYG -------------------------------------------------------------- >44420_44420_7_LDLR-ZGLP1_LDLR_chr19_11224438_ENST00000455727_ZGLP1_chr19_10415884_ENST00000403352_length(transcript)=1302nt_BP=1168nt GTGCAATCGCGGGAAGCCAGGGTTTCCAGCTAGGACACAGCAGGTCGTGATCCGGGTCGGGACACTGCCTGGCAGAGGCTGCGAGCATGG GGCCCTGGGGCTGGAAATTGCGCTGGACCGTCGCCTTGCTCCTCGCCGCGGCGGGGACTGCAGTGGGCGACAGATGCGAAAGAAACGAGT TCCAGTGCCAAGACGGGAAATGCATCTCCTACAAGTGGGTCTGCGATGGCAGCGCTGAGTGCCAGGATGGCTCTGATGAGTCCCAGGAGA CGTGCTTGTCTGTCACCTGCAAATCCGGGGACTTCAGCTGTGGGGGCCGTGTCAACCGCTGCATTCCTCAGTTCTGGAGGTGCGATGGCC AAGTGGACTGCGACAACGGCTCAGACGAGCAAGGCTGTCTGACACTCTGCGAGGGACCCAACAAGTTCAAGTGTCACAGCGGCGAATGCA TCACCCTGGACAAAGTCTGCAACATGGCTAGAGACTGCCGGGACTGGTCAGATGAACCCATCAAAGAGTGCGGGACCAACGAATGCTTGG ACAACAACGGCGGCTGTTCCCACGTCTGCAATGACCTTAAGATCGGCTACGAGTGCCTGTGCCCCGACGGCTTCCAGCTGGTGGCCCAGC GAAGATGCGAAGATATCGATGAGTGTCAGGATCCCGACACCTGCAGCCAGCTCTGCGTGAACCTGGAGGGTGGCTACAAGTGCCAGTGTG AGGAAGGCTTCCAGCTGGACCCCCACACGAAGGCCTGCAAGGCTGTGGGCTCCATCGCCTACCTCTTCTTCACCAACCGGCACGAGGTCA GGAAGATGACGCTGGACCGGAGCGAGTACACCAGCCTCATCCCCAACCTGAGGAACGTGGTCGCTCTGGACACGGAGGTGGCCAGCAATA GAATCTACTGGTCTGACCTGTCCCAGAGAATGATCTGCAGCACCCAGCTTGACAGAGCCCACGGCGTCTCTTCCTATGACACCGTCATCA GCAGAGACATCCAGGCCCCCGACGGGCTGGCTGTGGACTGGATCCACAGCAACATCTACTGGACCGACTCTGTCCTGGGCACTGTCTCTG TTGCGGATACCAAGGGCGTGAAGAGGAAAACGTTATTCAGGGAGAACGGCTCCAAGCCAAGGGCCATCGTGGTGGATCCTGTTCATGGGT ACAAGAAATACGGCACTCGCTGCTCCAGCTGCTGGCTGGTGCCCAGGAAAAATGTCCAGCCCAAGAGGCTATGTGGCAGATGTGGAGTGT >44420_44420_7_LDLR-ZGLP1_LDLR_chr19_11224438_ENST00000455727_ZGLP1_chr19_10415884_ENST00000403352_length(amino acids)=405AA_BP=367 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCLTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPD GFQLVAQRRCEDIDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGSIAYLFFTNRHEVRKMTLDRSEYTSLIPNLRNVVAL DTEVASNRIYWSDLSQRMICSTQLDRAHGVSSYDTVISRDIQAPDGLAVDWIHSNIYWTDSVLGTVSVADTKGVKRKTLFRENGSKPRAI -------------------------------------------------------------- >44420_44420_8_LDLR-ZGLP1_LDLR_chr19_11224438_ENST00000535915_ZGLP1_chr19_10415884_ENST00000403352_length(transcript)=1683nt_BP=1549nt GTGCAATCGCGGGAAGCCAGGGTTTCCAGCTAGGACACAGCAGGTCGTGATCCGGGTCGGGACACTGCCTGGCAGAGGCTGCGAGCATGG GGCCCTGGGGCTGGAAATTGCGCTGGACCGTCGCCTTGCTCCTCGCCGCGGCGGGGACTGCAGTGGGCGACAGATGCGAAAGAAACGAGT TCCAGTGCCAAGACGGGAAATGCATCTCCTACAAGTGGGTCTGCGATGGCAGCGCTGAGTGCCAGGATGGCTCTGATGAGTCCCAGGAGA CGTGCTCCCCCAAGACGTGCTCCCAGGACGAGTTTCGCTGCCACGATGGGAAGTGCATCTCTCGGCAGTTCGTCTGTGACTCAGACCGGG ACTGCTTGGACGGCTCAGACGAGGCCTCCTGCCCGGTGCTCACCTGTGGTCCCGCCAGCTTCCAGTGCAACAGCTCCACCTGCATCCCCC AGCTGTGGGCCTGCGACAACGACCCCGACTGCGAAGATGGCTCGGATGAGTGGCCGCAGCGCTGTAGGGGTCTTTACGTGTTCCAAGGGG ACAGTAGCCCCTGCTCGGCCTTCGAGTTCCACTGCCTAAGTGGCGAGTGCATCCACTCCAGCTGGCGCTGTGATGGTGGCCCCGACTGCA AGGACAAATCTGACGAGGAAAACTGCGCTGTGGCCACCTGTCGCCCTGACGAATTCCAGTGCTCTGATGGAAACTGCATCCATGGCAGCC GGCAGTGTGACCGGGAATATGACTGCAAGGACATGAGCGATGAAGTTGGCTGCGTTAATGTGACACTCTGCGAGGGACCCAACAAGTTCA AGTGTCACAGCGGCGAATGCATCACCCTGGACAAAGTCTGCAACATGGCTAGAGACTGCCGGGACTGGTCAGATGAACCCATCAAAGAGT GCGGGACCAACGAATGCTTGGACAACAACGGCGGCTGTTCCCACGTCTGCAATGACCTTAAGATCGGCTACGAGTGCCTGTGCCCCGACG GCTTCCAGCTGGTGGCCCAGCGAAGATGCGAAGATATCGATGAGTGTCAGGATCCCGACACCTGCAGCCAGCTCTGCGTGAACCTGGAGG GTGGCTACAAGTGCCAGTGTGAGGAAGGCTTCCAGCTGGACCCCCACACGAAGGCCTGCAAGGCTGTGGGCTCCATCGCCTACCTCTTCT TCACCAACCGGCACGAGGTCAGGAAGATGACGCTGGACCGGAGCGAGTACACCAGCCTCATCCCCAACCTGAGGAACGTGGTCGCTCTGG ACACGGAGGTGGCCAGCAATAGAATCTACTGGTCTGACCTGTCCCAGAGAATGATCTGCAGCACCCAGCTTGACAGAGCCCACGGCGTCT CTTCCTATGACACCGTCATCAGCAGAGACATCCAGGCCCCCGACGGGCTGGCTGTGGACTGGATCCACAGCAACATCTACTGGACCGACT CTGTCCTGGGCACTGTCTCTGTTGCGGATACCAAGGGCGTGAAGAGGAAAACGTTATTCAGGGAGAACGGCTCCAAGCCAAGGGCCATCG TGGTGGATCCTGTTCATGGGTACAAGAAATACGGCACTCGCTGCTCCAGCTGCTGGCTGGTGCCCAGGAAAAATGTCCAGCCCAAGAGGC >44420_44420_8_LDLR-ZGLP1_LDLR_chr19_11224438_ENST00000535915_ZGLP1_chr19_10415884_ENST00000403352_length(amino acids)=532AA_BP=494 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCSPKTCSQDEFRCHDGKCISRQ FVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCEDGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWR CDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDMSDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDW SDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCEDIDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAV GSIAYLFFTNRHEVRKMTLDRSEYTSLIPNLRNVVALDTEVASNRIYWSDLSQRMICSTQLDRAHGVSSYDTVISRDIQAPDGLAVDWIH -------------------------------------------------------------- >44420_44420_9_LDLR-ZGLP1_LDLR_chr19_11224438_ENST00000545707_ZGLP1_chr19_10415884_ENST00000403352_length(transcript)=1425nt_BP=1291nt GTGCAATCGCGGGAAGCCAGGGTTTCCAGCTAGGACACAGCAGGTCGTGATCCGGGTCGGGACACTGCCTGGCAGAGGCTGCGAGCATGG GGCCCTGGGGCTGGAAATTGCGCTGGACCGTCGCCTTGCTCCTCGCCGCGGCGGGGACTGCAGTGGGCGACAGATGCGAAAGAAACGAGT TCCAGTGCCAAGACGGGAAATGCATCTCCTACAAGTGGGTCTGCGATGGCAGCGCTGAGTGCCAGGATGGCTCTGATGAGTCCCAGGAGA CGTGCTTGTCTGTCACCTGCAAATCCGGGGACTTCAGCTGTGGGGGCCGTGTCAACCGCTGCATTCCTCAGTTCTGGAGGTGCGATGGCC AAGTGGACTGCGACAACGGCTCAGACGAGCAAGGCTGTCCTGTGGCCACCTGTCGCCCTGACGAATTCCAGTGCTCTGATGGAAACTGCA TCCATGGCAGCCGGCAGTGTGACCGGGAATATGACTGCAAGGACATGAGCGATGAAGTTGGCTGCGTTAATGTGACACTCTGCGAGGGAC CCAACAAGTTCAAGTGTCACAGCGGCGAATGCATCACCCTGGACAAAGTCTGCAACATGGCTAGAGACTGCCGGGACTGGTCAGATGAAC CCATCAAAGAGTGCGGGACCAACGAATGCTTGGACAACAACGGCGGCTGTTCCCACGTCTGCAATGACCTTAAGATCGGCTACGAGTGCC TGTGCCCCGACGGCTTCCAGCTGGTGGCCCAGCGAAGATGCGAAGATATCGATGAGTGTCAGGATCCCGACACCTGCAGCCAGCTCTGCG TGAACCTGGAGGGTGGCTACAAGTGCCAGTGTGAGGAAGGCTTCCAGCTGGACCCCCACACGAAGGCCTGCAAGGCTGTGGGCTCCATCG CCTACCTCTTCTTCACCAACCGGCACGAGGTCAGGAAGATGACGCTGGACCGGAGCGAGTACACCAGCCTCATCCCCAACCTGAGGAACG TGGTCGCTCTGGACACGGAGGTGGCCAGCAATAGAATCTACTGGTCTGACCTGTCCCAGAGAATGATCTGCAGCACCCAGCTTGACAGAG CCCACGGCGTCTCTTCCTATGACACCGTCATCAGCAGAGACATCCAGGCCCCCGACGGGCTGGCTGTGGACTGGATCCACAGCAACATCT ACTGGACCGACTCTGTCCTGGGCACTGTCTCTGTTGCGGATACCAAGGGCGTGAAGAGGAAAACGTTATTCAGGGAGAACGGCTCCAAGC CAAGGGCCATCGTGGTGGATCCTGTTCATGGGTACAAGAAATACGGCACTCGCTGCTCCAGCTGCTGGCTGGTGCCCAGGAAAAATGTCC >44420_44420_9_LDLR-ZGLP1_LDLR_chr19_11224438_ENST00000545707_ZGLP1_chr19_10415884_ENST00000403352_length(amino acids)=446AA_BP=408 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPVATCRPDEFQCSDGNCIHGSRQCDREYDCKDMSDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARD CRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCEDIDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKA CKAVGSIAYLFFTNRHEVRKMTLDRSEYTSLIPNLRNVVALDTEVASNRIYWSDLSQRMICSTQLDRAHGVSSYDTVISRDIQAPDGLAV -------------------------------------------------------------- >44420_44420_10_LDLR-ZGLP1_LDLR_chr19_11224438_ENST00000557933_ZGLP1_chr19_10415884_ENST00000403352_length(transcript)=1807nt_BP=1673nt AGTGCAATCGCGGGAAGCCAGGGTTTCCAGCTAGGACACAGCAGGTCGTGATCCGGGTCGGGACACTGCCTGGCAGAGGCTGCGAGCATG GGGCCCTGGGGCTGGAAATTGCGCTGGACCGTCGCCTTGCTCCTCGCCGCGGCGGGGACTGCAGTGGGCGACAGATGCGAAAGAAACGAG TTCCAGTGCCAAGACGGGAAATGCATCTCCTACAAGTGGGTCTGCGATGGCAGCGCTGAGTGCCAGGATGGCTCTGATGAGTCCCAGGAG ACGTGCTTGTCTGTCACCTGCAAATCCGGGGACTTCAGCTGTGGGGGCCGTGTCAACCGCTGCATTCCTCAGTTCTGGAGGTGCGATGGC CAAGTGGACTGCGACAACGGCTCAGACGAGCAAGGCTGTCCCCCCAAGACGTGCTCCCAGGACGAGTTTCGCTGCCACGATGGGAAGTGC ATCTCTCGGCAGTTCGTCTGTGACTCAGACCGGGACTGCTTGGACGGCTCAGACGAGGCCTCCTGCCCGGTGCTCACCTGTGGTCCCGCC AGCTTCCAGTGCAACAGCTCCACCTGCATCCCCCAGCTGTGGGCCTGCGACAACGACCCCGACTGCGAAGATGGCTCGGATGAGTGGCCG CAGCGCTGTAGGGGTCTTTACGTGTTCCAAGGGGACAGTAGCCCCTGCTCGGCCTTCGAGTTCCACTGCCTAAGTGGCGAGTGCATCCAC TCCAGCTGGCGCTGTGATGGTGGCCCCGACTGCAAGGACAAATCTGACGAGGAAAACTGCGCTGTGGCCACCTGTCGCCCTGACGAATTC CAGTGCTCTGATGGAAACTGCATCCATGGCAGCCGGCAGTGTGACCGGGAATATGACTGCAAGGACATGAGCGATGAAGTTGGCTGCGTT AATGTGACACTCTGCGAGGGACCCAACAAGTTCAAGTGTCACAGCGGCGAATGCATCACCCTGGACAAAGTCTGCAACATGGCTAGAGAC TGCCGGGACTGGTCAGATGAACCCATCAAAGAGTGCGGGACCAACGAATGCTTGGACAACAACGGCGGCTGTTCCCACGTCTGCAATGAC CTTAAGATCGGCTACGAGTGCCTGTGCCCCGACGGCTTCCAGCTGGTGGCCCAGCGAAGATGCGAAGATATCGATGAGTGTCAGGATCCC GACACCTGCAGCCAGCTCTGCGTGAACCTGGAGGGTGGCTACAAGTGCCAGTGTGAGGAAGGCTTCCAGCTGGACCCCCACACGAAGGCC TGCAAGGCTGTGGGCTCCATCGCCTACCTCTTCTTCACCAACCGGCACGAGGTCAGGAAGATGACGCTGGACCGGAGCGAGTACACCAGC CTCATCCCCAACCTGAGGAACGTGGTCGCTCTGGACACGGAGGTGGCCAGCAATAGAATCTACTGGTCTGACCTGTCCCAGAGAATGATC TGCAGCACCCAGCTTGACAGAGCCCACGGCGTCTCTTCCTATGACACCGTCATCAGCAGAGACATCCAGGCCCCCGACGGGCTGGCTGTG GACTGGATCCACAGCAACATCTACTGGACCGACTCTGTCCTGGGCACTGTCTCTGTTGCGGATACCAAGGGCGTGAAGAGGAAAACGTTA TTCAGGGAGAACGGCTCCAAGCCAAGGGCCATCGTGGTGGATCCTGTTCATGGGTACAAGAAATACGGCACTCGCTGCTCCAGCTGCTGG CTGGTGCCCAGGAAAAATGTCCAGCCCAAGAGGCTATGTGGCAGATGTGGAGTGTCCCTGGACCCCATTCAGGAAGGTTAAACCCAGCTT >44420_44420_10_LDLR-ZGLP1_LDLR_chr19_11224438_ENST00000557933_ZGLP1_chr19_10415884_ENST00000403352_length(amino acids)=573AA_BP=535 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPPKTCSQDEFRCHDGKCISRQFVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCE DGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWRCDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDM SDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCED IDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGSIAYLFFTNRHEVRKMTLDRSEYTSLIPNLRNVVALDTEVASNRIYWS DLSQRMICSTQLDRAHGVSSYDTVISRDIQAPDGLAVDWIHSNIYWTDSVLGTVSVADTKGVKRKTLFRENGSKPRAIVVDPVHGYKKYG -------------------------------------------------------------- >44420_44420_11_LDLR-ZGLP1_LDLR_chr19_11224438_ENST00000558013_ZGLP1_chr19_10415884_ENST00000403352_length(transcript)=1791nt_BP=1657nt GCCAGGGTTTCCAGCTAGGACACAGCAGGTCGTGATCCGGGTCGGGACACTGCCTGGCAGAGGCTGCGAGCATGGGGCCCTGGGGCTGGA AATTGCGCTGGACCGTCGCCTTGCTCCTCGCCGCGGCGGGGACTGCAGTGGGCGACAGATGCGAAAGAAACGAGTTCCAGTGCCAAGACG GGAAATGCATCTCCTACAAGTGGGTCTGCGATGGCAGCGCTGAGTGCCAGGATGGCTCTGATGAGTCCCAGGAGACGTGCTTGTCTGTCA CCTGCAAATCCGGGGACTTCAGCTGTGGGGGCCGTGTCAACCGCTGCATTCCTCAGTTCTGGAGGTGCGATGGCCAAGTGGACTGCGACA ACGGCTCAGACGAGCAAGGCTGTCCCCCCAAGACGTGCTCCCAGGACGAGTTTCGCTGCCACGATGGGAAGTGCATCTCTCGGCAGTTCG TCTGTGACTCAGACCGGGACTGCTTGGACGGCTCAGACGAGGCCTCCTGCCCGGTGCTCACCTGTGGTCCCGCCAGCTTCCAGTGCAACA GCTCCACCTGCATCCCCCAGCTGTGGGCCTGCGACAACGACCCCGACTGCGAAGATGGCTCGGATGAGTGGCCGCAGCGCTGTAGGGGTC TTTACGTGTTCCAAGGGGACAGTAGCCCCTGCTCGGCCTTCGAGTTCCACTGCCTAAGTGGCGAGTGCATCCACTCCAGCTGGCGCTGTG ATGGTGGCCCCGACTGCAAGGACAAATCTGACGAGGAAAACTGCGCTGTGGCCACCTGTCGCCCTGACGAATTCCAGTGCTCTGATGGAA ACTGCATCCATGGCAGCCGGCAGTGTGACCGGGAATATGACTGCAAGGACATGAGCGATGAAGTTGGCTGCGTTAATGTGACACTCTGCG AGGGACCCAACAAGTTCAAGTGTCACAGCGGCGAATGCATCACCCTGGACAAAGTCTGCAACATGGCTAGAGACTGCCGGGACTGGTCAG ATGAACCCATCAAAGAGTGCGGGACCAACGAATGCTTGGACAACAACGGCGGCTGTTCCCACGTCTGCAATGACCTTAAGATCGGCTACG AGTGCCTGTGCCCCGACGGCTTCCAGCTGGTGGCCCAGCGAAGATGCGAAGATATCGATGAGTGTCAGGATCCCGACACCTGCAGCCAGC TCTGCGTGAACCTGGAGGGTGGCTACAAGTGCCAGTGTGAGGAAGGCTTCCAGCTGGACCCCCACACGAAGGCCTGCAAGGCTGTGGGCT CCATCGCCTACCTCTTCTTCACCAACCGGCACGAGGTCAGGAAGATGACGCTGGACCGGAGCGAGTACACCAGCCTCATCCCCAACCTGA GGAACGTGGTCGCTCTGGACACGGAGGTGGCCAGCAATAGAATCTACTGGTCTGACCTGTCCCAGAGAATGATCTGCAGCACCCAGCTTG ACAGAGCCCACGGCGTCTCTTCCTATGACACCGTCATCAGCAGAGACATCCAGGCCCCCGACGGGCTGGCTGTGGACTGGATCCACAGCA ACATCTACTGGACCGACTCTGTCCTGGGCACTGTCTCTGTTGCGGATACCAAGGGCGTGAAGAGGAAAACGTTATTCAGGGAGAACGGCT CCAAGCCAAGGGCCATCGTGGTGGATCCTGTTCATGGGTACAAGAAATACGGCACTCGCTGCTCCAGCTGCTGGCTGGTGCCCAGGAAAA >44420_44420_11_LDLR-ZGLP1_LDLR_chr19_11224438_ENST00000558013_ZGLP1_chr19_10415884_ENST00000403352_length(amino acids)=573AA_BP=535 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPPKTCSQDEFRCHDGKCISRQFVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCE DGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWRCDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDM SDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCED IDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGSIAYLFFTNRHEVRKMTLDRSEYTSLIPNLRNVVALDTEVASNRIYWS DLSQRMICSTQLDRAHGVSSYDTVISRDIQAPDGLAVDWIHSNIYWTDSVLGTVSVADTKGVKRKTLFRENGSKPRAIVVDPVHGYKKYG -------------------------------------------------------------- >44420_44420_12_LDLR-ZGLP1_LDLR_chr19_11224438_ENST00000558518_ZGLP1_chr19_10415884_ENST00000403352_length(transcript)=1907nt_BP=1773nt CTCTTGCAGTGAGGTGAAGACATTTGAAAATCACCCCACTGCAAACTCCTCCCCCTGCTAGAAACCTCACATTGAAATGCTGTAAATGAC GTGGGCCCCGAGTGCAATCGCGGGAAGCCAGGGTTTCCAGCTAGGACACAGCAGGTCGTGATCCGGGTCGGGACACTGCCTGGCAGAGGC TGCGAGCATGGGGCCCTGGGGCTGGAAATTGCGCTGGACCGTCGCCTTGCTCCTCGCCGCGGCGGGGACTGCAGTGGGCGACAGATGCGA AAGAAACGAGTTCCAGTGCCAAGACGGGAAATGCATCTCCTACAAGTGGGTCTGCGATGGCAGCGCTGAGTGCCAGGATGGCTCTGATGA GTCCCAGGAGACGTGCTTGTCTGTCACCTGCAAATCCGGGGACTTCAGCTGTGGGGGCCGTGTCAACCGCTGCATTCCTCAGTTCTGGAG GTGCGATGGCCAAGTGGACTGCGACAACGGCTCAGACGAGCAAGGCTGTCCCCCCAAGACGTGCTCCCAGGACGAGTTTCGCTGCCACGA TGGGAAGTGCATCTCTCGGCAGTTCGTCTGTGACTCAGACCGGGACTGCTTGGACGGCTCAGACGAGGCCTCCTGCCCGGTGCTCACCTG TGGTCCCGCCAGCTTCCAGTGCAACAGCTCCACCTGCATCCCCCAGCTGTGGGCCTGCGACAACGACCCCGACTGCGAAGATGGCTCGGA TGAGTGGCCGCAGCGCTGTAGGGGTCTTTACGTGTTCCAAGGGGACAGTAGCCCCTGCTCGGCCTTCGAGTTCCACTGCCTAAGTGGCGA GTGCATCCACTCCAGCTGGCGCTGTGATGGTGGCCCCGACTGCAAGGACAAATCTGACGAGGAAAACTGCGCTGTGGCCACCTGTCGCCC TGACGAATTCCAGTGCTCTGATGGAAACTGCATCCATGGCAGCCGGCAGTGTGACCGGGAATATGACTGCAAGGACATGAGCGATGAAGT TGGCTGCGTTAATGTGACACTCTGCGAGGGACCCAACAAGTTCAAGTGTCACAGCGGCGAATGCATCACCCTGGACAAAGTCTGCAACAT GGCTAGAGACTGCCGGGACTGGTCAGATGAACCCATCAAAGAGTGCGGGACCAACGAATGCTTGGACAACAACGGCGGCTGTTCCCACGT CTGCAATGACCTTAAGATCGGCTACGAGTGCCTGTGCCCCGACGGCTTCCAGCTGGTGGCCCAGCGAAGATGCGAAGATATCGATGAGTG TCAGGATCCCGACACCTGCAGCCAGCTCTGCGTGAACCTGGAGGGTGGCTACAAGTGCCAGTGTGAGGAAGGCTTCCAGCTGGACCCCCA CACGAAGGCCTGCAAGGCTGTGGGCTCCATCGCCTACCTCTTCTTCACCAACCGGCACGAGGTCAGGAAGATGACGCTGGACCGGAGCGA GTACACCAGCCTCATCCCCAACCTGAGGAACGTGGTCGCTCTGGACACGGAGGTGGCCAGCAATAGAATCTACTGGTCTGACCTGTCCCA GAGAATGATCTGCAGCACCCAGCTTGACAGAGCCCACGGCGTCTCTTCCTATGACACCGTCATCAGCAGAGACATCCAGGCCCCCGACGG GCTGGCTGTGGACTGGATCCACAGCAACATCTACTGGACCGACTCTGTCCTGGGCACTGTCTCTGTTGCGGATACCAAGGGCGTGAAGAG GAAAACGTTATTCAGGGAGAACGGCTCCAAGCCAAGGGCCATCGTGGTGGATCCTGTTCATGGGTACAAGAAATACGGCACTCGCTGCTC CAGCTGCTGGCTGGTGCCCAGGAAAAATGTCCAGCCCAAGAGGCTATGTGGCAGATGTGGAGTGTCCCTGGACCCCATTCAGGAAGGTTA >44420_44420_12_LDLR-ZGLP1_LDLR_chr19_11224438_ENST00000558518_ZGLP1_chr19_10415884_ENST00000403352_length(amino acids)=573AA_BP=535 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPPKTCSQDEFRCHDGKCISRQFVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCE DGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWRCDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDM SDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCED IDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGSIAYLFFTNRHEVRKMTLDRSEYTSLIPNLRNVVALDTEVASNRIYWS DLSQRMICSTQLDRAHGVSSYDTVISRDIQAPDGLAVDWIHSNIYWTDSVLGTVSVADTKGVKRKTLFRENGSKPRAIVVDPVHGYKKYG -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for LDLR-ZGLP1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for LDLR-ZGLP1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for LDLR-ZGLP1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
