
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:LGMN-DGCR8 (FusionGDB2 ID:44573) |
Fusion Gene Summary for LGMN-DGCR8 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: LGMN-DGCR8 | Fusion gene ID: 44573 | Hgene | Tgene | Gene symbol | LGMN | DGCR8 | Gene ID | 5641 | 54487 |
| Gene name | legumain | DGCR8 microprocessor complex subunit | |
| Synonyms | AEP|LGMN1|PRSC1 | C22orf12|DGCRK6|Gy1|pasha | |
| Cytomap | 14q32.12 | 22q11.21 | |
| Type of gene | protein-coding | protein-coding | |
| Description | legumainasparaginyl endopeptidasecysteine protease 1protease, cysteine 1protease, cysteine, 1 (legumain) | microprocessor complex subunit DGCR8DiGeorge syndrome critical region 8DiGeorge syndrome critical region gene 8 | |
| Modification date | 20200313 | 20200320 | |
| UniProtAcc | Q99538 | Q8WYQ5 | |
| Ensembl transtripts involved in fusion gene | ENST00000334869, ENST00000555699, ENST00000557434, ENST00000393218, ENST00000557034, | ENST00000351989, ENST00000383024, ENST00000407755, | |
| Fusion gene scores | * DoF score | 6 X 6 X 3=108 | 9 X 8 X 6=432 |
| # samples | 6 | 9 | |
| ** MAII score | log2(6/108*10)=-0.84799690655495 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/432*10)=-2.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: LGMN [Title/Abstract] AND DGCR8 [Title/Abstract] AND fusion [Title/Abstract] | ||
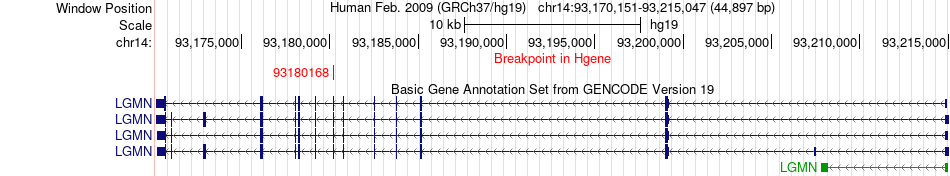
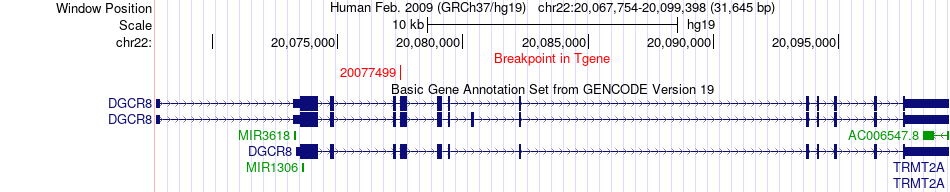
| Most frequent breakpoint | LGMN(93180168)-DGCR8(20077499), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | LGMN-DGCR8 seems lost the major protein functional domain in Hgene partner, which is a cell metabolism gene due to the frame-shifted ORF. LGMN-DGCR8 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. LGMN-DGCR8 seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. LGMN-DGCR8 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | LGMN | GO:0010447 | response to acidic pH | 18374643 |
| Hgene | LGMN | GO:0035729 | cellular response to hepatocyte growth factor stimulus | 21237226 |
| Hgene | LGMN | GO:0071277 | cellular response to calcium ion | 21237226 |
| Hgene | LGMN | GO:0090026 | positive regulation of monocyte chemotaxis | 18377911 |
| Hgene | LGMN | GO:0097202 | activation of cysteine-type endopeptidase activity | 9821970|18374643 |
| Hgene | LGMN | GO:0097264 | self proteolysis | 9821970|18374643 |
| Hgene | LGMN | GO:1904646 | cellular response to amyloid-beta | 25326800 |
| Hgene | LGMN | GO:2001028 | positive regulation of endothelial cell chemotaxis | 18377911 |
| Tgene | DGCR8 | GO:0031053 | primary miRNA processing | 15531877|15574589|24449907|24910438 |
 Fusion gene breakpoints across LGMN (5'-gene) Fusion gene breakpoints across LGMN (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across DGCR8 (3'-gene) Fusion gene breakpoints across DGCR8 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-D7-6820-01A | LGMN | chr14 | 93180168 | - | DGCR8 | chr22 | 20077499 | + |
Top |
Fusion Gene ORF analysis for LGMN-DGCR8 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| Frame-shift | ENST00000334869 | ENST00000351989 | LGMN | chr14 | 93180168 | - | DGCR8 | chr22 | 20077499 | + |
| Frame-shift | ENST00000334869 | ENST00000383024 | LGMN | chr14 | 93180168 | - | DGCR8 | chr22 | 20077499 | + |
| Frame-shift | ENST00000334869 | ENST00000407755 | LGMN | chr14 | 93180168 | - | DGCR8 | chr22 | 20077499 | + |
| Frame-shift | ENST00000555699 | ENST00000351989 | LGMN | chr14 | 93180168 | - | DGCR8 | chr22 | 20077499 | + |
| Frame-shift | ENST00000555699 | ENST00000383024 | LGMN | chr14 | 93180168 | - | DGCR8 | chr22 | 20077499 | + |
| Frame-shift | ENST00000555699 | ENST00000407755 | LGMN | chr14 | 93180168 | - | DGCR8 | chr22 | 20077499 | + |
| Frame-shift | ENST00000557434 | ENST00000351989 | LGMN | chr14 | 93180168 | - | DGCR8 | chr22 | 20077499 | + |
| Frame-shift | ENST00000557434 | ENST00000383024 | LGMN | chr14 | 93180168 | - | DGCR8 | chr22 | 20077499 | + |
| Frame-shift | ENST00000557434 | ENST00000407755 | LGMN | chr14 | 93180168 | - | DGCR8 | chr22 | 20077499 | + |
| In-frame | ENST00000393218 | ENST00000351989 | LGMN | chr14 | 93180168 | - | DGCR8 | chr22 | 20077499 | + |
| In-frame | ENST00000393218 | ENST00000383024 | LGMN | chr14 | 93180168 | - | DGCR8 | chr22 | 20077499 | + |
| In-frame | ENST00000393218 | ENST00000407755 | LGMN | chr14 | 93180168 | - | DGCR8 | chr22 | 20077499 | + |
| intron-3CDS | ENST00000557034 | ENST00000351989 | LGMN | chr14 | 93180168 | - | DGCR8 | chr22 | 20077499 | + |
| intron-3CDS | ENST00000557034 | ENST00000383024 | LGMN | chr14 | 93180168 | - | DGCR8 | chr22 | 20077499 | + |
| intron-3CDS | ENST00000557034 | ENST00000407755 | LGMN | chr14 | 93180168 | - | DGCR8 | chr22 | 20077499 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000393218 | LGMN | chr14 | 93180168 | - | ENST00000351989 | DGCR8 | chr22 | 20077499 | + | 3943 | 881 | 338 | 2179 | 613 |
| ENST00000393218 | LGMN | chr14 | 93180168 | - | ENST00000383024 | DGCR8 | chr22 | 20077499 | + | 3848 | 881 | 338 | 2080 | 580 |
| ENST00000393218 | LGMN | chr14 | 93180168 | - | ENST00000407755 | DGCR8 | chr22 | 20077499 | + | 3845 | 881 | 338 | 2080 | 580 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000393218 | ENST00000351989 | LGMN | chr14 | 93180168 | - | DGCR8 | chr22 | 20077499 | + | 0.001904641 | 0.99809533 |
| ENST00000393218 | ENST00000383024 | LGMN | chr14 | 93180168 | - | DGCR8 | chr22 | 20077499 | + | 0.002046708 | 0.9979533 |
| ENST00000393218 | ENST00000407755 | LGMN | chr14 | 93180168 | - | DGCR8 | chr22 | 20077499 | + | 0.002059775 | 0.99794024 |
Top |
Fusion Genomic Features for LGMN-DGCR8 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| LGMN | chr14 | 93180167 | - | DGCR8 | chr22 | 20077498 | + | 0.032113403 | 0.96788657 |
| LGMN | chr14 | 93180167 | - | DGCR8 | chr22 | 20077498 | + | 0.032113403 | 0.96788657 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for LGMN-DGCR8 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr14:93180168/chr22:20077499) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| LGMN | DGCR8 |
| FUNCTION: Has a strict specificity for hydrolysis of asparaginyl bonds. Can also cleave aspartyl bonds slowly, especially under acidic conditions. Required for normal lysosomal protein degradation in renal proximal tubules. Required for normal degradation of internalized EGFR. Plays a role in the regulation of cell proliferation via its role in EGFR degradation (By similarity). May be involved in the processing of proteins for MHC class II antigen presentation in the lysosomal/endosomal system. {ECO:0000250, ECO:0000269|PubMed:23776206}. | FUNCTION: Component of the microprocessor complex that acts as a RNA- and heme-binding protein that is involved in the initial step of microRNA (miRNA) biogenesis. Component of the microprocessor complex that is required to process primary miRNA transcripts (pri-miRNAs) to release precursor miRNA (pre-miRNA) in the nucleus. Within the microprocessor complex, DGCR8 function as a molecular anchor necessary for the recognition of pri-miRNA at dsRNA-ssRNA junction and directs DROSHA to cleave 11 bp away form the junction to release hairpin-shaped pre-miRNAs that are subsequently cut by the cytoplasmic DICER to generate mature miRNAs (PubMed:26027739, PubMed:26748718). The heme-bound DGCR8 dimer binds pri-miRNAs as a cooperative trimer (of dimers) and is active in triggering pri-miRNA cleavage, whereas the heme-free DGCR8 monomer binds pri-miRNAs as a dimer and is much less active. Both double-stranded and single-stranded regions of a pri-miRNA are required for its binding (PubMed:15531877, PubMed:15574589, PubMed:15589161, PubMed:16751099, PubMed:16906129, PubMed:16963499, PubMed:17159994). Specifically recognizes and binds N6-methyladenosine (m6A)-containing pri-miRNAs, a modification required for pri-miRNAs processing (PubMed:25799998). Involved in the silencing of embryonic stem cell self-renewal (By similarity). {ECO:0000250|UniProtKB:Q9EQM6, ECO:0000269|PubMed:15531877, ECO:0000269|PubMed:15574589, ECO:0000269|PubMed:15589161, ECO:0000269|PubMed:16751099, ECO:0000269|PubMed:16906129, ECO:0000269|PubMed:16963499, ECO:0000269|PubMed:17159994, ECO:0000269|PubMed:25799998, ECO:0000269|PubMed:26027739, ECO:0000269|PubMed:26748718}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | DGCR8 | chr14:93180168 | chr22:20077499 | ENST00000351989 | 3 | 14 | 511_578 | 341 | 774.0 | Domain | DRBM 1 | |
| Tgene | DGCR8 | chr14:93180168 | chr22:20077499 | ENST00000351989 | 3 | 14 | 620_685 | 341 | 774.0 | Domain | DRBM 2 | |
| Tgene | DGCR8 | chr14:93180168 | chr22:20077499 | ENST00000383024 | 3 | 13 | 511_578 | 341 | 741.0 | Domain | DRBM 1 | |
| Tgene | DGCR8 | chr14:93180168 | chr22:20077499 | ENST00000383024 | 3 | 13 | 620_685 | 341 | 741.0 | Domain | DRBM 2 | |
| Tgene | DGCR8 | chr14:93180168 | chr22:20077499 | ENST00000407755 | 2 | 12 | 511_578 | 341 | 741.0 | Domain | DRBM 1 | |
| Tgene | DGCR8 | chr14:93180168 | chr22:20077499 | ENST00000407755 | 2 | 12 | 620_685 | 341 | 741.0 | Domain | DRBM 2 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | DGCR8 | chr14:93180168 | chr22:20077499 | ENST00000351989 | 3 | 14 | 301_334 | 341 | 774.0 | Domain | WW | |
| Tgene | DGCR8 | chr14:93180168 | chr22:20077499 | ENST00000383024 | 3 | 13 | 301_334 | 341 | 741.0 | Domain | WW | |
| Tgene | DGCR8 | chr14:93180168 | chr22:20077499 | ENST00000407755 | 2 | 12 | 301_334 | 341 | 741.0 | Domain | WW | |
| Tgene | DGCR8 | chr14:93180168 | chr22:20077499 | ENST00000351989 | 3 | 14 | 1_275 | 341 | 774.0 | Region | Note=Necessary for nuclear localization and retention | |
| Tgene | DGCR8 | chr14:93180168 | chr22:20077499 | ENST00000351989 | 3 | 14 | 276_751 | 341 | 774.0 | Region | Note=Necessary for heme-binding and pri-miRNA processing | |
| Tgene | DGCR8 | chr14:93180168 | chr22:20077499 | ENST00000383024 | 3 | 13 | 1_275 | 341 | 741.0 | Region | Note=Necessary for nuclear localization and retention | |
| Tgene | DGCR8 | chr14:93180168 | chr22:20077499 | ENST00000383024 | 3 | 13 | 276_751 | 341 | 741.0 | Region | Note=Necessary for heme-binding and pri-miRNA processing | |
| Tgene | DGCR8 | chr14:93180168 | chr22:20077499 | ENST00000407755 | 2 | 12 | 1_275 | 341 | 741.0 | Region | Note=Necessary for nuclear localization and retention | |
| Tgene | DGCR8 | chr14:93180168 | chr22:20077499 | ENST00000407755 | 2 | 12 | 276_751 | 341 | 741.0 | Region | Note=Necessary for heme-binding and pri-miRNA processing |
Top |
Fusion Gene Sequence for LGMN-DGCR8 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >44573_44573_1_LGMN-DGCR8_LGMN_chr14_93180168_ENST00000393218_DGCR8_chr22_20077499_ENST00000351989_length(transcript)=3943nt_BP=881nt CCAGTCACCGCGGCACAGTGGCCCTTAAGCGAGGAGCGGCGGCGCCCGCAGCAATCACAGCAGTGCCGACGTCGTGGGTGTTTGGTGTGA GGCTGCGAGCCGCCGCGAGTTCTCACGGTCCCGCCGGCGCCACCACCGCGGTCACTCACCGCCGCCGCCGCCACCACTGCCACCACGGTC GCCTGCCACAGGGTCTTACTCTGTTGCCCAGGCTGGAGTGCAGTGGCACAATCTTGGCTCACTGCAACCTCTGCCTCCCGGGTTCAAGCA ATTCTCCTGCCTCAGCCTCCCGAGTAGCTGGGATTACAGGTGTCTGCAATTGAACTCCAAGGTGCAGAATGGTTTGGAAAGTAGCTGTAT TCCTCAGTGTGGCCCTGGGCATTGGTGCCGTTCCTATAGATGATCCTGAAGATGGAGGCAAGCACTGGGTGGTGATCGTGGCAGGTTCAA ATGGCTGGTATAATTATAGGCACCAGGCAGACGCGTGCCATGCCTACCAGATCATTCACCGCAATGGGATTCCTGACGAACAGATCGTTG TGATGATGTACGATGACATTGCTTACTCTGAAGACAATCCCACTCCAGGAATTGTGATCAACAGGCCCAATGGCACAGATGTCTATCAGG GAGTCCCGAAGGACTACACTGGAGAGGATGTTACCCCACAAAATTTCCTTGCTGTGTTGAGAGGCGATGCAGAAGCAGTGAAGGGCATAG GATCCGGCAAAGTCCTGAAGAGTGGCCCCCAGGATCACGTGTTCATTTACTTCACTGACCATGGATCTACTGGAATACTGGTTTTTCCCA ATGAAGATCTTCATGTAAAGGACCTGAATGAGACCATCCATTACATGTACAAACACAAAATGTACCGAAAGAAACACGACCCTCCTCTGA GTAGCATCCCTTGTCTGCATTATAAGAAAATGAAGGACAACGAGGAACGGGAGCAAAGCAGTGACCTCACCCCTAGTGGGGATGTGTCCC CCGTCAAGCCCCTGAGCCGATCTGCAGAGCTGGAGTTTCCCCTGGATGAGCCTGACTCTATGGGTGCTGACCCGGGGCCCCCGGACGAGA AAGACCCACTAGGGGCTGAGGCAGCCCCTGGGGCCCTGGGGCAGGTGAAGGCCAAAGTCGAGGTGTGCAAAGATGAATCCGTTGATCTCG AGGAATTTCGAAGCTACCTGGAGAAGCGTTTTGACTTTGAGCAAGTTACTGTGAAAAAATTCAGGACTTGGGCTGAGCGGCGGCAATTCA ATCGGGAAATGAAGCGGAAGCAGGCGGAGTCCGAGAGGCCCATCTTGCCAGCCAATCAGAAGCTCATTACTTTATCAGTGCAAGATGCAC CCACAAAGAAAGAGTTTGTTATTAACCCCAACGGGAAATCCGAGGTCTGCATCCTGCACGAGTACATGCAGCGTGTCCTCAAGGTCCGCC CTGTCTATAATTTCTTTGAATGTGAGAACCCAAGTGAGCCTTTTGGTGCCTCGGTGACCATTGATGGTGTGACTTACGGATCTGGAACTG CAAGCAGCAAAAAACTTGCGAAGAATAAAGCTGCCCGAGCTACACTGGAAATCCTCATCCCTGACTTTGTTAAACAGACCTCTGAAGAGA AGCCCAAAGACAGTGAAGAACTCGAGTATTTTAACCACATCAGCATCGAGGACTCGCGGGTCTACGAGCTGACCAGCAAGGCTGGGCTGT TGTCTCCATATCAGATCCTCCACGAGTGCCTTAAAAGAAACCATGGGATGGGTGACACGTCTATCAAGTTTGAAGTGGTTCCTGGGAAAA ACCAGAAGAGTGAATACGTCATGGCGTGTGGCAAGCACACAGTGCGCGGGTGGTGTAAGAACAAGAGAGTTGGAAAGCAGTTAGCCTCAC AGAAGATCCTTCAGCTGCTGCACCCACATGTCAAGAACTGGGGGTCTTTACTGCGCATGTATGGCCGTGAGAGCAGCAAGATGGTCAAGC AGGAGACATCGGACAAGAGTGTGATTGAGCTGCAGCAGTATGCCAAGAAGAACAAGCCCAACCTGCACATCCTCAGCAAGCTCCAAGAGG AGATGAAGAGGCTAGCTGAGGAAAGGGAGGAGACTCGAAAGAAGCCCAAGATGTCCATTGTGGCGTCCGCCCAGCCTGGCGGTGAGCCCC TGTGCACCGTGGACGTGTGAGGGAGGTGGCACGGGCCAGGGCGCGGGGGCCGCCAGCCGCACTTCTGAGGAGACCAGCAGTCATGCATCG TGCACCACAGTGTCAGGCCTCCAACCCACGCTCCTTCCCTGTGGCCAACCTGTGGGCCCGGCCTTAGGGTGGAGGCTTTAGTGTACAGGG ACAGCCATGGCCACACAGCACACATGTGGAGCAGCGGCTCTCCCTGGAAAGCTCCAGGCCTGAATGGATGGACTCAGCGACTGCACCAGT GGCAGCTGGTGACTGTGGACAGTGGTGGACCCTGCTTCTGTGCACCTGCTGCAGGCTCTTTTTATGAAGGCTTTCATGAATTTTAGTATG TAATACGCACTGACGACACATGATGCTTGGATGACAGATGAGAGGGGATGGCTGAGTCCTGTGGCTGGCCCGTGATGCCAGGTGGCCCAT GTGCCCAGGGCGCCTGCAGGGCTGCTACAGGGACCTGGTCAGGAGGTGCACATGGTGCCCTGCCCTCACCCACCCTCTGTGTTTCCCCTT CTTTGAAAAGGTAGAAGAGAAAGGAATATTTTAAACCTTTTTGGCTTAAACAGAATTTTAGCATCAGAACTAGCTTTCTGGGATTGGAGG CAAACCATCAAGGTGGTCCCTCTCCAGTCTGGACACGATGCCAGCAAGGATGACGTCCTGCCACCTCCTGGAGTTACCCTGGCCTCCTAG GGTCCCTTTTTCTGATGAAGTCTTAATTCCCTAAAAGCGCCTCTTTGGACACTGAGGCCCTCTCTGCCTTTCCTGGCCTCCGGCAACAGT TTTTTACAAAGATTTTTTGCAGTCGAGTCCATATGTCCACCCATTGATTTTTAAAGCTTTTGTGATATTTTAGCATTTTGAAAGACTTTC ACAGTGAGAGTAGAAGGTAGATTTGGAATCATGCATTTTAGCAAGTGGACTTGTTGAAACAGGAAGCAAGGGCCTTCAGTGTAGCCCATT CTTGATCCAGAGCTGTTGCCTGTGACAGCGGTTTCTCTGGATGTCAAAGGCAGCTGCCTGGTGCCCAGCTTGCTTCTCGACTGGTGGCCC CTATGGGTGGGTGTGCGATGGAAATGTGTTCCTGCCGGAGTCTGAGGCACCAGGGTGTGCTCAAAGGCTGGCCCTGGTGGTGGACTGGCA CCTGTGCAGAGTGCCGTGTGCTTGTGGTGCGCCATCTGAAGCAAGAGTCCAGCGTTCTGCCGTGTCTGTCCCCCACCATGCCCCCTACAG GCGGTACTGATGGCGCTTTTTTTTTTTTTTCTGTCAGGAAAACAATGTTGGCCTGTGGGCCGCCCACAACATATCCTTCCCTCACTACCT GTGTGACCAAGGTTGGCTTCTGTTGACCTTTAAAAAAGAAACCCTCAACTCAAATTGCTATAATTAGACACTTGCTTCTGTCTTGCCTCC TGTCTGCAGCTGTGAATAGTCATTTGACTGTGACTGTTGCCCTTAGCCAGCCAGATGCGCCTGTGAACCAAAGCTTCGTGCACATGTGTT CCCCTAAAGGTTGGGGAGCCTCGCTGTGTCTTGCTGTTCCCAGGCACCACCACAGCAGGTGCTGCCATACTCTTGTGGTCTCTGTGCGCC CCCCCCCCCCCCCCACCCGTCTGCCAAGCATGGGTATGAATCGTGCACACAGCCATGCTTCAAGGCCGGGGCAGGGGAGCCTGTGCTGAT >44573_44573_1_LGMN-DGCR8_LGMN_chr14_93180168_ENST00000393218_DGCR8_chr22_20077499_ENST00000351989_length(amino acids)=613AA_BP=181 MVWKVAVFLSVALGIGAVPIDDPEDGGKHWVVIVAGSNGWYNYRHQADACHAYQIIHRNGIPDEQIVVMMYDDIAYSEDNPTPGIVINRP NGTDVYQGVPKDYTGEDVTPQNFLAVLRGDAEAVKGIGSGKVLKSGPQDHVFIYFTDHGSTGILVFPNEDLHVKDLNETIHYMYKHKMYR KKHDPPLSSIPCLHYKKMKDNEEREQSSDLTPSGDVSPVKPLSRSAELEFPLDEPDSMGADPGPPDEKDPLGAEAAPGALGQVKAKVEVC KDESVDLEEFRSYLEKRFDFEQVTVKKFRTWAERRQFNREMKRKQAESERPILPANQKLITLSVQDAPTKKEFVINPNGKSEVCILHEYM QRVLKVRPVYNFFECENPSEPFGASVTIDGVTYGSGTASSKKLAKNKAARATLEILIPDFVKQTSEEKPKDSEELEYFNHISIEDSRVYE LTSKAGLLSPYQILHECLKRNHGMGDTSIKFEVVPGKNQKSEYVMACGKHTVRGWCKNKRVGKQLASQKILQLLHPHVKNWGSLLRMYGR -------------------------------------------------------------- >44573_44573_2_LGMN-DGCR8_LGMN_chr14_93180168_ENST00000393218_DGCR8_chr22_20077499_ENST00000383024_length(transcript)=3848nt_BP=881nt CCAGTCACCGCGGCACAGTGGCCCTTAAGCGAGGAGCGGCGGCGCCCGCAGCAATCACAGCAGTGCCGACGTCGTGGGTGTTTGGTGTGA GGCTGCGAGCCGCCGCGAGTTCTCACGGTCCCGCCGGCGCCACCACCGCGGTCACTCACCGCCGCCGCCGCCACCACTGCCACCACGGTC GCCTGCCACAGGGTCTTACTCTGTTGCCCAGGCTGGAGTGCAGTGGCACAATCTTGGCTCACTGCAACCTCTGCCTCCCGGGTTCAAGCA ATTCTCCTGCCTCAGCCTCCCGAGTAGCTGGGATTACAGGTGTCTGCAATTGAACTCCAAGGTGCAGAATGGTTTGGAAAGTAGCTGTAT TCCTCAGTGTGGCCCTGGGCATTGGTGCCGTTCCTATAGATGATCCTGAAGATGGAGGCAAGCACTGGGTGGTGATCGTGGCAGGTTCAA ATGGCTGGTATAATTATAGGCACCAGGCAGACGCGTGCCATGCCTACCAGATCATTCACCGCAATGGGATTCCTGACGAACAGATCGTTG TGATGATGTACGATGACATTGCTTACTCTGAAGACAATCCCACTCCAGGAATTGTGATCAACAGGCCCAATGGCACAGATGTCTATCAGG GAGTCCCGAAGGACTACACTGGAGAGGATGTTACCCCACAAAATTTCCTTGCTGTGTTGAGAGGCGATGCAGAAGCAGTGAAGGGCATAG GATCCGGCAAAGTCCTGAAGAGTGGCCCCCAGGATCACGTGTTCATTTACTTCACTGACCATGGATCTACTGGAATACTGGTTTTTCCCA ATGAAGATCTTCATGTAAAGGACCTGAATGAGACCATCCATTACATGTACAAACACAAAATGTACCGAAAGAAACACGACCCTCCTCTGA GTAGCATCCCTTGTCTGCATTATAAGAAAATGAAGGACAACGAGGAACGGGAGCAAAGCAGTGACCTCACCCCTAGTGGGGATGTGTCCC CCGTCAAGCCCCTGAGCCGATCTGCAGAGCTGGAGTTTCCCCTGGATGAGCCTGACTCTATGGGTGCTGACCCGGGGCCCCCGGACGAGA AAGACCCACTAGGGGCTGAGGCAGCCCCTGGGGCCCTGGGGCAGGTGAAGGCCAAAGTCGAGGTGTGCAAAGATGAATCCGTTGATCTCG AGGAATTTCGAAGCTACCTGGAGAAGCGTTTTGACTTTGAGCAAGTTACTGTGAAAAAATTCAGGACTTGGGCTGAGCGGCGGCAATTCA ATCGGGAAATGAAGCGGAAGCAGGCGGAGTCCGAGAGGCCCATCTTGCCAGCCAATCAGAAGCTCATTACTTTATCAGTGCAAGATGCAC CCACAAAGAAAGAGTTTGTTATTAACCCCAACGGGAAATCCGAGGTCTGCATCCTGCACGAGTACATGCAGCGTGTCCTCAAGGTCCGCC CTGTCTATAATTTCTTTGAATGTGCCCGAGCTACACTGGAAATCCTCATCCCTGACTTTGTTAAACAGACCTCTGAAGAGAAGCCCAAAG ACAGTGAAGAACTCGAGTATTTTAACCACATCAGCATCGAGGACTCGCGGGTCTACGAGCTGACCAGCAAGGCTGGGCTGTTGTCTCCAT ATCAGATCCTCCACGAGTGCCTTAAAAGAAACCATGGGATGGGTGACACGTCTATCAAGTTTGAAGTGGTTCCTGGGAAAAACCAGAAGA GTGAATACGTCATGGCGTGTGGCAAGCACACAGTGCGCGGGTGGTGTAAGAACAAGAGAGTTGGAAAGCAGTTAGCCTCACAGAAGATCC TTCAGCTGCTGCACCCACATGTCAAGAACTGGGGGTCTTTACTGCGCATGTATGGCCGTGAGAGCAGCAAGATGGTCAAGCAGGAGACAT CGGACAAGAGTGTGATTGAGCTGCAGCAGTATGCCAAGAAGAACAAGCCCAACCTGCACATCCTCAGCAAGCTCCAAGAGGAGATGAAGA GGCTAGCTGAGGAAAGGGAGGAGACTCGAAAGAAGCCCAAGATGTCCATTGTGGCGTCCGCCCAGCCTGGCGGTGAGCCCCTGTGCACCG TGGACGTGTGAGGGAGGTGGCACGGGCCAGGGCGCGGGGGCCGCCAGCCGCACTTCTGAGGAGACCAGCAGTCATGCATCGTGCACCACA GTGTCAGGCCTCCAACCCACGCTCCTTCCCTGTGGCCAACCTGTGGGCCCGGCCTTAGGGTGGAGGCTTTAGTGTACAGGGACAGCCATG GCCACACAGCACACATGTGGAGCAGCGGCTCTCCCTGGAAAGCTCCAGGCCTGAATGGATGGACTCAGCGACTGCACCAGTGGCAGCTGG TGACTGTGGACAGTGGTGGACCCTGCTTCTGTGCACCTGCTGCAGGCTCTTTTTATGAAGGCTTTCATGAATTTTAGTATGTAATACGCA CTGACGACACATGATGCTTGGATGACAGATGAGAGGGGATGGCTGAGTCCTGTGGCTGGCCCGTGATGCCAGGTGGCCCATGTGCCCAGG GCGCCTGCAGGGCTGCTACAGGGACCTGGTCAGGAGGTGCACATGGTGCCCTGCCCTCACCCACCCTCTGTGTTTCCCCTTCTTTGAAAA GGTAGAAGAGAAAGGAATATTTTAAACCTTTTTGGCTTAAACAGAATTTTAGCATCAGAACTAGCTTTCTGGGATTGGAGGCAAACCATC AAGGTGGTCCCTCTCCAGTCTGGACACGATGCCAGCAAGGATGACGTCCTGCCACCTCCTGGAGTTACCCTGGCCTCCTAGGGTCCCTTT TTCTGATGAAGTCTTAATTCCCTAAAAGCGCCTCTTTGGACACTGAGGCCCTCTCTGCCTTTCCTGGCCTCCGGCAACAGTTTTTTACAA AGATTTTTTGCAGTCGAGTCCATATGTCCACCCATTGATTTTTAAAGCTTTTGTGATATTTTAGCATTTTGAAAGACTTTCACAGTGAGA GTAGAAGGTAGATTTGGAATCATGCATTTTAGCAAGTGGACTTGTTGAAACAGGAAGCAAGGGCCTTCAGTGTAGCCCATTCTTGATCCA GAGCTGTTGCCTGTGACAGCGGTTTCTCTGGATGTCAAAGGCAGCTGCCTGGTGCCCAGCTTGCTTCTCGACTGGTGGCCCCTATGGGTG GGTGTGCGATGGAAATGTGTTCCTGCCGGAGTCTGAGGCACCAGGGTGTGCTCAAAGGCTGGCCCTGGTGGTGGACTGGCACCTGTGCAG AGTGCCGTGTGCTTGTGGTGCGCCATCTGAAGCAAGAGTCCAGCGTTCTGCCGTGTCTGTCCCCCACCATGCCCCCTACAGGCGGTACTG ATGGCGCTTTTTTTTTTTTTTCTGTCAGGAAAACAATGTTGGCCTGTGGGCCGCCCACAACATATCCTTCCCTCACTACCTGTGTGACCA AGGTTGGCTTCTGTTGACCTTTAAAAAAGAAACCCTCAACTCAAATTGCTATAATTAGACACTTGCTTCTGTCTTGCCTCCTGTCTGCAG CTGTGAATAGTCATTTGACTGTGACTGTTGCCCTTAGCCAGCCAGATGCGCCTGTGAACCAAAGCTTCGTGCACATGTGTTCCCCTAAAG GTTGGGGAGCCTCGCTGTGTCTTGCTGTTCCCAGGCACCACCACAGCAGGTGCTGCCATACTCTTGTGGTCTCTGTGCGCCCCCCCCCCC CCCCCACCCGTCTGCCAAGCATGGGTATGAATCGTGCACACAGCCATGCTTCAAGGCCGGGGCAGGGGAGCCTGTGCTGATGCCATCCAG >44573_44573_2_LGMN-DGCR8_LGMN_chr14_93180168_ENST00000393218_DGCR8_chr22_20077499_ENST00000383024_length(amino acids)=580AA_BP=181 MVWKVAVFLSVALGIGAVPIDDPEDGGKHWVVIVAGSNGWYNYRHQADACHAYQIIHRNGIPDEQIVVMMYDDIAYSEDNPTPGIVINRP NGTDVYQGVPKDYTGEDVTPQNFLAVLRGDAEAVKGIGSGKVLKSGPQDHVFIYFTDHGSTGILVFPNEDLHVKDLNETIHYMYKHKMYR KKHDPPLSSIPCLHYKKMKDNEEREQSSDLTPSGDVSPVKPLSRSAELEFPLDEPDSMGADPGPPDEKDPLGAEAAPGALGQVKAKVEVC KDESVDLEEFRSYLEKRFDFEQVTVKKFRTWAERRQFNREMKRKQAESERPILPANQKLITLSVQDAPTKKEFVINPNGKSEVCILHEYM QRVLKVRPVYNFFECARATLEILIPDFVKQTSEEKPKDSEELEYFNHISIEDSRVYELTSKAGLLSPYQILHECLKRNHGMGDTSIKFEV VPGKNQKSEYVMACGKHTVRGWCKNKRVGKQLASQKILQLLHPHVKNWGSLLRMYGRESSKMVKQETSDKSVIELQQYAKKNKPNLHILS -------------------------------------------------------------- >44573_44573_3_LGMN-DGCR8_LGMN_chr14_93180168_ENST00000393218_DGCR8_chr22_20077499_ENST00000407755_length(transcript)=3845nt_BP=881nt CCAGTCACCGCGGCACAGTGGCCCTTAAGCGAGGAGCGGCGGCGCCCGCAGCAATCACAGCAGTGCCGACGTCGTGGGTGTTTGGTGTGA GGCTGCGAGCCGCCGCGAGTTCTCACGGTCCCGCCGGCGCCACCACCGCGGTCACTCACCGCCGCCGCCGCCACCACTGCCACCACGGTC GCCTGCCACAGGGTCTTACTCTGTTGCCCAGGCTGGAGTGCAGTGGCACAATCTTGGCTCACTGCAACCTCTGCCTCCCGGGTTCAAGCA ATTCTCCTGCCTCAGCCTCCCGAGTAGCTGGGATTACAGGTGTCTGCAATTGAACTCCAAGGTGCAGAATGGTTTGGAAAGTAGCTGTAT TCCTCAGTGTGGCCCTGGGCATTGGTGCCGTTCCTATAGATGATCCTGAAGATGGAGGCAAGCACTGGGTGGTGATCGTGGCAGGTTCAA ATGGCTGGTATAATTATAGGCACCAGGCAGACGCGTGCCATGCCTACCAGATCATTCACCGCAATGGGATTCCTGACGAACAGATCGTTG TGATGATGTACGATGACATTGCTTACTCTGAAGACAATCCCACTCCAGGAATTGTGATCAACAGGCCCAATGGCACAGATGTCTATCAGG GAGTCCCGAAGGACTACACTGGAGAGGATGTTACCCCACAAAATTTCCTTGCTGTGTTGAGAGGCGATGCAGAAGCAGTGAAGGGCATAG GATCCGGCAAAGTCCTGAAGAGTGGCCCCCAGGATCACGTGTTCATTTACTTCACTGACCATGGATCTACTGGAATACTGGTTTTTCCCA ATGAAGATCTTCATGTAAAGGACCTGAATGAGACCATCCATTACATGTACAAACACAAAATGTACCGAAAGAAACACGACCCTCCTCTGA GTAGCATCCCTTGTCTGCATTATAAGAAAATGAAGGACAACGAGGAACGGGAGCAAAGCAGTGACCTCACCCCTAGTGGGGATGTGTCCC CCGTCAAGCCCCTGAGCCGATCTGCAGAGCTGGAGTTTCCCCTGGATGAGCCTGACTCTATGGGTGCTGACCCGGGGCCCCCGGACGAGA AAGACCCACTAGGGGCTGAGGCAGCCCCTGGGGCCCTGGGGCAGGTGAAGGCCAAAGTCGAGGTGTGCAAAGATGAATCCGTTGATCTCG AGGAATTTCGAAGCTACCTGGAGAAGCGTTTTGACTTTGAGCAAGTTACTGTGAAAAAATTCAGGACTTGGGCTGAGCGGCGGCAATTCA ATCGGGAAATGAAGCGGAAGCAGGCGGAGTCCGAGAGGCCCATCTTGCCAGCCAATCAGAAGCTCATTACTTTATCAGTGCAAGATGCAC CCACAAAGAAAGAGTTTGTTATTAACCCCAACGGGAAATCCGAGGTCTGCATCCTGCACGAGTACATGCAGCGTGTCCTCAAGGTCCGCC CTGTCTATAATTTCTTTGAATGTGCCCGAGCTACACTGGAAATCCTCATCCCTGACTTTGTTAAACAGACCTCTGAAGAGAAGCCCAAAG ACAGTGAAGAACTCGAGTATTTTAACCACATCAGCATCGAGGACTCGCGGGTCTACGAGCTGACCAGCAAGGCTGGGCTGTTGTCTCCAT ATCAGATCCTCCACGAGTGCCTTAAAAGAAACCATGGGATGGGTGACACGTCTATCAAGTTTGAAGTGGTTCCTGGGAAAAACCAGAAGA GTGAATACGTCATGGCGTGTGGCAAGCACACAGTGCGCGGGTGGTGTAAGAACAAGAGAGTTGGAAAGCAGTTAGCCTCACAGAAGATCC TTCAGCTGCTGCACCCACATGTCAAGAACTGGGGGTCTTTACTGCGCATGTATGGCCGTGAGAGCAGCAAGATGGTCAAGCAGGAGACAT CGGACAAGAGTGTGATTGAGCTGCAGCAGTATGCCAAGAAGAACAAGCCCAACCTGCACATCCTCAGCAAGCTCCAAGAGGAGATGAAGA GGCTAGCTGAGGAAAGGGAGGAGACTCGAAAGAAGCCCAAGATGTCCATTGTGGCGTCCGCCCAGCCTGGCGGTGAGCCCCTGTGCACCG TGGACGTGTGAGGGAGGTGGCACGGGCCAGGGCGCGGGGGCCGCCAGCCGCACTTCTGAGGAGACCAGCAGTCATGCATCGTGCACCACA GTGTCAGGCCTCCAACCCACGCTCCTTCCCTGTGGCCAACCTGTGGGCCCGGCCTTAGGGTGGAGGCTTTAGTGTACAGGGACAGCCATG GCCACACAGCACACATGTGGAGCAGCGGCTCTCCCTGGAAAGCTCCAGGCCTGAATGGATGGACTCAGCGACTGCACCAGTGGCAGCTGG TGACTGTGGACAGTGGTGGACCCTGCTTCTGTGCACCTGCTGCAGGCTCTTTTTATGAAGGCTTTCATGAATTTTAGTATGTAATACGCA CTGACGACACATGATGCTTGGATGACAGATGAGAGGGGATGGCTGAGTCCTGTGGCTGGCCCGTGATGCCAGGTGGCCCATGTGCCCAGG GCGCCTGCAGGGCTGCTACAGGGACCTGGTCAGGAGGTGCACATGGTGCCCTGCCCTCACCCACCCTCTGTGTTTCCCCTTCTTTGAAAA GGTAGAAGAGAAAGGAATATTTTAAACCTTTTTGGCTTAAACAGAATTTTAGCATCAGAACTAGCTTTCTGGGATTGGAGGCAAACCATC AAGGTGGTCCCTCTCCAGTCTGGACACGATGCCAGCAAGGATGACGTCCTGCCACCTCCTGGAGTTACCCTGGCCTCCTAGGGTCCCTTT TTCTGATGAAGTCTTAATTCCCTAAAAGCGCCTCTTTGGACACTGAGGCCCTCTCTGCCTTTCCTGGCCTCCGGCAACAGTTTTTTACAA AGATTTTTTGCAGTCGAGTCCATATGTCCACCCATTGATTTTTAAAGCTTTTGTGATATTTTAGCATTTTGAAAGACTTTCACAGTGAGA GTAGAAGGTAGATTTGGAATCATGCATTTTAGCAAGTGGACTTGTTGAAACAGGAAGCAAGGGCCTTCAGTGTAGCCCATTCTTGATCCA GAGCTGTTGCCTGTGACAGCGGTTTCTCTGGATGTCAAAGGCAGCTGCCTGGTGCCCAGCTTGCTTCTCGACTGGTGGCCCCTATGGGTG GGTGTGCGATGGAAATGTGTTCCTGCCGGAGTCTGAGGCACCAGGGTGTGCTCAAAGGCTGGCCCTGGTGGTGGACTGGCACCTGTGCAG AGTGCCGTGTGCTTGTGGTGCGCCATCTGAAGCAAGAGTCCAGCGTTCTGCCGTGTCTGTCCCCCACCATGCCCCCTACAGGCGGTACTG ATGGCGCTTTTTTTTTTTTTTCTGTCAGGAAAACAATGTTGGCCTGTGGGCCGCCCACAACATATCCTTCCCTCACTACCTGTGTGACCA AGGTTGGCTTCTGTTGACCTTTAAAAAAGAAACCCTCAACTCAAATTGCTATAATTAGACACTTGCTTCTGTCTTGCCTCCTGTCTGCAG CTGTGAATAGTCATTTGACTGTGACTGTTGCCCTTAGCCAGCCAGATGCGCCTGTGAACCAAAGCTTCGTGCACATGTGTTCCCCTAAAG GTTGGGGAGCCTCGCTGTGTCTTGCTGTTCCCAGGCACCACCACAGCAGGTGCTGCCATACTCTTGTGGTCTCTGTGCGCCCCCCCCCCC CCCCCACCCGTCTGCCAAGCATGGGTATGAATCGTGCACACAGCCATGCTTCAAGGCCGGGGCAGGGGAGCCTGTGCTGATGCCATCCAG >44573_44573_3_LGMN-DGCR8_LGMN_chr14_93180168_ENST00000393218_DGCR8_chr22_20077499_ENST00000407755_length(amino acids)=580AA_BP=181 MVWKVAVFLSVALGIGAVPIDDPEDGGKHWVVIVAGSNGWYNYRHQADACHAYQIIHRNGIPDEQIVVMMYDDIAYSEDNPTPGIVINRP NGTDVYQGVPKDYTGEDVTPQNFLAVLRGDAEAVKGIGSGKVLKSGPQDHVFIYFTDHGSTGILVFPNEDLHVKDLNETIHYMYKHKMYR KKHDPPLSSIPCLHYKKMKDNEEREQSSDLTPSGDVSPVKPLSRSAELEFPLDEPDSMGADPGPPDEKDPLGAEAAPGALGQVKAKVEVC KDESVDLEEFRSYLEKRFDFEQVTVKKFRTWAERRQFNREMKRKQAESERPILPANQKLITLSVQDAPTKKEFVINPNGKSEVCILHEYM QRVLKVRPVYNFFECARATLEILIPDFVKQTSEEKPKDSEELEYFNHISIEDSRVYELTSKAGLLSPYQILHECLKRNHGMGDTSIKFEV VPGKNQKSEYVMACGKHTVRGWCKNKRVGKQLASQKILQLLHPHVKNWGSLLRMYGRESSKMVKQETSDKSVIELQQYAKKNKPNLHILS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for LGMN-DGCR8 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | DGCR8 | chr14:93180168 | chr22:20077499 | ENST00000351989 | 3 | 14 | 701_773 | 341.0 | 774.0 | DROSHA | |
| Tgene | DGCR8 | chr14:93180168 | chr22:20077499 | ENST00000383024 | 3 | 13 | 701_773 | 341.0 | 741.0 | DROSHA | |
| Tgene | DGCR8 | chr14:93180168 | chr22:20077499 | ENST00000407755 | 2 | 12 | 701_773 | 341.0 | 741.0 | DROSHA |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for LGMN-DGCR8 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for LGMN-DGCR8 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
