
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:LIMK1-TYW1 (FusionGDB2 ID:44717) |
Fusion Gene Summary for LIMK1-TYW1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: LIMK1-TYW1 | Fusion gene ID: 44717 | Hgene | Tgene | Gene symbol | LIMK1 | TYW1 | Gene ID | 3984 | 55253 |
| Gene name | LIM domain kinase 1 | tRNA-yW synthesizing protein 1 homolog | |
| Synonyms | LIMK|LIMK-1 | RSAFD1|TYW1A|YPL207W | |
| Cytomap | 7q11.23 | 7q11.21 | |
| Type of gene | protein-coding | protein-coding | |
| Description | LIM domain kinase 1LIM motif-containing protein kinase | S-adenosyl-L-methionine-dependent tRNA 4-demethylwyosine synthase TYW1S-adenosyl-L-methionine-dependent tRNA 4-demethylwyosine synthaseradical S-adenosyl methionine and flavodoxin domain-containing protein 1radical S-adenosyl methionine and flavodoxin | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | P53667 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000336180, ENST00000418310, ENST00000538333, ENST00000491052, | ENST00000491969, ENST00000359626, | |
| Fusion gene scores | * DoF score | 7 X 6 X 4=168 | 11 X 9 X 5=495 |
| # samples | 6 | 11 | |
| ** MAII score | log2(6/168*10)=-1.48542682717024 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(11/495*10)=-2.16992500144231 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: LIMK1 [Title/Abstract] AND TYW1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | LIMK1(73526328)-TYW1(66489887), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | LIMK1 | GO:0006468 | protein phosphorylation | 17512523|22328514 |
| Hgene | LIMK1 | GO:0032233 | positive regulation of actin filament bundle assembly | 17512523 |
| Hgene | LIMK1 | GO:0051444 | negative regulation of ubiquitin-protein transferase activity | 17512523 |
 Fusion gene breakpoints across LIMK1 (5'-gene) Fusion gene breakpoints across LIMK1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
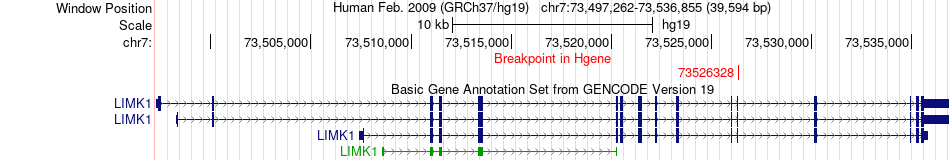 |
 Fusion gene breakpoints across TYW1 (3'-gene) Fusion gene breakpoints across TYW1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
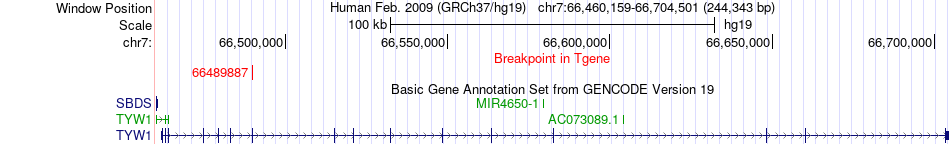 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-DX-A1L1-01A | LIMK1 | chr7 | 73526328 | + | TYW1 | chr7 | 66489887 | + |
Top |
Fusion Gene ORF analysis for LIMK1-TYW1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000336180 | ENST00000491969 | LIMK1 | chr7 | 73526328 | + | TYW1 | chr7 | 66489887 | + |
| 5CDS-intron | ENST00000418310 | ENST00000491969 | LIMK1 | chr7 | 73526328 | + | TYW1 | chr7 | 66489887 | + |
| 5CDS-intron | ENST00000538333 | ENST00000491969 | LIMK1 | chr7 | 73526328 | + | TYW1 | chr7 | 66489887 | + |
| In-frame | ENST00000336180 | ENST00000359626 | LIMK1 | chr7 | 73526328 | + | TYW1 | chr7 | 66489887 | + |
| In-frame | ENST00000418310 | ENST00000359626 | LIMK1 | chr7 | 73526328 | + | TYW1 | chr7 | 66489887 | + |
| In-frame | ENST00000538333 | ENST00000359626 | LIMK1 | chr7 | 73526328 | + | TYW1 | chr7 | 66489887 | + |
| intron-3CDS | ENST00000491052 | ENST00000359626 | LIMK1 | chr7 | 73526328 | + | TYW1 | chr7 | 66489887 | + |
| intron-intron | ENST00000491052 | ENST00000491969 | LIMK1 | chr7 | 73526328 | + | TYW1 | chr7 | 66489887 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000418310 | LIMK1 | chr7 | 73526328 | + | ENST00000359626 | TYW1 | chr7 | 66489887 | + | 3925 | 1602 | 96 | 2939 | 947 |
| ENST00000336180 | LIMK1 | chr7 | 73526328 | + | ENST00000359626 | TYW1 | chr7 | 66489887 | + | 3784 | 1461 | 51 | 2798 | 915 |
| ENST00000538333 | LIMK1 | chr7 | 73526328 | + | ENST00000359626 | TYW1 | chr7 | 66489887 | + | 3802 | 1479 | 171 | 2816 | 881 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000418310 | ENST00000359626 | LIMK1 | chr7 | 73526328 | + | TYW1 | chr7 | 66489887 | + | 0.000645182 | 0.9993548 |
| ENST00000336180 | ENST00000359626 | LIMK1 | chr7 | 73526328 | + | TYW1 | chr7 | 66489887 | + | 0.000659872 | 0.9993401 |
| ENST00000538333 | ENST00000359626 | LIMK1 | chr7 | 73526328 | + | TYW1 | chr7 | 66489887 | + | 0.001030561 | 0.99896944 |
Top |
Fusion Genomic Features for LIMK1-TYW1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| LIMK1 | chr7 | 73526328 | + | TYW1 | chr7 | 66489886 | + | 6.07E-11 | 1 |
| LIMK1 | chr7 | 73526328 | + | TYW1 | chr7 | 66489886 | + | 6.07E-11 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
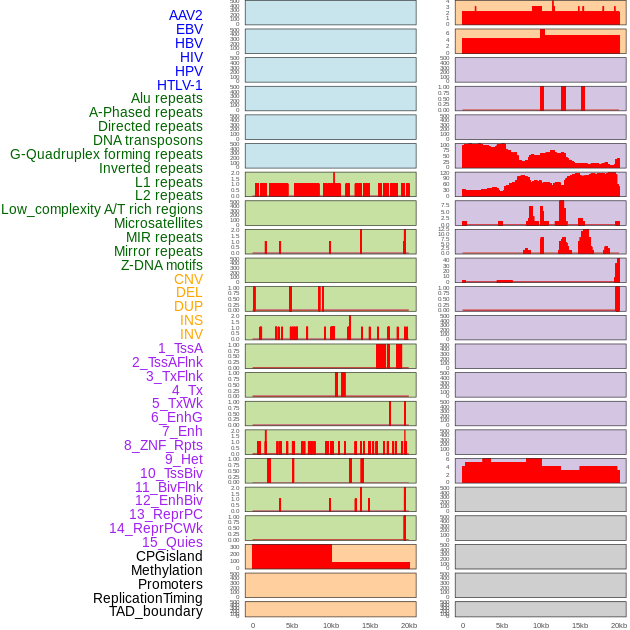 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
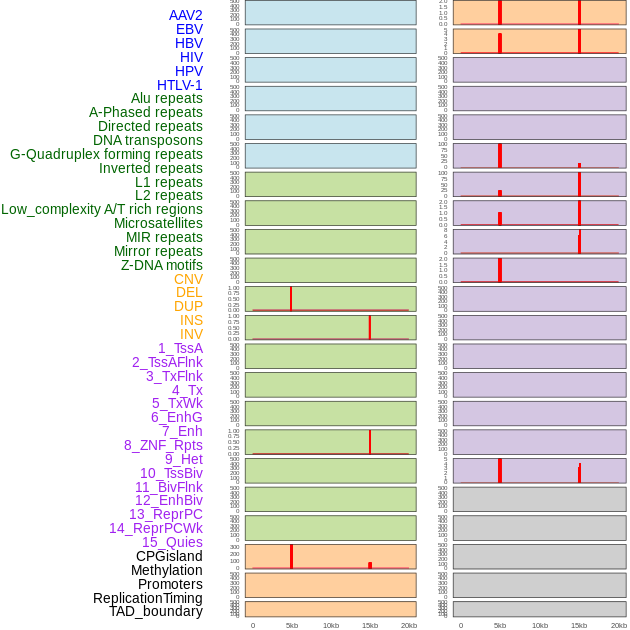 |
Top |
Fusion Protein Features for LIMK1-TYW1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr7:73526328/chr7:66489887) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| LIMK1 | . |
| FUNCTION: Serine/threonine-protein kinase that plays an essential role in the regulation of actin filament dynamics. Acts downstream of several Rho family GTPase signal transduction pathways (PubMed:10436159, PubMed:11832213, PubMed:12807904, PubMed:15660133, PubMed:16230460, PubMed:18028908, PubMed:22328514, PubMed:23633677). Activated by upstream kinases including ROCK1, PAK1 and PAK4, which phosphorylate LIMK1 on a threonine residue located in its activation loop (PubMed:10436159). LIMK1 subsequently phosphorylates and inactivates the actin binding/depolymerizing factors cofilin-1/CFL1, cofilin-2/CFL2 and destrin/DSTN, thereby preventing the cleavage of filamentous actin (F-actin), and stabilizing the actin cytoskeleton (PubMed:11832213, PubMed:15660133, PubMed:16230460, PubMed:23633677). In this way LIMK1 regulates several actin-dependent biological processes including cell motility, cell cycle progression, and differentiation (PubMed:11832213, PubMed:15660133, PubMed:16230460, PubMed:23633677). Phosphorylates TPPP on serine residues, thereby promoting microtubule disassembly (PubMed:18028908). Stimulates axonal outgrowth and may be involved in brain development (PubMed:18028908). {ECO:0000269|PubMed:10436159, ECO:0000269|PubMed:11832213, ECO:0000269|PubMed:12807904, ECO:0000269|PubMed:15660133, ECO:0000269|PubMed:16230460, ECO:0000269|PubMed:18028908, ECO:0000269|PubMed:22328514, ECO:0000269|PubMed:23633677}.; FUNCTION: [Isoform 3]: Has a dominant negative effect on actin cytoskeletal changes. Required for atypical chemokine receptor ACKR2-induced phosphorylation of cofilin (CFL1). {ECO:0000269|PubMed:10196227}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LIMK1 | chr7:73526328 | chr7:66489887 | ENST00000336180 | + | 12 | 16 | 165_258 | 470 | 648.0 | Domain | PDZ |
| Hgene | LIMK1 | chr7:73526328 | chr7:66489887 | ENST00000336180 | + | 12 | 16 | 25_75 | 470 | 648.0 | Domain | LIM zinc-binding 1 |
| Hgene | LIMK1 | chr7:73526328 | chr7:66489887 | ENST00000336180 | + | 12 | 16 | 84_137 | 470 | 648.0 | Domain | LIM zinc-binding 2 |
| Hgene | LIMK1 | chr7:73526328 | chr7:66489887 | ENST00000538333 | + | 11 | 15 | 165_258 | 436 | 614.0 | Domain | PDZ |
| Hgene | LIMK1 | chr7:73526328 | chr7:66489887 | ENST00000538333 | + | 11 | 15 | 25_75 | 436 | 614.0 | Domain | LIM zinc-binding 1 |
| Hgene | LIMK1 | chr7:73526328 | chr7:66489887 | ENST00000538333 | + | 11 | 15 | 84_137 | 436 | 614.0 | Domain | LIM zinc-binding 2 |
| Hgene | LIMK1 | chr7:73526328 | chr7:66489887 | ENST00000336180 | + | 12 | 16 | 345_353 | 470 | 648.0 | Nucleotide binding | ATP |
| Hgene | LIMK1 | chr7:73526328 | chr7:66489887 | ENST00000538333 | + | 11 | 15 | 345_353 | 436 | 614.0 | Nucleotide binding | ATP |
| Tgene | TYW1 | chr7:73526328 | chr7:66489887 | ENST00000359626 | 5 | 16 | 400_644 | 287 | 733.0 | Domain | Radical SAM core |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LIMK1 | chr7:73526328 | chr7:66489887 | ENST00000336180 | + | 12 | 16 | 339_604 | 470 | 648.0 | Domain | Protein kinase |
| Hgene | LIMK1 | chr7:73526328 | chr7:66489887 | ENST00000538333 | + | 11 | 15 | 339_604 | 436 | 614.0 | Domain | Protein kinase |
| Tgene | TYW1 | chr7:73526328 | chr7:66489887 | ENST00000359626 | 5 | 16 | 79_237 | 287 | 733.0 | Domain | Flavodoxin-like | |
| Tgene | TYW1 | chr7:73526328 | chr7:66489887 | ENST00000359626 | 5 | 16 | 176_208 | 287 | 733.0 | Nucleotide binding | FMN | |
| Tgene | TYW1 | chr7:73526328 | chr7:66489887 | ENST00000359626 | 5 | 16 | 85_89 | 287 | 733.0 | Nucleotide binding | FMN |
Top |
Fusion Gene Sequence for LIMK1-TYW1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >44717_44717_1_LIMK1-TYW1_LIMK1_chr7_73526328_ENST00000336180_TYW1_chr7_66489887_ENST00000359626_length(transcript)=3784nt_BP=1461nt CCCGGCCGCCCCCAGCCCCAGCCCCGCCGGGCCCCGCCCCCCGTCGAGTGCATGAGGTTGACGCTACTTTGTTGCACCTGGAGGGAAGAA CGTATGGGAGAGGAAGGAAGCGAGTTGCCCGTGTGTGCAAGCTGCGGCCAGAGGATCTATGATGGCCAGTACCTCCAGGCCCTGAACGCG GACTGGCACGCAGACTGCTTCAGGTGTTGTGACTGCAGTGCCTCCCTGTCGCACCAGTACTATGAGAAGGATGGGCAGCTCTTCTGCAAG AAGGACTACTGGGCCCGCTATGGCGAGTCCTGCCATGGGTGCTCTGAGCAAATCACCAAGGGACTGGTTATGGTGGCTGGGGAGCTGAAG TACCACCCCGAGTGTTTCATCTGCCTCACGTGTGGGACCTTTATCGGTGACGGGGACACCTACACGCTGGTGGAGCACTCCAAGCTGTAC TGCGGGCACTGCTACTACCAGACTGTGGTGACCCCCGTCATCGAGCAGATCCTGCCTGACTCCCCTGGCTCCCACCTGCCCCACACCGTC ACCCTGGTGTCCATCCCAGCCTCATCTCATGGCAAGCGTGGACTTTCAGTCTCCATTGACCCCCCGCACGGCCCACCGGGCTGTGGCACC GAGCACTCACACACCGTCCGCGTCCAGGGAGTGGATCCGGGCTGCATGAGCCCAGATGTGAAGAATTCCATCCACGTCGGAGACCGGATC TTGGAAATCAATGGCACGCCCATCCGAAATGTGCCCCTGGACGAGATTGACCTGCTGATTCAGGAAACCAGCCGCCTGCTCCAGCTGACC CTCGAGCATGACCCTCACGATACACTGGGCCACGGGCTGGGGCCTGAGACCAGCCCCCTGAGCTCTCCGGCTTATACTCCCAGCGGGGAG GCGGGCAGCTCTGCCCGGCAGAAACCTGTCTTGAGGAGCTGCAGCATCGACAGGTCTCCGGGCGCTGGCTCACTGGGCTCCCCGGCCTCC CAGCGCAAGGACCTGGGTCGCTCTGAGTCCCTCCGCGTAGTCTGCCGGCCACACCGCATCTTCCGGCCGTCGGACCTCATCCACGGGGAG GTGCTGGGCAAGGGCTGCTTCGGCCAGGCTATCAAGGTGACACACCGTGAGACAGGTGAGGTGATGGTGATGAAGGAGCTGATCCGGTTC GACGAGGAGACCCAGAGGACGTTCCTCAAGGAGGTGAAGGTCATGCGATGCCTGGAACACCCCAACGTGCTCAAGTTCATCGGGGTGCTC TACAAGGACAAGAGGCTCAACTTCATCACTGAGTACATCAAGGGCGGCACGCTCCGGGGCATCATCAAGAGCATGGACAGCCAGTACCCA TGGAGCCAGAGAGTGAGCTTTGCCAAGGACATCGCATCAGGGATGGCCTACCTCCACTCCATGAACATCATCCACCGAGACCTCAACTCC CACAACTGCCTGGTCCGCGAGGAGGAAGAACCCTTTGAGAGCTCCAGTGAAGAAGAGTTTGGTGGTGAGGACCATCAGAGCCTAAATTCC ATTGTTGATGTTGAAGATTTGGGCAAAATTATGGATCATGTGAAGAAAGAAAAGAGAGAAAAGGAACAGCAGGAAGAGAAGTCTGGTTTG TTCAGGAACATGGGGAGGAATGAAGATGGTGAAAGAAGAGCTATGATAACTCCTGCTCTCCGAGAAGCCCTTACTAAACAAGGTTATCAG TTGATTGGGAGCCACTCGGGGGTGAAGCTTTGCAGGTGGACAAAGTCCATGCTCCGAGGGAGAGGAGGTTGTTACAAACACACATTCTAT GGAATTGAGAGCCATCGCTGCATGGAAACCACCCCGAGCTTGGCGTGTGCTAATAAATGTGTCTTCTGTTGGCGGCACCACACCAACCCC GTGGGCACTGAGTGGCGGTGGAAGATGGACCAGCCTGAAATGATCTTGAAGGAAGCCATTGAAAACCATCAGAACATGATTAAGCAGTTT AAAGGAGTACCGGGCGTCAAAGCAGAACGCTTTGAAGAAGGAATGACGGTAAAGCACTGTGCATTGTCCCTCGTGGGAGAACCAATAATG TACCCAGAGATCAACAGGTTTTTGAAGCTACTCCACCAGTGTAAAATTTCCAGCTTCCTGGTCACAAACGCACAATTTCCTGCGGAAATC AGGAACCTCGAGCCGGTTACTCAGCTGTATGTCAGTGTGGATGCCAGTACCAAAGACAGCCTGAAGAAAATCGACCGCCCACTCTTCAAG GATTTCTGGCAGAGATTCCTTGACAGTTTAAAAGCCTTGGCAGTCAAGCAACAACGAACTGTCTACAGACTGACGCTCGTGAAAGCATGG AACGTGGACGAGCTCCAGGCCTACGCGCAGCTCGTGTCCCTGGGGAATCCTGACTTCATCGAAGTGAAGGGCGTTACCTACTGCGGAGAA AGTTCAGCAAGCAGTCTTACCATGGCCCACGTGCCCTGGCATGAGGAAGTGGTACAGTTTGTCCACGAGTTGGTGGATCTGATCCCCGAA TATGAAATTGCATGTGAACACGAACACTCTAATTGCCTCCTGATAGCACACAGAAAGTTTAAAATTGGTGGTGAATGGTGGACATGGATC GATTATAACCGCTTCCAGGAGCTCATCCAGGAATATGAAGATAGTGGTGGATCAAAAACGTTCAGCGCAAAGGATTATATGGCCAGAACT CCTCACTGGGCATTATTTGGTGCCAGTGAAAGAGGCTTTGATCCCAAGGACACAAGACATCAGAGAAAGAACAAATCAAAGGCTATTTCT GGATGTTGAGATTATCTGATTTCAAGGTACTGAAGGACAAAAACTTGGATGGCCTCAAAAGGTTCTTGAACACCACTGTGATTCTCCAAG GACGAATTACGTAAATTATACTTTCATACAAAGGAGACGATAAGGCAGTAAACATGGAGACACGGGGGACAGCGTCCACACTCAGAGGGC CTGGGCCACAGCCCCGATGTTTCTTTTCAGAACTCAGCCCCTTTCCTGATTTTACTTCTAAGAGGAAAATTATTTTGGGGAGGAACTACA CAGTCGTGATTAGAATTTATCTGATGGTTTTGTATTATAACTTGTAAGACCTGCCAGAATGCTAGTCCCGAGAGTGTCAGACAAGGAAGA AGTCCCTGGGCCTCTTCCCCTTACCCGGCCCTTAGATTTCATGGAGCAGCCACTTAGCATTGAATTGCACTACCCTGAGCTAAACGTGTC TGTGCTTTCTAAGATAAGAGCTTGATCCCTTTCTTCTATCTTAAGACAGCACCTCCTGAAAAGAATCGAAGTTGTCACAACTCTCAATTA TTTTTTAAATACTGCATAGATTGAGTTTTGGTTTATTACCAACCCTTCCCAGAATTGCGTTGGATCTAAAACTACTAGATCTCATCCCAT TCCCATGTAAATTACCACAGACCGCAGTACCGGGGCTGGAGCGGAGTGAAGCTGTCTGCTGTAAGAGGAGTGGCCATGTGAGGGCATGGA GTCATTAGTCTCACAAACACACTTTGGACTGAAGAGGATCATTTCTTTTTGTTCGTGAGGTCACTGTCCAGGCCTCTCATATCATGACCA GACGGCGGGTCTCCATCTTCTTTCACTCCTGTGGCCCTGGCTGCTTTACACAATCTGTTCTATAAGGTTCAGGTGTTTTCAAGTTGGAAA GATCATAAATACTCAAAATTGTTTTCAAGTTAGCAAGTTCTTTTAACAGTCTTTTATGCAAAAATTGAATTAATAAAATAATCTTTTGTA >44717_44717_1_LIMK1-TYW1_LIMK1_chr7_73526328_ENST00000336180_TYW1_chr7_66489887_ENST00000359626_length(amino acids)=915AA_BP=470 MRLTLLCCTWREERMGEEGSELPVCASCGQRIYDGQYLQALNADWHADCFRCCDCSASLSHQYYEKDGQLFCKKDYWARYGESCHGCSEQ ITKGLVMVAGELKYHPECFICLTCGTFIGDGDTYTLVEHSKLYCGHCYYQTVVTPVIEQILPDSPGSHLPHTVTLVSIPASSHGKRGLSV SIDPPHGPPGCGTEHSHTVRVQGVDPGCMSPDVKNSIHVGDRILEINGTPIRNVPLDEIDLLIQETSRLLQLTLEHDPHDTLGHGLGPET SPLSSPAYTPSGEAGSSARQKPVLRSCSIDRSPGAGSLGSPASQRKDLGRSESLRVVCRPHRIFRPSDLIHGEVLGKGCFGQAIKVTHRE TGEVMVMKELIRFDEETQRTFLKEVKVMRCLEHPNVLKFIGVLYKDKRLNFITEYIKGGTLRGIIKSMDSQYPWSQRVSFAKDIASGMAY LHSMNIIHRDLNSHNCLVREEEEPFESSSEEEFGGEDHQSLNSIVDVEDLGKIMDHVKKEKREKEQQEEKSGLFRNMGRNEDGERRAMIT PALREALTKQGYQLIGSHSGVKLCRWTKSMLRGRGGCYKHTFYGIESHRCMETTPSLACANKCVFCWRHHTNPVGTEWRWKMDQPEMILK EAIENHQNMIKQFKGVPGVKAERFEEGMTVKHCALSLVGEPIMYPEINRFLKLLHQCKISSFLVTNAQFPAEIRNLEPVTQLYVSVDAST KDSLKKIDRPLFKDFWQRFLDSLKALAVKQQRTVYRLTLVKAWNVDELQAYAQLVSLGNPDFIEVKGVTYCGESSASSLTMAHVPWHEEV VQFVHELVDLIPEYEIACEHEHSNCLLIAHRKFKIGGEWWTWIDYNRFQELIQEYEDSGGSKTFSAKDYMARTPHWALFGASERGFDPKD -------------------------------------------------------------- >44717_44717_2_LIMK1-TYW1_LIMK1_chr7_73526328_ENST00000418310_TYW1_chr7_66489887_ENST00000359626_length(transcript)=3925nt_BP=1602nt ACTGGGTTCACGTGGGCACGTGGCTCCACTCCGGTCACTGCTTCCTGGGCAGAGGGATGACGTGGAAGGTAGACCTAGGGGGGACAGGGT TGCTTCCTGCCCATGTCCAGCATTCTAGAATTGTCCACATCTAGGCAGGGCGTTCCACCATCAGTGCTGGACTGGACCACCCAATTCCTT TGGACGCTTGACTCAGCCCAAACCCGTGCTCCCTCCAGGACCCTGGGCAGGGCAGCCCCAGCTCCAGGAAGCGAGTTGCCCGTGTGTGCA AGCTGCGGCCAGAGGATCTATGATGGCCAGTACCTCCAGGCCCTGAACGCGGACTGGCACGCAGACTGCTTCAGGTGTTGTGACTGCAGT GCCTCCCTGTCGCACCAGTACTATGAGAAGGATGGGCAGCTCTTCTGCAAGAAGGACTACTGGGCCCGCTATGGCGAGTCCTGCCATGGG TGCTCTGAGCAAATCACCAAGGGACTGGTTATGGTGGCTGGGGAGCTGAAGTACCACCCCGAGTGTTTCATCTGCCTCACGTGTGGGACC TTTATCGGTGACGGGGACACCTACACGCTGGTGGAGCACTCCAAGCTGTACTGCGGGCACTGCTACTACCAGACTGTGGTGACCCCCGTC ATCGAGCAGATCCTGCCTGACTCCCCTGGCTCCCACCTGCCCCACACCGTCACCCTGGTGTCCATCCCAGCCTCATCTCATGGCAAGCGT GGACTTTCAGTCTCCATTGACCCCCCGCACGGCCCACCGGGCTGTGGCACCGAGCACTCACACACCGTCCGCGTCCAGGGAGTGGATCCG GGCTGCATGAGCCCAGATGTGAAGAATTCCATCCACGTCGGAGACCGGATCTTGGAAATCAATGGCACGCCCATCCGAAATGTGCCCCTG GACGAGATTGACCTGCTGATTCAGGAAACCAGCCGCCTGCTCCAGCTGACCCTCGAGCATGACCCTCACGATACACTGGGCCACGGGCTG GGGCCTGAGACCAGCCCCCTGAGCTCTCCGGCTTATACTCCCAGCGGGGAGGCGGGCAGCTCTGCCCGGCAGAAACCTGTCTTGAGGAGC TGCAGCATCGACAGGTCTCCGGGCGCTGGCTCACTGGGCTCCCCGGCCTCCCAGCGCAAGGACCTGGGTCGCTCTGAGTCCCTCCGCGTA GTCTGCCGGCCACACCGCATCTTCCGGCCGTCGGACCTCATCCACGGGGAGGTGCTGGGCAAGGGCTGCTTCGGCCAGGCTATCAAGGTG ACACACCGTGAGACAGGTGAGGTGATGGTGATGAAGGAGCTGATCCGGTTCGACGAGGAGACCCAGAGGACGTTCCTCAAGGAGGTGAAG GTCATGCGATGCCTGGAACACCCCAACGTGCTCAAGTTCATCGGGGTGCTCTACAAGGACAAGAGGCTCAACTTCATCACTGAGTACATC AAGGGCGGCACGCTCCGGGGCATCATCAAGAGCATGGACAGCCAGTACCCATGGAGCCAGAGAGTGAGCTTTGCCAAGGACATCGCATCA GGGATGGCCTACCTCCACTCCATGAACATCATCCACCGAGACCTCAACTCCCACAACTGCCTGGTCCGCGAGGAGGAAGAACCCTTTGAG AGCTCCAGTGAAGAAGAGTTTGGTGGTGAGGACCATCAGAGCCTAAATTCCATTGTTGATGTTGAAGATTTGGGCAAAATTATGGATCAT GTGAAGAAAGAAAAGAGAGAAAAGGAACAGCAGGAAGAGAAGTCTGGTTTGTTCAGGAACATGGGGAGGAATGAAGATGGTGAAAGAAGA GCTATGATAACTCCTGCTCTCCGAGAAGCCCTTACTAAACAAGGTTATCAGTTGATTGGGAGCCACTCGGGGGTGAAGCTTTGCAGGTGG ACAAAGTCCATGCTCCGAGGGAGAGGAGGTTGTTACAAACACACATTCTATGGAATTGAGAGCCATCGCTGCATGGAAACCACCCCGAGC TTGGCGTGTGCTAATAAATGTGTCTTCTGTTGGCGGCACCACACCAACCCCGTGGGCACTGAGTGGCGGTGGAAGATGGACCAGCCTGAA ATGATCTTGAAGGAAGCCATTGAAAACCATCAGAACATGATTAAGCAGTTTAAAGGAGTACCGGGCGTCAAAGCAGAACGCTTTGAAGAA GGAATGACGGTAAAGCACTGTGCATTGTCCCTCGTGGGAGAACCAATAATGTACCCAGAGATCAACAGGTTTTTGAAGCTACTCCACCAG TGTAAAATTTCCAGCTTCCTGGTCACAAACGCACAATTTCCTGCGGAAATCAGGAACCTCGAGCCGGTTACTCAGCTGTATGTCAGTGTG GATGCCAGTACCAAAGACAGCCTGAAGAAAATCGACCGCCCACTCTTCAAGGATTTCTGGCAGAGATTCCTTGACAGTTTAAAAGCCTTG GCAGTCAAGCAACAACGAACTGTCTACAGACTGACGCTCGTGAAAGCATGGAACGTGGACGAGCTCCAGGCCTACGCGCAGCTCGTGTCC CTGGGGAATCCTGACTTCATCGAAGTGAAGGGCGTTACCTACTGCGGAGAAAGTTCAGCAAGCAGTCTTACCATGGCCCACGTGCCCTGG CATGAGGAAGTGGTACAGTTTGTCCACGAGTTGGTGGATCTGATCCCCGAATATGAAATTGCATGTGAACACGAACACTCTAATTGCCTC CTGATAGCACACAGAAAGTTTAAAATTGGTGGTGAATGGTGGACATGGATCGATTATAACCGCTTCCAGGAGCTCATCCAGGAATATGAA GATAGTGGTGGATCAAAAACGTTCAGCGCAAAGGATTATATGGCCAGAACTCCTCACTGGGCATTATTTGGTGCCAGTGAAAGAGGCTTT GATCCCAAGGACACAAGACATCAGAGAAAGAACAAATCAAAGGCTATTTCTGGATGTTGAGATTATCTGATTTCAAGGTACTGAAGGACA AAAACTTGGATGGCCTCAAAAGGTTCTTGAACACCACTGTGATTCTCCAAGGACGAATTACGTAAATTATACTTTCATACAAAGGAGACG ATAAGGCAGTAAACATGGAGACACGGGGGACAGCGTCCACACTCAGAGGGCCTGGGCCACAGCCCCGATGTTTCTTTTCAGAACTCAGCC CCTTTCCTGATTTTACTTCTAAGAGGAAAATTATTTTGGGGAGGAACTACACAGTCGTGATTAGAATTTATCTGATGGTTTTGTATTATA ACTTGTAAGACCTGCCAGAATGCTAGTCCCGAGAGTGTCAGACAAGGAAGAAGTCCCTGGGCCTCTTCCCCTTACCCGGCCCTTAGATTT CATGGAGCAGCCACTTAGCATTGAATTGCACTACCCTGAGCTAAACGTGTCTGTGCTTTCTAAGATAAGAGCTTGATCCCTTTCTTCTAT CTTAAGACAGCACCTCCTGAAAAGAATCGAAGTTGTCACAACTCTCAATTATTTTTTAAATACTGCATAGATTGAGTTTTGGTTTATTAC CAACCCTTCCCAGAATTGCGTTGGATCTAAAACTACTAGATCTCATCCCATTCCCATGTAAATTACCACAGACCGCAGTACCGGGGCTGG AGCGGAGTGAAGCTGTCTGCTGTAAGAGGAGTGGCCATGTGAGGGCATGGAGTCATTAGTCTCACAAACACACTTTGGACTGAAGAGGAT CATTTCTTTTTGTTCGTGAGGTCACTGTCCAGGCCTCTCATATCATGACCAGACGGCGGGTCTCCATCTTCTTTCACTCCTGTGGCCCTG GCTGCTTTACACAATCTGTTCTATAAGGTTCAGGTGTTTTCAAGTTGGAAAGATCATAAATACTCAAAATTGTTTTCAAGTTAGCAAGTT >44717_44717_2_LIMK1-TYW1_LIMK1_chr7_73526328_ENST00000418310_TYW1_chr7_66489887_ENST00000359626_length(amino acids)=947AA_BP=502 MPMSSILELSTSRQGVPPSVLDWTTQFLWTLDSAQTRAPSRTLGRAAPAPGSELPVCASCGQRIYDGQYLQALNADWHADCFRCCDCSAS LSHQYYEKDGQLFCKKDYWARYGESCHGCSEQITKGLVMVAGELKYHPECFICLTCGTFIGDGDTYTLVEHSKLYCGHCYYQTVVTPVIE QILPDSPGSHLPHTVTLVSIPASSHGKRGLSVSIDPPHGPPGCGTEHSHTVRVQGVDPGCMSPDVKNSIHVGDRILEINGTPIRNVPLDE IDLLIQETSRLLQLTLEHDPHDTLGHGLGPETSPLSSPAYTPSGEAGSSARQKPVLRSCSIDRSPGAGSLGSPASQRKDLGRSESLRVVC RPHRIFRPSDLIHGEVLGKGCFGQAIKVTHRETGEVMVMKELIRFDEETQRTFLKEVKVMRCLEHPNVLKFIGVLYKDKRLNFITEYIKG GTLRGIIKSMDSQYPWSQRVSFAKDIASGMAYLHSMNIIHRDLNSHNCLVREEEEPFESSSEEEFGGEDHQSLNSIVDVEDLGKIMDHVK KEKREKEQQEEKSGLFRNMGRNEDGERRAMITPALREALTKQGYQLIGSHSGVKLCRWTKSMLRGRGGCYKHTFYGIESHRCMETTPSLA CANKCVFCWRHHTNPVGTEWRWKMDQPEMILKEAIENHQNMIKQFKGVPGVKAERFEEGMTVKHCALSLVGEPIMYPEINRFLKLLHQCK ISSFLVTNAQFPAEIRNLEPVTQLYVSVDASTKDSLKKIDRPLFKDFWQRFLDSLKALAVKQQRTVYRLTLVKAWNVDELQAYAQLVSLG NPDFIEVKGVTYCGESSASSLTMAHVPWHEEVVQFVHELVDLIPEYEIACEHEHSNCLLIAHRKFKIGGEWWTWIDYNRFQELIQEYEDS -------------------------------------------------------------- >44717_44717_3_LIMK1-TYW1_LIMK1_chr7_73526328_ENST00000538333_TYW1_chr7_66489887_ENST00000359626_length(transcript)=3802nt_BP=1479nt AAGTCCCCTGGCACCCACGCGGCTGCAGCCTCCTCCTGTGCGCTGCAGCCACGCTGGCCCCACCCTCTGCAGCCTCCAATCCTGAGCCCC TGAGGGAGGATGGGGAAGCAGCTGGTCTGGCCACCCCTGCCCTCCCTTAGACCTCCAGAGCCCCCAGTGTAGCCACAGAGGATGCTGTTG GCTTCAGCCCCAAGAAGACGCCGCTTCCTCCAGAGGGCTAAGTGTTGTGACTGCAGTGCCTCCCTGTCGCACCAGTACTATGAGAAGGAT GGGCAGCTCTTCTGCAAGAAGGACTACTGGGCCCGCTATGGCGAGTCCTGCCATGGGTGCTCTGAGCAAATCACCAAGGGACTGGTTATG GTGGCTGGGGAGCTGAAGTACCACCCCGAGTGTTTCATCTGCCTCACGTGTGGGACCTTTATCGGTGACGGGGACACCTACACGCTGGTG GAGCACTCCAAGCTGTACTGCGGGCACTGCTACTACCAGACTGTGGTGACCCCCGTCATCGAGCAGATCCTGCCTGACTCCCCTGGCTCC CACCTGCCCCACACCGTCACCCTGGTGTCCATCCCAGCCTCATCTCATGGCAAGCGTGGACTTTCAGTCTCCATTGACCCCCCGCACGGC CCACCGGGCTGTGGCACCGAGCACTCACACACCGTCCGCGTCCAGGGAGTGGATCCGGGCTGCATGAGCCCAGATGTGAAGAATTCCATC CACGTCGGAGACCGGATCTTGGAAATCAATGGCACGCCCATCCGAAATGTGCCCCTGGACGAGATTGACCTGCTGATTCAGGAAACCAGC CGCCTGCTCCAGCTGACCCTCGAGCATGACCCTCACGATACACTGGGCCACGGGCTGGGGCCTGAGACCAGCCCCCTGAGCTCTCCGGCT TATACTCCCAGCGGGGAGGCGGGCAGCTCTGCCCGGCAGAAACCTGTCTTGAGGAGCTGCAGCATCGACAGGTCTCCGGGCGCTGGCTCA CTGGGCTCCCCGGCCTCCCAGCGCAAGGACCTGGGTCGCTCTGAGTCCCTCCGCGTAGTCTGCCGGCCACACCGCATCTTCCGGCCGTCG GACCTCATCCACGGGGAGGTGCTGGGCAAGGGCTGCTTCGGCCAGGCTATCAAGGTGACACACCGTGAGACAGGTGAGGTGATGGTGATG AAGGAGCTGATCCGGTTCGACGAGGAGACCCAGAGGACGTTCCTCAAGGAGGTGAAGGTCATGCGATGCCTGGAACACCCCAACGTGCTC AAGTTCATCGGGGTGCTCTACAAGGACAAGAGGCTCAACTTCATCACTGAGTACATCAAGGGCGGCACGCTCCGGGGCATCATCAAGAGC ATGGACAGCCAGTACCCATGGAGCCAGAGAGTGAGCTTTGCCAAGGACATCGCATCAGGGATGGCCTACCTCCACTCCATGAACATCATC CACCGAGACCTCAACTCCCACAACTGCCTGGTCCGCGAGGAGGAAGAACCCTTTGAGAGCTCCAGTGAAGAAGAGTTTGGTGGTGAGGAC CATCAGAGCCTAAATTCCATTGTTGATGTTGAAGATTTGGGCAAAATTATGGATCATGTGAAGAAAGAAAAGAGAGAAAAGGAACAGCAG GAAGAGAAGTCTGGTTTGTTCAGGAACATGGGGAGGAATGAAGATGGTGAAAGAAGAGCTATGATAACTCCTGCTCTCCGAGAAGCCCTT ACTAAACAAGGTTATCAGTTGATTGGGAGCCACTCGGGGGTGAAGCTTTGCAGGTGGACAAAGTCCATGCTCCGAGGGAGAGGAGGTTGT TACAAACACACATTCTATGGAATTGAGAGCCATCGCTGCATGGAAACCACCCCGAGCTTGGCGTGTGCTAATAAATGTGTCTTCTGTTGG CGGCACCACACCAACCCCGTGGGCACTGAGTGGCGGTGGAAGATGGACCAGCCTGAAATGATCTTGAAGGAAGCCATTGAAAACCATCAG AACATGATTAAGCAGTTTAAAGGAGTACCGGGCGTCAAAGCAGAACGCTTTGAAGAAGGAATGACGGTAAAGCACTGTGCATTGTCCCTC GTGGGAGAACCAATAATGTACCCAGAGATCAACAGGTTTTTGAAGCTACTCCACCAGTGTAAAATTTCCAGCTTCCTGGTCACAAACGCA CAATTTCCTGCGGAAATCAGGAACCTCGAGCCGGTTACTCAGCTGTATGTCAGTGTGGATGCCAGTACCAAAGACAGCCTGAAGAAAATC GACCGCCCACTCTTCAAGGATTTCTGGCAGAGATTCCTTGACAGTTTAAAAGCCTTGGCAGTCAAGCAACAACGAACTGTCTACAGACTG ACGCTCGTGAAAGCATGGAACGTGGACGAGCTCCAGGCCTACGCGCAGCTCGTGTCCCTGGGGAATCCTGACTTCATCGAAGTGAAGGGC GTTACCTACTGCGGAGAAAGTTCAGCAAGCAGTCTTACCATGGCCCACGTGCCCTGGCATGAGGAAGTGGTACAGTTTGTCCACGAGTTG GTGGATCTGATCCCCGAATATGAAATTGCATGTGAACACGAACACTCTAATTGCCTCCTGATAGCACACAGAAAGTTTAAAATTGGTGGT GAATGGTGGACATGGATCGATTATAACCGCTTCCAGGAGCTCATCCAGGAATATGAAGATAGTGGTGGATCAAAAACGTTCAGCGCAAAG GATTATATGGCCAGAACTCCTCACTGGGCATTATTTGGTGCCAGTGAAAGAGGCTTTGATCCCAAGGACACAAGACATCAGAGAAAGAAC AAATCAAAGGCTATTTCTGGATGTTGAGATTATCTGATTTCAAGGTACTGAAGGACAAAAACTTGGATGGCCTCAAAAGGTTCTTGAACA CCACTGTGATTCTCCAAGGACGAATTACGTAAATTATACTTTCATACAAAGGAGACGATAAGGCAGTAAACATGGAGACACGGGGGACAG CGTCCACACTCAGAGGGCCTGGGCCACAGCCCCGATGTTTCTTTTCAGAACTCAGCCCCTTTCCTGATTTTACTTCTAAGAGGAAAATTA TTTTGGGGAGGAACTACACAGTCGTGATTAGAATTTATCTGATGGTTTTGTATTATAACTTGTAAGACCTGCCAGAATGCTAGTCCCGAG AGTGTCAGACAAGGAAGAAGTCCCTGGGCCTCTTCCCCTTACCCGGCCCTTAGATTTCATGGAGCAGCCACTTAGCATTGAATTGCACTA CCCTGAGCTAAACGTGTCTGTGCTTTCTAAGATAAGAGCTTGATCCCTTTCTTCTATCTTAAGACAGCACCTCCTGAAAAGAATCGAAGT TGTCACAACTCTCAATTATTTTTTAAATACTGCATAGATTGAGTTTTGGTTTATTACCAACCCTTCCCAGAATTGCGTTGGATCTAAAAC TACTAGATCTCATCCCATTCCCATGTAAATTACCACAGACCGCAGTACCGGGGCTGGAGCGGAGTGAAGCTGTCTGCTGTAAGAGGAGTG GCCATGTGAGGGCATGGAGTCATTAGTCTCACAAACACACTTTGGACTGAAGAGGATCATTTCTTTTTGTTCGTGAGGTCACTGTCCAGG CCTCTCATATCATGACCAGACGGCGGGTCTCCATCTTCTTTCACTCCTGTGGCCCTGGCTGCTTTACACAATCTGTTCTATAAGGTTCAG GTGTTTTCAAGTTGGAAAGATCATAAATACTCAAAATTGTTTTCAAGTTAGCAAGTTCTTTTAACAGTCTTTTATGCAAAAATTGAATTA >44717_44717_3_LIMK1-TYW1_LIMK1_chr7_73526328_ENST00000538333_TYW1_chr7_66489887_ENST00000359626_length(amino acids)=881AA_BP=436 MLLASAPRRRRFLQRAKCCDCSASLSHQYYEKDGQLFCKKDYWARYGESCHGCSEQITKGLVMVAGELKYHPECFICLTCGTFIGDGDTY TLVEHSKLYCGHCYYQTVVTPVIEQILPDSPGSHLPHTVTLVSIPASSHGKRGLSVSIDPPHGPPGCGTEHSHTVRVQGVDPGCMSPDVK NSIHVGDRILEINGTPIRNVPLDEIDLLIQETSRLLQLTLEHDPHDTLGHGLGPETSPLSSPAYTPSGEAGSSARQKPVLRSCSIDRSPG AGSLGSPASQRKDLGRSESLRVVCRPHRIFRPSDLIHGEVLGKGCFGQAIKVTHRETGEVMVMKELIRFDEETQRTFLKEVKVMRCLEHP NVLKFIGVLYKDKRLNFITEYIKGGTLRGIIKSMDSQYPWSQRVSFAKDIASGMAYLHSMNIIHRDLNSHNCLVREEEEPFESSSEEEFG GEDHQSLNSIVDVEDLGKIMDHVKKEKREKEQQEEKSGLFRNMGRNEDGERRAMITPALREALTKQGYQLIGSHSGVKLCRWTKSMLRGR GGCYKHTFYGIESHRCMETTPSLACANKCVFCWRHHTNPVGTEWRWKMDQPEMILKEAIENHQNMIKQFKGVPGVKAERFEEGMTVKHCA LSLVGEPIMYPEINRFLKLLHQCKISSFLVTNAQFPAEIRNLEPVTQLYVSVDASTKDSLKKIDRPLFKDFWQRFLDSLKALAVKQQRTV YRLTLVKAWNVDELQAYAQLVSLGNPDFIEVKGVTYCGESSASSLTMAHVPWHEEVVQFVHELVDLIPEYEIACEHEHSNCLLIAHRKFK -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for LIMK1-TYW1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for LIMK1-TYW1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for LIMK1-TYW1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
