
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:LLGL2-TMEM104 (FusionGDB2 ID:45705) |
Fusion Gene Summary for LLGL2-TMEM104 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: LLGL2-TMEM104 | Fusion gene ID: 45705 | Hgene | Tgene | Gene symbol | LLGL2 | TMEM104 | Gene ID | 3993 | 54868 |
| Gene name | LLGL scribble cell polarity complex component 2 | transmembrane protein 104 | |
| Synonyms | HGL|Hugl-2|LGL2 | - | |
| Cytomap | 17q25.1 | 17q25.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | LLGL scribble cell polarity complex component 2LLGL2, scribble cell polarity complex componenthuman giant larvae homologlethal giant larvae homolog 2, scribble cell polarity complex componentlethal(2) giant larvae protein homolog 2 | transmembrane protein 104 | |
| Modification date | 20200320 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000167462, ENST00000392550, ENST00000577200, ENST00000375227, ENST00000578363, | ENST00000417024, ENST00000582330, ENST00000582773, ENST00000584171, ENST00000335464, | |
| Fusion gene scores | * DoF score | 17 X 14 X 10=2380 | 8 X 7 X 7=392 |
| # samples | 20 | 9 | |
| ** MAII score | log2(20/2380*10)=-3.57288966842058 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/392*10)=-2.12285674778553 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: LLGL2 [Title/Abstract] AND TMEM104 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | LLGL2(73568145)-TMEM104(72832066), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | LLGL2-TMEM104 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | LLGL2 | GO:0032878 | regulation of establishment or maintenance of cell polarity | 12725730 |
 Fusion gene breakpoints across LLGL2 (5'-gene) Fusion gene breakpoints across LLGL2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
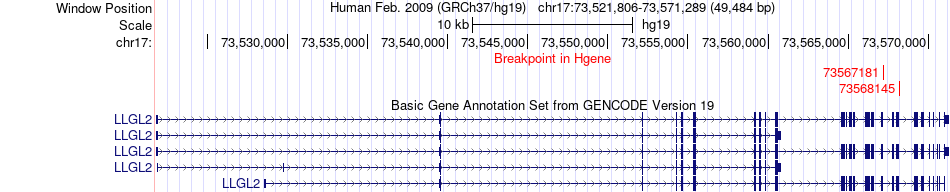 |
 Fusion gene breakpoints across TMEM104 (3'-gene) Fusion gene breakpoints across TMEM104 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
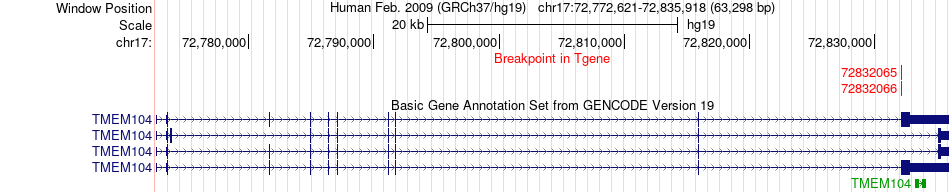 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-24-0970-01B | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + |
| ChimerDB4 | OV | TCGA-24-0970-01B | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832066 | + |
| ChimerDB4 | OV | TCGA-24-0970 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + |
Top |
Fusion Gene ORF analysis for LLGL2-TMEM104 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000167462 | ENST00000417024 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + |
| 5CDS-intron | ENST00000167462 | ENST00000417024 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832066 | + |
| 5CDS-intron | ENST00000167462 | ENST00000417024 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + |
| 5CDS-intron | ENST00000167462 | ENST00000582330 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + |
| 5CDS-intron | ENST00000167462 | ENST00000582330 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832066 | + |
| 5CDS-intron | ENST00000167462 | ENST00000582330 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + |
| 5CDS-intron | ENST00000167462 | ENST00000582773 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + |
| 5CDS-intron | ENST00000167462 | ENST00000582773 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832066 | + |
| 5CDS-intron | ENST00000167462 | ENST00000582773 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + |
| 5CDS-intron | ENST00000167462 | ENST00000584171 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + |
| 5CDS-intron | ENST00000167462 | ENST00000584171 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832066 | + |
| 5CDS-intron | ENST00000167462 | ENST00000584171 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + |
| 5CDS-intron | ENST00000392550 | ENST00000417024 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + |
| 5CDS-intron | ENST00000392550 | ENST00000417024 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832066 | + |
| 5CDS-intron | ENST00000392550 | ENST00000417024 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + |
| 5CDS-intron | ENST00000392550 | ENST00000582330 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + |
| 5CDS-intron | ENST00000392550 | ENST00000582330 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832066 | + |
| 5CDS-intron | ENST00000392550 | ENST00000582330 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + |
| 5CDS-intron | ENST00000392550 | ENST00000582773 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + |
| 5CDS-intron | ENST00000392550 | ENST00000582773 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832066 | + |
| 5CDS-intron | ENST00000392550 | ENST00000582773 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + |
| 5CDS-intron | ENST00000392550 | ENST00000584171 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + |
| 5CDS-intron | ENST00000392550 | ENST00000584171 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832066 | + |
| 5CDS-intron | ENST00000392550 | ENST00000584171 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + |
| 5CDS-intron | ENST00000577200 | ENST00000417024 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + |
| 5CDS-intron | ENST00000577200 | ENST00000417024 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832066 | + |
| 5CDS-intron | ENST00000577200 | ENST00000417024 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + |
| 5CDS-intron | ENST00000577200 | ENST00000582330 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + |
| 5CDS-intron | ENST00000577200 | ENST00000582330 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832066 | + |
| 5CDS-intron | ENST00000577200 | ENST00000582330 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + |
| 5CDS-intron | ENST00000577200 | ENST00000582773 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + |
| 5CDS-intron | ENST00000577200 | ENST00000582773 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832066 | + |
| 5CDS-intron | ENST00000577200 | ENST00000582773 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + |
| 5CDS-intron | ENST00000577200 | ENST00000584171 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + |
| 5CDS-intron | ENST00000577200 | ENST00000584171 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832066 | + |
| 5CDS-intron | ENST00000577200 | ENST00000584171 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + |
| Frame-shift | ENST00000167462 | ENST00000335464 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832066 | + |
| Frame-shift | ENST00000167462 | ENST00000335464 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + |
| Frame-shift | ENST00000392550 | ENST00000335464 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832066 | + |
| Frame-shift | ENST00000392550 | ENST00000335464 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + |
| Frame-shift | ENST00000577200 | ENST00000335464 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832066 | + |
| Frame-shift | ENST00000577200 | ENST00000335464 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + |
| In-frame | ENST00000167462 | ENST00000335464 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + |
| In-frame | ENST00000392550 | ENST00000335464 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + |
| In-frame | ENST00000577200 | ENST00000335464 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + |
| intron-3CDS | ENST00000375227 | ENST00000335464 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + |
| intron-3CDS | ENST00000375227 | ENST00000335464 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832066 | + |
| intron-3CDS | ENST00000375227 | ENST00000335464 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + |
| intron-3CDS | ENST00000578363 | ENST00000335464 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + |
| intron-3CDS | ENST00000578363 | ENST00000335464 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832066 | + |
| intron-3CDS | ENST00000578363 | ENST00000335464 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + |
| intron-intron | ENST00000375227 | ENST00000417024 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + |
| intron-intron | ENST00000375227 | ENST00000417024 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832066 | + |
| intron-intron | ENST00000375227 | ENST00000417024 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + |
| intron-intron | ENST00000375227 | ENST00000582330 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + |
| intron-intron | ENST00000375227 | ENST00000582330 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832066 | + |
| intron-intron | ENST00000375227 | ENST00000582330 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + |
| intron-intron | ENST00000375227 | ENST00000582773 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + |
| intron-intron | ENST00000375227 | ENST00000582773 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832066 | + |
| intron-intron | ENST00000375227 | ENST00000582773 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + |
| intron-intron | ENST00000375227 | ENST00000584171 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + |
| intron-intron | ENST00000375227 | ENST00000584171 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832066 | + |
| intron-intron | ENST00000375227 | ENST00000584171 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + |
| intron-intron | ENST00000578363 | ENST00000417024 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + |
| intron-intron | ENST00000578363 | ENST00000417024 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832066 | + |
| intron-intron | ENST00000578363 | ENST00000417024 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + |
| intron-intron | ENST00000578363 | ENST00000582330 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + |
| intron-intron | ENST00000578363 | ENST00000582330 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832066 | + |
| intron-intron | ENST00000578363 | ENST00000582330 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + |
| intron-intron | ENST00000578363 | ENST00000582773 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + |
| intron-intron | ENST00000578363 | ENST00000582773 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832066 | + |
| intron-intron | ENST00000578363 | ENST00000582773 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + |
| intron-intron | ENST00000578363 | ENST00000584171 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + |
| intron-intron | ENST00000578363 | ENST00000584171 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832066 | + |
| intron-intron | ENST00000578363 | ENST00000584171 | LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000167462 | LLGL2 | chr17 | 73567181 | + | ENST00000335464 | TMEM104 | chr17 | 72832066 | + | 6159 | 2306 | 130 | 3066 | 978 |
| ENST00000392550 | LLGL2 | chr17 | 73567181 | + | ENST00000335464 | TMEM104 | chr17 | 72832066 | + | 6146 | 2293 | 117 | 3053 | 978 |
| ENST00000577200 | LLGL2 | chr17 | 73567181 | + | ENST00000335464 | TMEM104 | chr17 | 72832066 | + | 6133 | 2280 | 62 | 3040 | 992 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000167462 | ENST00000335464 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + | 0.003549249 | 0.9964508 |
| ENST00000392550 | ENST00000335464 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + | 0.003500445 | 0.99649954 |
| ENST00000577200 | ENST00000335464 | LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832066 | + | 0.003698092 | 0.9963019 |
Top |
Fusion Genomic Features for LLGL2-TMEM104 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + | 6.85E-05 | 0.99993145 |
| LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + | 6.85E-05 | 0.99993145 |
| LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832065 | + | 1.73E-06 | 0.99999833 |
| LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + | 6.85E-05 | 0.99993145 |
| LLGL2 | chr17 | 73568145 | + | TMEM104 | chr17 | 72832065 | + | 6.85E-05 | 0.99993145 |
| LLGL2 | chr17 | 73567181 | + | TMEM104 | chr17 | 72832065 | + | 1.73E-06 | 0.99999833 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
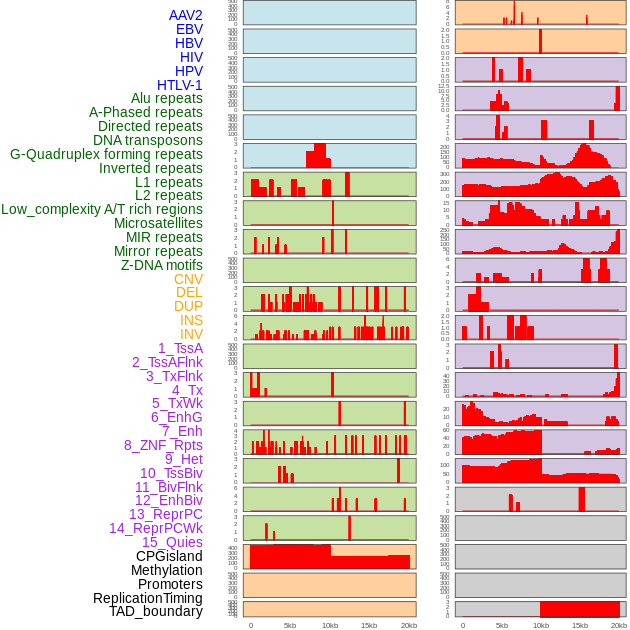 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
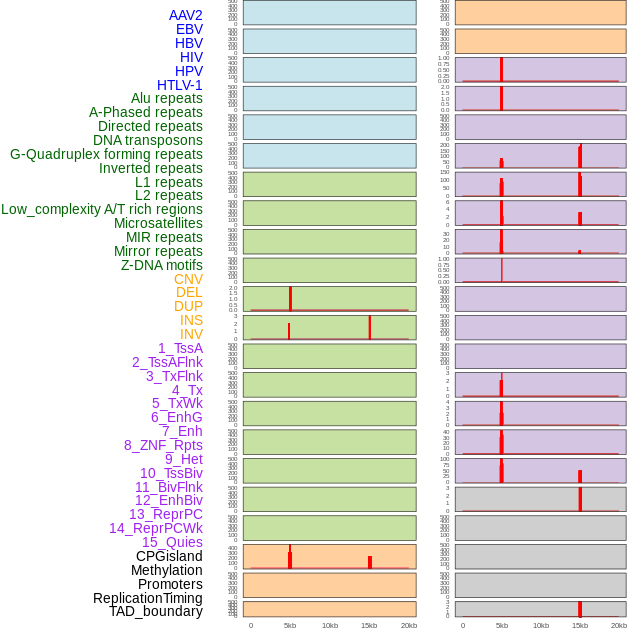 |
Top |
Fusion Protein Features for LLGL2-TMEM104 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:73568145/chr17:72832066) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000167462 | + | 17 | 25 | 132_169 | 725 | 1016.0 | Repeat | Note=WD 3 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000167462 | + | 17 | 25 | 193_227 | 725 | 1016.0 | Repeat | Note=WD 4 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000167462 | + | 17 | 25 | 233_268 | 725 | 1016.0 | Repeat | Note=WD 5 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000167462 | + | 17 | 25 | 282_324 | 725 | 1016.0 | Repeat | Note=WD 6 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000167462 | + | 17 | 25 | 332_366 | 725 | 1016.0 | Repeat | Note=WD 7 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000167462 | + | 17 | 25 | 36_69 | 725 | 1016.0 | Repeat | Note=WD 1 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000167462 | + | 17 | 25 | 388_464 | 725 | 1016.0 | Repeat | Note=WD 8 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000167462 | + | 17 | 25 | 508_583 | 725 | 1016.0 | Repeat | Note=WD 9 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000167462 | + | 17 | 25 | 592_653 | 725 | 1016.0 | Repeat | Note=WD 10 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000167462 | + | 17 | 25 | 76_117 | 725 | 1016.0 | Repeat | Note=WD 2 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000392550 | + | 17 | 26 | 132_169 | 725 | 1021.0 | Repeat | Note=WD 3 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000392550 | + | 17 | 26 | 193_227 | 725 | 1021.0 | Repeat | Note=WD 4 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000392550 | + | 17 | 26 | 233_268 | 725 | 1021.0 | Repeat | Note=WD 5 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000392550 | + | 17 | 26 | 282_324 | 725 | 1021.0 | Repeat | Note=WD 6 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000392550 | + | 17 | 26 | 332_366 | 725 | 1021.0 | Repeat | Note=WD 7 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000392550 | + | 17 | 26 | 36_69 | 725 | 1021.0 | Repeat | Note=WD 1 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000392550 | + | 17 | 26 | 388_464 | 725 | 1021.0 | Repeat | Note=WD 8 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000392550 | + | 17 | 26 | 508_583 | 725 | 1021.0 | Repeat | Note=WD 9 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000392550 | + | 17 | 26 | 592_653 | 725 | 1021.0 | Repeat | Note=WD 10 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000392550 | + | 17 | 26 | 76_117 | 725 | 1021.0 | Repeat | Note=WD 2 |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000335464 | 8 | 10 | 255_276 | 243 | 497.0 | Topological domain | Extracellular | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000335464 | 8 | 10 | 298_306 | 243 | 497.0 | Topological domain | Cytoplasmic | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000335464 | 8 | 10 | 328_354 | 243 | 497.0 | Topological domain | Extracellular | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000335464 | 8 | 10 | 376_397 | 243 | 497.0 | Topological domain | Cytoplasmic | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000335464 | 8 | 10 | 419_421 | 243 | 497.0 | Topological domain | Extracellular | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000335464 | 8 | 10 | 443_470 | 243 | 497.0 | Topological domain | Cytoplasmic | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000335464 | 8 | 10 | 492_496 | 243 | 497.0 | Topological domain | Extracellular | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582330 | 8 | 10 | 255_276 | 243 | 497.0 | Topological domain | Extracellular | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582330 | 8 | 10 | 298_306 | 243 | 497.0 | Topological domain | Cytoplasmic | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582330 | 8 | 10 | 328_354 | 243 | 497.0 | Topological domain | Extracellular | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582330 | 8 | 10 | 376_397 | 243 | 497.0 | Topological domain | Cytoplasmic | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582330 | 8 | 10 | 419_421 | 243 | 497.0 | Topological domain | Extracellular | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582330 | 8 | 10 | 443_470 | 243 | 497.0 | Topological domain | Cytoplasmic | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582330 | 8 | 10 | 492_496 | 243 | 497.0 | Topological domain | Extracellular | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582773 | 0 | 10 | 168_204 | 0 | 315.0 | Topological domain | Extracellular | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582773 | 0 | 10 | 1_10 | 0 | 315.0 | Topological domain | Cytoplasmic | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582773 | 0 | 10 | 226_233 | 0 | 315.0 | Topological domain | Cytoplasmic | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582773 | 0 | 10 | 255_276 | 0 | 315.0 | Topological domain | Extracellular | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582773 | 0 | 10 | 298_306 | 0 | 315.0 | Topological domain | Cytoplasmic | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582773 | 0 | 10 | 328_354 | 0 | 315.0 | Topological domain | Extracellular | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582773 | 0 | 10 | 32_36 | 0 | 315.0 | Topological domain | Extracellular | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582773 | 0 | 10 | 376_397 | 0 | 315.0 | Topological domain | Cytoplasmic | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582773 | 0 | 10 | 419_421 | 0 | 315.0 | Topological domain | Extracellular | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582773 | 0 | 10 | 443_470 | 0 | 315.0 | Topological domain | Cytoplasmic | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582773 | 0 | 10 | 492_496 | 0 | 315.0 | Topological domain | Extracellular | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582773 | 0 | 10 | 58_146 | 0 | 315.0 | Topological domain | Cytoplasmic | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000335464 | 8 | 10 | 277_297 | 243 | 497.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000335464 | 8 | 10 | 307_327 | 243 | 497.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000335464 | 8 | 10 | 355_375 | 243 | 497.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000335464 | 8 | 10 | 398_418 | 243 | 497.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000335464 | 8 | 10 | 422_442 | 243 | 497.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000335464 | 8 | 10 | 471_491 | 243 | 497.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582330 | 8 | 10 | 277_297 | 243 | 497.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582330 | 8 | 10 | 307_327 | 243 | 497.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582330 | 8 | 10 | 355_375 | 243 | 497.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582330 | 8 | 10 | 398_418 | 243 | 497.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582330 | 8 | 10 | 422_442 | 243 | 497.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582330 | 8 | 10 | 471_491 | 243 | 497.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582773 | 0 | 10 | 11_31 | 0 | 315.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582773 | 0 | 10 | 147_167 | 0 | 315.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582773 | 0 | 10 | 205_225 | 0 | 315.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582773 | 0 | 10 | 234_254 | 0 | 315.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582773 | 0 | 10 | 277_297 | 0 | 315.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582773 | 0 | 10 | 307_327 | 0 | 315.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582773 | 0 | 10 | 355_375 | 0 | 315.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582773 | 0 | 10 | 37_57 | 0 | 315.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582773 | 0 | 10 | 398_418 | 0 | 315.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582773 | 0 | 10 | 422_442 | 0 | 315.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582773 | 0 | 10 | 471_491 | 0 | 315.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000167462 | + | 17 | 25 | 713_769 | 725 | 1016.0 | Repeat | Note=WD 11 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000167462 | + | 17 | 25 | 778_830 | 725 | 1016.0 | Repeat | Note=WD 12 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000167462 | + | 17 | 25 | 835_888 | 725 | 1016.0 | Repeat | Note=WD 13 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000167462 | + | 17 | 25 | 902_925 | 725 | 1016.0 | Repeat | Note=WD 14 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000375227 | + | 1 | 10 | 132_169 | 0 | 357.0 | Repeat | Note=WD 3 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000375227 | + | 1 | 10 | 193_227 | 0 | 357.0 | Repeat | Note=WD 4 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000375227 | + | 1 | 10 | 233_268 | 0 | 357.0 | Repeat | Note=WD 5 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000375227 | + | 1 | 10 | 282_324 | 0 | 357.0 | Repeat | Note=WD 6 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000375227 | + | 1 | 10 | 332_366 | 0 | 357.0 | Repeat | Note=WD 7 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000375227 | + | 1 | 10 | 36_69 | 0 | 357.0 | Repeat | Note=WD 1 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000375227 | + | 1 | 10 | 388_464 | 0 | 357.0 | Repeat | Note=WD 8 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000375227 | + | 1 | 10 | 508_583 | 0 | 357.0 | Repeat | Note=WD 9 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000375227 | + | 1 | 10 | 592_653 | 0 | 357.0 | Repeat | Note=WD 10 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000375227 | + | 1 | 10 | 713_769 | 0 | 357.0 | Repeat | Note=WD 11 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000375227 | + | 1 | 10 | 76_117 | 0 | 357.0 | Repeat | Note=WD 2 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000375227 | + | 1 | 10 | 778_830 | 0 | 357.0 | Repeat | Note=WD 12 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000375227 | + | 1 | 10 | 835_888 | 0 | 357.0 | Repeat | Note=WD 13 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000375227 | + | 1 | 10 | 902_925 | 0 | 357.0 | Repeat | Note=WD 14 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000392550 | + | 17 | 26 | 713_769 | 725 | 1021.0 | Repeat | Note=WD 11 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000392550 | + | 17 | 26 | 778_830 | 725 | 1021.0 | Repeat | Note=WD 12 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000392550 | + | 17 | 26 | 835_888 | 725 | 1021.0 | Repeat | Note=WD 13 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000392550 | + | 17 | 26 | 902_925 | 725 | 1021.0 | Repeat | Note=WD 14 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000578363 | + | 1 | 11 | 132_169 | 0 | 357.0 | Repeat | Note=WD 3 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000578363 | + | 1 | 11 | 193_227 | 0 | 357.0 | Repeat | Note=WD 4 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000578363 | + | 1 | 11 | 233_268 | 0 | 357.0 | Repeat | Note=WD 5 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000578363 | + | 1 | 11 | 282_324 | 0 | 357.0 | Repeat | Note=WD 6 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000578363 | + | 1 | 11 | 332_366 | 0 | 357.0 | Repeat | Note=WD 7 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000578363 | + | 1 | 11 | 36_69 | 0 | 357.0 | Repeat | Note=WD 1 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000578363 | + | 1 | 11 | 388_464 | 0 | 357.0 | Repeat | Note=WD 8 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000578363 | + | 1 | 11 | 508_583 | 0 | 357.0 | Repeat | Note=WD 9 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000578363 | + | 1 | 11 | 592_653 | 0 | 357.0 | Repeat | Note=WD 10 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000578363 | + | 1 | 11 | 713_769 | 0 | 357.0 | Repeat | Note=WD 11 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000578363 | + | 1 | 11 | 76_117 | 0 | 357.0 | Repeat | Note=WD 2 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000578363 | + | 1 | 11 | 778_830 | 0 | 357.0 | Repeat | Note=WD 12 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000578363 | + | 1 | 11 | 835_888 | 0 | 357.0 | Repeat | Note=WD 13 |
| Hgene | LLGL2 | chr17:73567181 | chr17:72832066 | ENST00000578363 | + | 1 | 11 | 902_925 | 0 | 357.0 | Repeat | Note=WD 14 |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000335464 | 8 | 10 | 168_204 | 243 | 497.0 | Topological domain | Extracellular | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000335464 | 8 | 10 | 1_10 | 243 | 497.0 | Topological domain | Cytoplasmic | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000335464 | 8 | 10 | 226_233 | 243 | 497.0 | Topological domain | Cytoplasmic | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000335464 | 8 | 10 | 32_36 | 243 | 497.0 | Topological domain | Extracellular | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000335464 | 8 | 10 | 58_146 | 243 | 497.0 | Topological domain | Cytoplasmic | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582330 | 8 | 10 | 168_204 | 243 | 497.0 | Topological domain | Extracellular | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582330 | 8 | 10 | 1_10 | 243 | 497.0 | Topological domain | Cytoplasmic | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582330 | 8 | 10 | 226_233 | 243 | 497.0 | Topological domain | Cytoplasmic | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582330 | 8 | 10 | 32_36 | 243 | 497.0 | Topological domain | Extracellular | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582330 | 8 | 10 | 58_146 | 243 | 497.0 | Topological domain | Cytoplasmic | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000335464 | 8 | 10 | 11_31 | 243 | 497.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000335464 | 8 | 10 | 147_167 | 243 | 497.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000335464 | 8 | 10 | 205_225 | 243 | 497.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000335464 | 8 | 10 | 234_254 | 243 | 497.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000335464 | 8 | 10 | 37_57 | 243 | 497.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582330 | 8 | 10 | 11_31 | 243 | 497.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582330 | 8 | 10 | 147_167 | 243 | 497.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582330 | 8 | 10 | 205_225 | 243 | 497.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582330 | 8 | 10 | 234_254 | 243 | 497.0 | Transmembrane | Helical | |
| Tgene | TMEM104 | chr17:73567181 | chr17:72832066 | ENST00000582330 | 8 | 10 | 37_57 | 243 | 497.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for LLGL2-TMEM104 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >45705_45705_1_LLGL2-TMEM104_LLGL2_chr17_73567181_ENST00000167462_TMEM104_chr17_72832066_ENST00000335464_length(transcript)=6159nt_BP=2306nt CGCCCAGCAGCCCGTGGGCAGGCGCGGCGGAGCGAGCGGGGCCGGCGGCGGGCGCCGAGGGACGCCGAGGCCTCGGGCGGGGGCTGGCCC GGGGTTCCAGGTCTCCAGTGGGGGCTGCAGACTAAGCAAAATGAGGCGGTTCCTGAGGCCAGGGCATGACCCTGTGCGGGAGAGGCTCAA GCGGGACCTGTTCCAGTTTAACAAGACGGTGGAGCATGGCTTCCCGCACCAGCCCAGCGCCCTCGGCTACAGCCCGTCCCTGCGCATCCT GGCCATCGGCACCCGTTCTGGAGCCATCAAGCTCTACGGAGCCCCAGGCGTGGAGTTCATGGGGCTGCACCAGGAGAACAACGCTGTGAC GCAGATCCACCTCCTGCCCGGCCAGTGCCAGCTGGTCACCCTGCTGGATGACAACAGCCTGCACCTTTGGAGCCTGAAGGTCAAGGGCGG GGCATCGGAGCTGCAGGAGGATGAGAGCTTCACACTGCGTGGACCCCCAGGGGCTGCCCCCAGTGCCACACAGATCACCGTGGTCCTGCC ACATTCCTCCTGCGAGCTGCTCTACCTGGGCACCGAGAGTGGCAACGTGTTTGTGGTGCAGCTGCCAGCTTTTCGTGCGCTGGAGGACCG GACCATCAGCTCGGACGCGGTGCTGCAGCGGTTGCCAGAGGAGGCCCGCCACCGGCGTGTGTTCGAGATGGTGGAGGCACTGCAGGAGCA CCCTCGAGACCCCAACCAGATCCTGATCGGCTACAGCCGAGGCCTCGTTGTCATCTGGGACCTACAGGGCAGCCGCGTGCTCTACCACTT CCTCAGCAGCCAGCAACTGGAGAACATCTGGTGGCAGCGGGACGGCCGCCTGCTCGTCAGCTGTCACTCTGACGGCAGCTACTGCCAGTG GCCCGTGTCCAGCGAAGCCCAGCAACCAGAGCCCCTCCGCAGCCTCGTGCCTTACGGTCCCTTTCCTTGCAAAGCGATTACCAGAATCCT CTGGCTGACCACTAGGCAGGGGTTGCCCTTCACCATCTTCCAGGGTGGCATGCCACGGGCCAGCTACGGGGACCGCCACTGCATCTCAGT GATCCACGATGGCCAGCAGACGGCCTTCGACTTCACCTCCCGTGTCATCGGCTTCACTGTCCTCACAGAGGCAGACCCTGCAGCCACCTT TGACGACCCCTATGCCCTGGTGGTGCTGGCTGAGGAGGAGCTGGTGGTGATTGACCTGCAGACAGCAGGCTGGCCACCGGTCCAGCTGCC CTACCTGGCTTCTCTGCACTGTTCCGCCATCACCTGCTCTCACCACGTCTCCAACATCCCGCTGAAGCTGTGGGAGCGGATCATTGCCGC CGGCAGCCGGCAGAACGCACACTTCTCCACCATGGAGTGGCCAATTGATGGTGGCACCAGCCTGACCCCAGCCCCACCCCAGAGGGACCT GCTGCTCACAGGGCACGAGGACGGCACGGTGCGGTTCTGGGATGCCTCGGGTGTCTGCCTGCGGCTGCTCTACAAACTCAGCACTGTGCG CGTGTTCCTCACCGACACGGACCCCAACGAGAACTTCAGTGCCCAGGGCGAGGACGAGTGGCCCCCACTCCGCAAGGTGGGCTCCTTTGA CCCCTACAGTGATGACCCCCGGCTGGGCATCCAGAAGATCTTCCTCTGCAAGTACAGCGGCTACCTGGCTGTGGCAGGCACGGCAGGGCA GGTGCTGGTACTGGAACTGAATGACGAGGCAGCGGAGCAGGCTGTGGAGCAGGTGGAGGCCGACCTGCTGCAGGACCAAGAGGGCTACCG CTGGAAGGGGCACGAGCGCCTGGCAGCCCGCTCAGGGCCCGTGCGCTTTGAGCCTGGCTTTCAGCCCTTCGTGTTGGTGCAGTGTCAGCC CCCGGCTGTGGTCACCTCCTTGGCCCTGCACTCTGAGTGGCGGCTCGTGGCCTTCGGCACCAGCCATGGCTTTGGCCTCTTTGACCACCA GCAGCGGCGGCAGGTCTTTGTTAAGTGCACACTGCACCCCAGTGACCAGCTGGCCTTGGAGGGCCCACTCTCCCGCGTCAAGTCCCTCAA GAAGTCCTTGCGTCAGTCATTCCGCCGGATGCGTCGGAGCCGGGTGTCCAGCCGGAAGCGGCACCCGGCTGGCCCCCCAGGAGAGGCACA GGAGGGGAGTGCCAAGGCTGAGCGGCCAGGCCTCCAGAACATGGAGCTGGCGCCTGTGCAGCGCAAGATCGAGGCTCGCTCGGCAGAGGA CTCCTTCACAGGCTTCGTCCGGACCCTGTACTTTGCTGACACCTACCTGAAGGACACTTTCGCCGTCATGATTGTGCTGGCCCTGATCCG CATCGGGCACGGACAAGGGGAGGGGCACCCGCCCCTGGCTGACTTCTCGGGGGTCCGGAACCTGTTTGGGGTGTGCGTCTACTCCTTCAT GTGCCAGCACTCTCTGCCATCCCTCATTACCCCCGTCTCCTCCAAGCGCCACCTCACAAGGCTGGTGTTCCTGGACTACGTGCTGATCCT GGCCTTCTACGGCCTCCTCTCCTTCACCGCCATCTTCTGCTTCCGCGGCGACAGCCTCATGGACATGTACACCCTCAACTTCGCGCGCTG TGACGTCGTGGGCCTGGCCGCTGTGCGCTTTTTCCTGGGCCTCTTCCCCGTCTTCACCATCAGCACCAACTTCCCCATCATTGCCGTGAC CCTGCGCAACAACTGGAAGACACTCTTCCACCGCGAGGGCGGCACGTACCCGTGGGTGGTGGACCGCGTCGTGTTTCCCACCATCACCCT GGTGCCGCCTGTGCTGGTGGCCTTCTGCACCCACGACCTGGAGTCCCTGGTGGGCATCACAGGGGCCTACGCGGGCACCGGCATCCAGTA CGTCATCCCCGCCTTCCTGGTGTACCACTGCCGCAGGGACACCCAGCTGGCCTTTGGCTGTGGGGTCAGCAACAAGCACAGGTCCCCTTT CCGCCACACCTTCTGGGTGGGCTTCGTGCTGCTCTGGGCTTTCTCCTGCTTCATTTTCGTCACGGCCAATATCATCCTCAGCGAGACCAA GCTCTGATGGCAGGACAGGCAGGTCTAGTGACAGGAGGCATAGGGGCCCCAGCCCCACCCCTCGCCCGCAGAGCCCAGATTTAGTTGCTC CCAGGTCTTCATGGGGCAGGTGGGACTGTGGCTCTAGGGGAAACCCCCTGTCCAGCTGGGGTTGTTCTCCCCACCCACCTTCCACCCAGT CTGCACCTGTCCTCACTCCCCCGGGCAGCTCTCCCACCAGGTCCTTCTGCCTGGGTGACGCGTGGCTGCTCCCAAGTTCTAAGACTCATT ACTGGAATCACACCTCTCCCACATCTTCCAACACGCTTCACCCCCAACCCTACTTCCAGGGCGGGCTACTCCCTGCACAATGGGAGATGC AGTGCTGGCACTGCCATCATTTGTCCCAGACCCATGCCCCTCCCCTCTCCTGCCCCATCCTGCGTCCTTGTCTAAGAAACCTCCACTAGC AGCTGCTGCCTCTTCAGAGGCGAATCCCAGGCTGGAAGTATCCTAGCCAGGTCTGCCTGTGACGGCAGGGCCCACCCTCCCACCAGGCCT GGGAGGCGGTAGGTGCCACCTCAGAGAGGGCCTGGCTGGCCCCATGACAAGAACAAGAAGTGCTAACTGGTACCAGAGTCCCCAGATAGA GCTGCGAGGGCCCTCGCTGTGGGCGCGAGCTCCGGAGTAGCAGGCTGGATGGAGCATCCCTTCTACAGGGACACAGCCGCCTGCCAGCTG GGCCTAAAGTCTGGAGCTTGTCTCTGAAAGGGACCGTCTGCTGCCCTCGCTGCCCCACTGGAGCCCTCAGGGCCTCCTACAGTGCAGGTG GTCCTACTAGAGGGAAGCCATCCCCACCTGGCACCTCAGCGGCTTGGCAGTGACATGGCAGTGGGAGGGCTCTGAGGTTCCCACCCTGAC AATGGTCAGGCTGCCCGTGCTCCCACTCCAACACTAAAAGCGGCTCCCTTAGGGGCTGAGCGTGGGTGCTCAGTGTTCACGCTATATCCC TTGGGCAATGTGGGGTTGGATGGGGCCCCCACTTCCATTCCCATGGAAGGAGGCCAGGTCCCCAGCCACCTCCCACTCAGCCATGCACGC ACTTGCTGGGCTGGCCTCCTGGGAAACACAGGTGACTCGAATGAACTCTGCATTTTCAACGTGCCTTCTACTGCTTCAGGACCTGGGGGT CCCCCTGACCCTCACTGGCTTGCCCCCAGCCCTGGGCCTGGCCCCACCTGTCCTGGAGCCCAGAGCCCCTGGCCTGGAGCTACCTCTCTG GGGTGGGTCTCAGGCCCCACCCCTCCCTCTTTTGAGTTCAGTGCCTTGCTCAGCCCCTCCCCTGTATCTCAGCGTCTTCAGACCTCTGAC AGAGCGACGATGTAGGGTCTCCCGGGGCCCAAGGTGGTCTCAGGGTCAGGGGTGGGATTTGCAGGGAACTCGGGGAGCCCACGGGCTGCG CCACCTCTGCCCTGGCAGCTGAAGCCTGGGAGAGTCCCTGCGTGGTGTAATTGGCCTCAGCCCGCTTTCTCTGTGCCGTCGCACCTCAGT GTTTCTCATAGCTTGTCCCCACGTGTCACTTTCCATCCACGGGAAAAACAAATGCCCCTTCTCCATCTATCATTGCGACTTCCTCCCAGG AGGCCTCTCAGGTTGGGTAGAGCAGGGGCCTGCAGTGGTCAGGCCAAGAGCAAGGAAGACCTGGCTGCCCCACTGTGGCTGAAAACTCAA AACATCTGGAACTACCCTTCTCCAATTACGTTCCCTCTTGCTTAAAGACAAAGCTAATTAAATCATCCTGCCACCCGAGGCTCCAAACCA AATCTTGGACGCAAATTCATTAGCTTCATAAAGCCCAGGTTGATTTATGTGACAGTCTGGAACCGCCCAGCCATGGAGTCGGTGGCCGTG TTCCCTGCCCTGGAGGGAACTGGCCAGCAGTGACCACATCGCTGATGCCAAGTGGGGACCCTCACCACTGCCGTTGTCTAGCTGCTTTGC TCCTCAGTGTCCTCCTGCTGTAAGTCAGATGAGGTCACCTTCCTCTCCATCGATGCCAGGCTTCTGCCGCTGACGGCCAATGGGGCTTGG GGCCAGGGGCAGAGAACCCCAACAGCAAGAGAGCCCCAGGTGATGCTGGCTCTCCACGTGCATCCGGCACGTTTCACTGCAGCCATCATC AACTTGGCAAAGTAGGCAAGCTCAGCTTGTGGTTTCCATTTTGCAGATGGGCCCCCGGGGTCAGTGATGGGAAGAGCAGAAGAGGATGCC AGCCCAGCCTGGCCCAGGAGCAGGTGTCGTTGTGGAGGAAAGCTTAAACTCAACAGCCTCTACCCAACATCTGCCACCTGCATGGCCCCA GATGCCCCCGGTGCAGGCCAGGTCAAGGTGGGAGTAAACTTTCTTTGCCTCCCTCCCCACAGAAGCACCAGACCCACCTGAGCCCCAGAG CCTCATGCCAGCAGCTCCTGGCTGTTCCTCACCTGAGGCTAGAGCAGCAGCTGCCAGCTTATAGATGGGGCGGATGGCAGGTGATAGACT GGGAAGCACTGCTGGGTGGTGGAGGTGGGCAGTGGCACCATGACGGGCCCCTAGCCACACAGCTGTCCTCACTACGGGGCAGGGAGCAGC CTCGGCAACAGGCAAAGGCCAGAGTCAGAGTGGTGGGGAGGACAGGGGCTGCTGCCCCGCTCCTGGAAAGCCACTGCAGAGGGGCAGTGG CTGGCAGTGCCAGGCCTGGGGGAAGTGGAAGCGTCCTGTCTGGGGGCAGATTCCCAAGCAAGGGTGACCCATGGTTAAGGGCCACTGGAA AGCTGGAGAGAGCTTGGGATCCCTTCCACCTGGGGCCAATGGTGTTGATTGCAGACTGGAGGGGTAACCTCCGCTGAGGGTCATTATGTG CCAGGCATGGCATTGGGTACTTTCTGCATTGGGACCAGGCAGCCGGGCCTGCCATTTGAGGCAGCGAGCCTCCGTGACCTGTCCTCCCTC ATTTGTAAAGTGGGGTAACAGCTATGTCACTGGCCAGTTGTGGGGATTAAAGTGCTGAGCTTAGTGCCCAGCCCAAAATGCTCAATAAAG >45705_45705_1_LLGL2-TMEM104_LLGL2_chr17_73567181_ENST00000167462_TMEM104_chr17_72832066_ENST00000335464_length(amino acids)=978AA_BP=726 MRRFLRPGHDPVRERLKRDLFQFNKTVEHGFPHQPSALGYSPSLRILAIGTRSGAIKLYGAPGVEFMGLHQENNAVTQIHLLPGQCQLVT LLDDNSLHLWSLKVKGGASELQEDESFTLRGPPGAAPSATQITVVLPHSSCELLYLGTESGNVFVVQLPAFRALEDRTISSDAVLQRLPE EARHRRVFEMVEALQEHPRDPNQILIGYSRGLVVIWDLQGSRVLYHFLSSQQLENIWWQRDGRLLVSCHSDGSYCQWPVSSEAQQPEPLR SLVPYGPFPCKAITRILWLTTRQGLPFTIFQGGMPRASYGDRHCISVIHDGQQTAFDFTSRVIGFTVLTEADPAATFDDPYALVVLAEEE LVVIDLQTAGWPPVQLPYLASLHCSAITCSHHVSNIPLKLWERIIAAGSRQNAHFSTMEWPIDGGTSLTPAPPQRDLLLTGHEDGTVRFW DASGVCLRLLYKLSTVRVFLTDTDPNENFSAQGEDEWPPLRKVGSFDPYSDDPRLGIQKIFLCKYSGYLAVAGTAGQVLVLELNDEAAEQ AVEQVEADLLQDQEGYRWKGHERLAARSGPVRFEPGFQPFVLVQCQPPAVVTSLALHSEWRLVAFGTSHGFGLFDHQQRRQVFVKCTLHP SDQLALEGPLSRVKSLKKSLRQSFRRMRRSRVSSRKRHPAGPPGEAQEGSAKAERPGLQNMELAPVQRKIEARSAEDSFTGFVRTLYFAD TYLKDTFAVMIVLALIRIGHGQGEGHPPLADFSGVRNLFGVCVYSFMCQHSLPSLITPVSSKRHLTRLVFLDYVLILAFYGLLSFTAIFC FRGDSLMDMYTLNFARCDVVGLAAVRFFLGLFPVFTISTNFPIIAVTLRNNWKTLFHREGGTYPWVVDRVVFPTITLVPPVLVAFCTHDL -------------------------------------------------------------- >45705_45705_2_LLGL2-TMEM104_LLGL2_chr17_73567181_ENST00000392550_TMEM104_chr17_72832066_ENST00000335464_length(transcript)=6146nt_BP=2293nt GTGGGCAGGCGCGGCGGAGCGAGCGGGGCCGGCGGCGGGCGCCGAGGGACGCCGAGGCCTCGGGCGGGGGCTGGCCCGGGGTTCCAGGTC TCCAGTGGGGGCTGCAGACTAAGCAAAATGAGGCGGTTCCTGAGGCCAGGGCATGACCCTGTGCGGGAGAGGCTCAAGCGGGACCTGTTC CAGTTTAACAAGACGGTGGAGCATGGCTTCCCGCACCAGCCCAGCGCCCTCGGCTACAGCCCGTCCCTGCGCATCCTGGCCATCGGCACC CGTTCTGGAGCCATCAAGCTCTACGGAGCCCCAGGCGTGGAGTTCATGGGGCTGCACCAGGAGAACAACGCTGTGACGCAGATCCACCTC CTGCCCGGCCAGTGCCAGCTGGTCACCCTGCTGGATGACAACAGCCTGCACCTTTGGAGCCTGAAGGTCAAGGGCGGGGCATCGGAGCTG CAGGAGGATGAGAGCTTCACACTGCGTGGACCCCCAGGGGCTGCCCCCAGTGCCACACAGATCACCGTGGTCCTGCCACATTCCTCCTGC GAGCTGCTCTACCTGGGCACCGAGAGTGGCAACGTGTTTGTGGTGCAGCTGCCAGCTTTTCGTGCGCTGGAGGACCGGACCATCAGCTCG GACGCGGTGCTGCAGCGGTTGCCAGAGGAGGCCCGCCACCGGCGTGTGTTCGAGATGGTGGAGGCACTGCAGGAGCACCCTCGAGACCCC AACCAGATCCTGATCGGCTACAGCCGAGGCCTCGTTGTCATCTGGGACCTACAGGGCAGCCGCGTGCTCTACCACTTCCTCAGCAGCCAG CAACTGGAGAACATCTGGTGGCAGCGGGACGGCCGCCTGCTCGTCAGCTGTCACTCTGACGGCAGCTACTGCCAGTGGCCCGTGTCCAGC GAAGCCCAGCAACCAGAGCCCCTCCGCAGCCTCGTGCCTTACGGTCCCTTTCCTTGCAAAGCGATTACCAGAATCCTCTGGCTGACCACT AGGCAGGGGTTGCCCTTCACCATCTTCCAGGGTGGCATGCCACGGGCCAGCTACGGGGACCGCCACTGCATCTCAGTGATCCACGATGGC CAGCAGACGGCCTTCGACTTCACCTCCCGTGTCATCGGCTTCACTGTCCTCACAGAGGCAGACCCTGCAGCCACCTTTGACGACCCCTAT GCCCTGGTGGTGCTGGCTGAGGAGGAGCTGGTGGTGATTGACCTGCAGACAGCAGGCTGGCCACCGGTCCAGCTGCCCTACCTGGCTTCT CTGCACTGTTCCGCCATCACCTGCTCTCACCACGTCTCCAACATCCCGCTGAAGCTGTGGGAGCGGATCATTGCCGCCGGCAGCCGGCAG AACGCACACTTCTCCACCATGGAGTGGCCAATTGATGGTGGCACCAGCCTGACCCCAGCCCCACCCCAGAGGGACCTGCTGCTCACAGGG CACGAGGACGGCACGGTGCGGTTCTGGGATGCCTCGGGTGTCTGCCTGCGGCTGCTCTACAAACTCAGCACTGTGCGCGTGTTCCTCACC GACACGGACCCCAACGAGAACTTCAGTGCCCAGGGCGAGGACGAGTGGCCCCCACTCCGCAAGGTGGGCTCCTTTGACCCCTACAGTGAT GACCCCCGGCTGGGCATCCAGAAGATCTTCCTCTGCAAGTACAGCGGCTACCTGGCTGTGGCAGGCACGGCAGGGCAGGTGCTGGTACTG GAACTGAATGACGAGGCAGCGGAGCAGGCTGTGGAGCAGGTGGAGGCCGACCTGCTGCAGGACCAAGAGGGCTACCGCTGGAAGGGGCAC GAGCGCCTGGCAGCCCGCTCAGGGCCCGTGCGCTTTGAGCCTGGCTTTCAGCCCTTCGTGTTGGTGCAGTGTCAGCCCCCGGCTGTGGTC ACCTCCTTGGCCCTGCACTCTGAGTGGCGGCTCGTGGCCTTCGGCACCAGCCATGGCTTTGGCCTCTTTGACCACCAGCAGCGGCGGCAG GTCTTTGTTAAGTGCACACTGCACCCCAGTGACCAGCTGGCCTTGGAGGGCCCACTCTCCCGCGTCAAGTCCCTCAAGAAGTCCTTGCGT CAGTCATTCCGCCGGATGCGTCGGAGCCGGGTGTCCAGCCGGAAGCGGCACCCGGCTGGCCCCCCAGGAGAGGCACAGGAGGGGAGTGCC AAGGCTGAGCGGCCAGGCCTCCAGAACATGGAGCTGGCGCCTGTGCAGCGCAAGATCGAGGCTCGCTCGGCAGAGGACTCCTTCACAGGC TTCGTCCGGACCCTGTACTTTGCTGACACCTACCTGAAGGACACTTTCGCCGTCATGATTGTGCTGGCCCTGATCCGCATCGGGCACGGA CAAGGGGAGGGGCACCCGCCCCTGGCTGACTTCTCGGGGGTCCGGAACCTGTTTGGGGTGTGCGTCTACTCCTTCATGTGCCAGCACTCT CTGCCATCCCTCATTACCCCCGTCTCCTCCAAGCGCCACCTCACAAGGCTGGTGTTCCTGGACTACGTGCTGATCCTGGCCTTCTACGGC CTCCTCTCCTTCACCGCCATCTTCTGCTTCCGCGGCGACAGCCTCATGGACATGTACACCCTCAACTTCGCGCGCTGTGACGTCGTGGGC CTGGCCGCTGTGCGCTTTTTCCTGGGCCTCTTCCCCGTCTTCACCATCAGCACCAACTTCCCCATCATTGCCGTGACCCTGCGCAACAAC TGGAAGACACTCTTCCACCGCGAGGGCGGCACGTACCCGTGGGTGGTGGACCGCGTCGTGTTTCCCACCATCACCCTGGTGCCGCCTGTG CTGGTGGCCTTCTGCACCCACGACCTGGAGTCCCTGGTGGGCATCACAGGGGCCTACGCGGGCACCGGCATCCAGTACGTCATCCCCGCC TTCCTGGTGTACCACTGCCGCAGGGACACCCAGCTGGCCTTTGGCTGTGGGGTCAGCAACAAGCACAGGTCCCCTTTCCGCCACACCTTC TGGGTGGGCTTCGTGCTGCTCTGGGCTTTCTCCTGCTTCATTTTCGTCACGGCCAATATCATCCTCAGCGAGACCAAGCTCTGATGGCAG GACAGGCAGGTCTAGTGACAGGAGGCATAGGGGCCCCAGCCCCACCCCTCGCCCGCAGAGCCCAGATTTAGTTGCTCCCAGGTCTTCATG GGGCAGGTGGGACTGTGGCTCTAGGGGAAACCCCCTGTCCAGCTGGGGTTGTTCTCCCCACCCACCTTCCACCCAGTCTGCACCTGTCCT CACTCCCCCGGGCAGCTCTCCCACCAGGTCCTTCTGCCTGGGTGACGCGTGGCTGCTCCCAAGTTCTAAGACTCATTACTGGAATCACAC CTCTCCCACATCTTCCAACACGCTTCACCCCCAACCCTACTTCCAGGGCGGGCTACTCCCTGCACAATGGGAGATGCAGTGCTGGCACTG CCATCATTTGTCCCAGACCCATGCCCCTCCCCTCTCCTGCCCCATCCTGCGTCCTTGTCTAAGAAACCTCCACTAGCAGCTGCTGCCTCT TCAGAGGCGAATCCCAGGCTGGAAGTATCCTAGCCAGGTCTGCCTGTGACGGCAGGGCCCACCCTCCCACCAGGCCTGGGAGGCGGTAGG TGCCACCTCAGAGAGGGCCTGGCTGGCCCCATGACAAGAACAAGAAGTGCTAACTGGTACCAGAGTCCCCAGATAGAGCTGCGAGGGCCC TCGCTGTGGGCGCGAGCTCCGGAGTAGCAGGCTGGATGGAGCATCCCTTCTACAGGGACACAGCCGCCTGCCAGCTGGGCCTAAAGTCTG GAGCTTGTCTCTGAAAGGGACCGTCTGCTGCCCTCGCTGCCCCACTGGAGCCCTCAGGGCCTCCTACAGTGCAGGTGGTCCTACTAGAGG GAAGCCATCCCCACCTGGCACCTCAGCGGCTTGGCAGTGACATGGCAGTGGGAGGGCTCTGAGGTTCCCACCCTGACAATGGTCAGGCTG CCCGTGCTCCCACTCCAACACTAAAAGCGGCTCCCTTAGGGGCTGAGCGTGGGTGCTCAGTGTTCACGCTATATCCCTTGGGCAATGTGG GGTTGGATGGGGCCCCCACTTCCATTCCCATGGAAGGAGGCCAGGTCCCCAGCCACCTCCCACTCAGCCATGCACGCACTTGCTGGGCTG GCCTCCTGGGAAACACAGGTGACTCGAATGAACTCTGCATTTTCAACGTGCCTTCTACTGCTTCAGGACCTGGGGGTCCCCCTGACCCTC ACTGGCTTGCCCCCAGCCCTGGGCCTGGCCCCACCTGTCCTGGAGCCCAGAGCCCCTGGCCTGGAGCTACCTCTCTGGGGTGGGTCTCAG GCCCCACCCCTCCCTCTTTTGAGTTCAGTGCCTTGCTCAGCCCCTCCCCTGTATCTCAGCGTCTTCAGACCTCTGACAGAGCGACGATGT AGGGTCTCCCGGGGCCCAAGGTGGTCTCAGGGTCAGGGGTGGGATTTGCAGGGAACTCGGGGAGCCCACGGGCTGCGCCACCTCTGCCCT GGCAGCTGAAGCCTGGGAGAGTCCCTGCGTGGTGTAATTGGCCTCAGCCCGCTTTCTCTGTGCCGTCGCACCTCAGTGTTTCTCATAGCT TGTCCCCACGTGTCACTTTCCATCCACGGGAAAAACAAATGCCCCTTCTCCATCTATCATTGCGACTTCCTCCCAGGAGGCCTCTCAGGT TGGGTAGAGCAGGGGCCTGCAGTGGTCAGGCCAAGAGCAAGGAAGACCTGGCTGCCCCACTGTGGCTGAAAACTCAAAACATCTGGAACT ACCCTTCTCCAATTACGTTCCCTCTTGCTTAAAGACAAAGCTAATTAAATCATCCTGCCACCCGAGGCTCCAAACCAAATCTTGGACGCA AATTCATTAGCTTCATAAAGCCCAGGTTGATTTATGTGACAGTCTGGAACCGCCCAGCCATGGAGTCGGTGGCCGTGTTCCCTGCCCTGG AGGGAACTGGCCAGCAGTGACCACATCGCTGATGCCAAGTGGGGACCCTCACCACTGCCGTTGTCTAGCTGCTTTGCTCCTCAGTGTCCT CCTGCTGTAAGTCAGATGAGGTCACCTTCCTCTCCATCGATGCCAGGCTTCTGCCGCTGACGGCCAATGGGGCTTGGGGCCAGGGGCAGA GAACCCCAACAGCAAGAGAGCCCCAGGTGATGCTGGCTCTCCACGTGCATCCGGCACGTTTCACTGCAGCCATCATCAACTTGGCAAAGT AGGCAAGCTCAGCTTGTGGTTTCCATTTTGCAGATGGGCCCCCGGGGTCAGTGATGGGAAGAGCAGAAGAGGATGCCAGCCCAGCCTGGC CCAGGAGCAGGTGTCGTTGTGGAGGAAAGCTTAAACTCAACAGCCTCTACCCAACATCTGCCACCTGCATGGCCCCAGATGCCCCCGGTG CAGGCCAGGTCAAGGTGGGAGTAAACTTTCTTTGCCTCCCTCCCCACAGAAGCACCAGACCCACCTGAGCCCCAGAGCCTCATGCCAGCA GCTCCTGGCTGTTCCTCACCTGAGGCTAGAGCAGCAGCTGCCAGCTTATAGATGGGGCGGATGGCAGGTGATAGACTGGGAAGCACTGCT GGGTGGTGGAGGTGGGCAGTGGCACCATGACGGGCCCCTAGCCACACAGCTGTCCTCACTACGGGGCAGGGAGCAGCCTCGGCAACAGGC AAAGGCCAGAGTCAGAGTGGTGGGGAGGACAGGGGCTGCTGCCCCGCTCCTGGAAAGCCACTGCAGAGGGGCAGTGGCTGGCAGTGCCAG GCCTGGGGGAAGTGGAAGCGTCCTGTCTGGGGGCAGATTCCCAAGCAAGGGTGACCCATGGTTAAGGGCCACTGGAAAGCTGGAGAGAGC TTGGGATCCCTTCCACCTGGGGCCAATGGTGTTGATTGCAGACTGGAGGGGTAACCTCCGCTGAGGGTCATTATGTGCCAGGCATGGCAT TGGGTACTTTCTGCATTGGGACCAGGCAGCCGGGCCTGCCATTTGAGGCAGCGAGCCTCCGTGACCTGTCCTCCCTCATTTGTAAAGTGG GGTAACAGCTATGTCACTGGCCAGTTGTGGGGATTAAAGTGCTGAGCTTAGTGCCCAGCCCAAAATGCTCAATAAAGCTATTCAGTGATA >45705_45705_2_LLGL2-TMEM104_LLGL2_chr17_73567181_ENST00000392550_TMEM104_chr17_72832066_ENST00000335464_length(amino acids)=978AA_BP=726 MRRFLRPGHDPVRERLKRDLFQFNKTVEHGFPHQPSALGYSPSLRILAIGTRSGAIKLYGAPGVEFMGLHQENNAVTQIHLLPGQCQLVT LLDDNSLHLWSLKVKGGASELQEDESFTLRGPPGAAPSATQITVVLPHSSCELLYLGTESGNVFVVQLPAFRALEDRTISSDAVLQRLPE EARHRRVFEMVEALQEHPRDPNQILIGYSRGLVVIWDLQGSRVLYHFLSSQQLENIWWQRDGRLLVSCHSDGSYCQWPVSSEAQQPEPLR SLVPYGPFPCKAITRILWLTTRQGLPFTIFQGGMPRASYGDRHCISVIHDGQQTAFDFTSRVIGFTVLTEADPAATFDDPYALVVLAEEE LVVIDLQTAGWPPVQLPYLASLHCSAITCSHHVSNIPLKLWERIIAAGSRQNAHFSTMEWPIDGGTSLTPAPPQRDLLLTGHEDGTVRFW DASGVCLRLLYKLSTVRVFLTDTDPNENFSAQGEDEWPPLRKVGSFDPYSDDPRLGIQKIFLCKYSGYLAVAGTAGQVLVLELNDEAAEQ AVEQVEADLLQDQEGYRWKGHERLAARSGPVRFEPGFQPFVLVQCQPPAVVTSLALHSEWRLVAFGTSHGFGLFDHQQRRQVFVKCTLHP SDQLALEGPLSRVKSLKKSLRQSFRRMRRSRVSSRKRHPAGPPGEAQEGSAKAERPGLQNMELAPVQRKIEARSAEDSFTGFVRTLYFAD TYLKDTFAVMIVLALIRIGHGQGEGHPPLADFSGVRNLFGVCVYSFMCQHSLPSLITPVSSKRHLTRLVFLDYVLILAFYGLLSFTAIFC FRGDSLMDMYTLNFARCDVVGLAAVRFFLGLFPVFTISTNFPIIAVTLRNNWKTLFHREGGTYPWVVDRVVFPTITLVPPVLVAFCTHDL -------------------------------------------------------------- >45705_45705_3_LLGL2-TMEM104_LLGL2_chr17_73567181_ENST00000577200_TMEM104_chr17_72832066_ENST00000335464_length(transcript)=6133nt_BP=2280nt GGCTCACTGCAACCTCTGCCTCCCGGGTTCAAGCAATTCTCCTGCCTCGGCCTCCTGAGTAGCTGGGATTACAGGTCTCCAGTGGGGGCT GCAGACTAAGCAAAATGAGGCGGTTCCTGAGGCCAGGGCATGACCCTGTGCGGGAGAGGCTCAAGCGGGACCTGTTCCAGTTTAACAAGA CGGTGGAGCATGGCTTCCCGCACCAGCCCAGCGCCCTCGGCTACAGCCCGTCCCTGCGCATCCTGGCCATCGGCACCCGTTCTGGAGCCA TCAAGCTCTACGGAGCCCCAGGCGTGGAGTTCATGGGGCTGCACCAGGAGAACAACGCTGTGACGCAGATCCACCTCCTGCCCGGCCAGT GCCAGCTGGTCACCCTGCTGGATGACAACAGCCTGCACCTTTGGAGCCTGAAGGTCAAGGGCGGGGCATCGGAGCTGCAGGAGGATGAGA GCTTCACACTGCGTGGACCCCCAGGGGCTGCCCCCAGTGCCACACAGATCACCGTGGTCCTGCCACATTCCTCCTGCGAGCTGCTCTACC TGGGCACCGAGAGTGGCAACGTGTTTGTGGTGCAGCTGCCAGCTTTTCGTGCGCTGGAGGACCGGACCATCAGCTCGGACGCGGTGCTGC AGCGGTTGCCAGAGGAGGCCCGCCACCGGCGTGTGTTCGAGATGGTGGAGGCACTGCAGGAGCACCCTCGAGACCCCAACCAGATCCTGA TCGGCTACAGCCGAGGCCTCGTTGTCATCTGGGACCTACAGGGCAGCCGCGTGCTCTACCACTTCCTCAGCAGCCAGCAACTGGAGAACA TCTGGTGGCAGCGGGACGGCCGCCTGCTCGTCAGCTGTCACTCTGACGGCAGCTACTGCCAGTGGCCCGTGTCCAGCGAAGCCCAGCAAC CAGAGCCCCTCCGCAGCCTCGTGCCTTACGGTCCCTTTCCTTGCAAAGCGATTACCAGAATCCTCTGGCTGACCACTAGGCAGGGGTTGC CCTTCACCATCTTCCAGGGTGGCATGCCACGGGCCAGCTACGGGGACCGCCACTGCATCTCAGTGATCCACGATGGCCAGCAGACGGCCT TCGACTTCACCTCCCGTGTCATCGGCTTCACTGTCCTCACAGAGGCAGACCCTGCAGCCACCTTTGACGACCCCTATGCCCTGGTGGTGC TGGCTGAGGAGGAGCTGGTGGTGATTGACCTGCAGACAGCAGGCTGGCCACCGGTCCAGCTGCCCTACCTGGCTTCTCTGCACTGTTCCG CCATCACCTGCTCTCACCACGTCTCCAACATCCCGCTGAAGCTGTGGGAGCGGATCATTGCCGCCGGCAGCCGGCAGAACGCACACTTCT CCACCATGGAGTGGCCAATTGATGGTGGCACCAGCCTGACCCCAGCCCCACCCCAGAGGGACCTGCTGCTCACAGGGCACGAGGACGGCA CGGTGCGGTTCTGGGATGCCTCGGGTGTCTGCCTGCGGCTGCTCTACAAACTCAGCACTGTGCGCGTGTTCCTCACCGACACGGACCCCA ACGAGAACTTCAGTGCCCAGGGCGAGGACGAGTGGCCCCCACTCCGCAAGGTGGGCTCCTTTGACCCCTACAGTGATGACCCCCGGCTGG GCATCCAGAAGATCTTCCTCTGCAAGTACAGCGGCTACCTGGCTGTGGCAGGCACGGCAGGGCAGGTGCTGGTACTGGAACTGAATGACG AGGCAGCGGAGCAGGCTGTGGAGCAGGTGGAGGCCGACCTGCTGCAGGACCAAGAGGGCTACCGCTGGAAGGGGCACGAGCGCCTGGCAG CCCGCTCAGGGCCCGTGCGCTTTGAGCCTGGCTTTCAGCCCTTCGTGTTGGTGCAGTGTCAGCCCCCGGCTGTGGTCACCTCCTTGGCCC TGCACTCTGAGTGGCGGCTCGTGGCCTTCGGCACCAGCCATGGCTTTGGCCTCTTTGACCACCAGCAGCGGCGGCAGGTCTTTGTTAAGT GCACACTGCACCCCAGTGACCAGCTGGCCTTGGAGGGCCCACTCTCCCGCGTCAAGTCCCTCAAGAAGTCCTTGCGTCAGTCATTCCGCC GGATGCGTCGGAGCCGGGTGTCCAGCCGGAAGCGGCACCCGGCTGGCCCCCCAGGAGAGGCACAGGAGGGGAGTGCCAAGGCTGAGCGGC CAGGCCTCCAGAACATGGAGCTGGCGCCTGTGCAGCGCAAGATCGAGGCTCGCTCGGCAGAGGACTCCTTCACAGGCTTCGTCCGGACCC TGTACTTTGCTGACACCTACCTGAAGGACACTTTCGCCGTCATGATTGTGCTGGCCCTGATCCGCATCGGGCACGGACAAGGGGAGGGGC ACCCGCCCCTGGCTGACTTCTCGGGGGTCCGGAACCTGTTTGGGGTGTGCGTCTACTCCTTCATGTGCCAGCACTCTCTGCCATCCCTCA TTACCCCCGTCTCCTCCAAGCGCCACCTCACAAGGCTGGTGTTCCTGGACTACGTGCTGATCCTGGCCTTCTACGGCCTCCTCTCCTTCA CCGCCATCTTCTGCTTCCGCGGCGACAGCCTCATGGACATGTACACCCTCAACTTCGCGCGCTGTGACGTCGTGGGCCTGGCCGCTGTGC GCTTTTTCCTGGGCCTCTTCCCCGTCTTCACCATCAGCACCAACTTCCCCATCATTGCCGTGACCCTGCGCAACAACTGGAAGACACTCT TCCACCGCGAGGGCGGCACGTACCCGTGGGTGGTGGACCGCGTCGTGTTTCCCACCATCACCCTGGTGCCGCCTGTGCTGGTGGCCTTCT GCACCCACGACCTGGAGTCCCTGGTGGGCATCACAGGGGCCTACGCGGGCACCGGCATCCAGTACGTCATCCCCGCCTTCCTGGTGTACC ACTGCCGCAGGGACACCCAGCTGGCCTTTGGCTGTGGGGTCAGCAACAAGCACAGGTCCCCTTTCCGCCACACCTTCTGGGTGGGCTTCG TGCTGCTCTGGGCTTTCTCCTGCTTCATTTTCGTCACGGCCAATATCATCCTCAGCGAGACCAAGCTCTGATGGCAGGACAGGCAGGTCT AGTGACAGGAGGCATAGGGGCCCCAGCCCCACCCCTCGCCCGCAGAGCCCAGATTTAGTTGCTCCCAGGTCTTCATGGGGCAGGTGGGAC TGTGGCTCTAGGGGAAACCCCCTGTCCAGCTGGGGTTGTTCTCCCCACCCACCTTCCACCCAGTCTGCACCTGTCCTCACTCCCCCGGGC AGCTCTCCCACCAGGTCCTTCTGCCTGGGTGACGCGTGGCTGCTCCCAAGTTCTAAGACTCATTACTGGAATCACACCTCTCCCACATCT TCCAACACGCTTCACCCCCAACCCTACTTCCAGGGCGGGCTACTCCCTGCACAATGGGAGATGCAGTGCTGGCACTGCCATCATTTGTCC CAGACCCATGCCCCTCCCCTCTCCTGCCCCATCCTGCGTCCTTGTCTAAGAAACCTCCACTAGCAGCTGCTGCCTCTTCAGAGGCGAATC CCAGGCTGGAAGTATCCTAGCCAGGTCTGCCTGTGACGGCAGGGCCCACCCTCCCACCAGGCCTGGGAGGCGGTAGGTGCCACCTCAGAG AGGGCCTGGCTGGCCCCATGACAAGAACAAGAAGTGCTAACTGGTACCAGAGTCCCCAGATAGAGCTGCGAGGGCCCTCGCTGTGGGCGC GAGCTCCGGAGTAGCAGGCTGGATGGAGCATCCCTTCTACAGGGACACAGCCGCCTGCCAGCTGGGCCTAAAGTCTGGAGCTTGTCTCTG AAAGGGACCGTCTGCTGCCCTCGCTGCCCCACTGGAGCCCTCAGGGCCTCCTACAGTGCAGGTGGTCCTACTAGAGGGAAGCCATCCCCA CCTGGCACCTCAGCGGCTTGGCAGTGACATGGCAGTGGGAGGGCTCTGAGGTTCCCACCCTGACAATGGTCAGGCTGCCCGTGCTCCCAC TCCAACACTAAAAGCGGCTCCCTTAGGGGCTGAGCGTGGGTGCTCAGTGTTCACGCTATATCCCTTGGGCAATGTGGGGTTGGATGGGGC CCCCACTTCCATTCCCATGGAAGGAGGCCAGGTCCCCAGCCACCTCCCACTCAGCCATGCACGCACTTGCTGGGCTGGCCTCCTGGGAAA CACAGGTGACTCGAATGAACTCTGCATTTTCAACGTGCCTTCTACTGCTTCAGGACCTGGGGGTCCCCCTGACCCTCACTGGCTTGCCCC CAGCCCTGGGCCTGGCCCCACCTGTCCTGGAGCCCAGAGCCCCTGGCCTGGAGCTACCTCTCTGGGGTGGGTCTCAGGCCCCACCCCTCC CTCTTTTGAGTTCAGTGCCTTGCTCAGCCCCTCCCCTGTATCTCAGCGTCTTCAGACCTCTGACAGAGCGACGATGTAGGGTCTCCCGGG GCCCAAGGTGGTCTCAGGGTCAGGGGTGGGATTTGCAGGGAACTCGGGGAGCCCACGGGCTGCGCCACCTCTGCCCTGGCAGCTGAAGCC TGGGAGAGTCCCTGCGTGGTGTAATTGGCCTCAGCCCGCTTTCTCTGTGCCGTCGCACCTCAGTGTTTCTCATAGCTTGTCCCCACGTGT CACTTTCCATCCACGGGAAAAACAAATGCCCCTTCTCCATCTATCATTGCGACTTCCTCCCAGGAGGCCTCTCAGGTTGGGTAGAGCAGG GGCCTGCAGTGGTCAGGCCAAGAGCAAGGAAGACCTGGCTGCCCCACTGTGGCTGAAAACTCAAAACATCTGGAACTACCCTTCTCCAAT TACGTTCCCTCTTGCTTAAAGACAAAGCTAATTAAATCATCCTGCCACCCGAGGCTCCAAACCAAATCTTGGACGCAAATTCATTAGCTT CATAAAGCCCAGGTTGATTTATGTGACAGTCTGGAACCGCCCAGCCATGGAGTCGGTGGCCGTGTTCCCTGCCCTGGAGGGAACTGGCCA GCAGTGACCACATCGCTGATGCCAAGTGGGGACCCTCACCACTGCCGTTGTCTAGCTGCTTTGCTCCTCAGTGTCCTCCTGCTGTAAGTC AGATGAGGTCACCTTCCTCTCCATCGATGCCAGGCTTCTGCCGCTGACGGCCAATGGGGCTTGGGGCCAGGGGCAGAGAACCCCAACAGC AAGAGAGCCCCAGGTGATGCTGGCTCTCCACGTGCATCCGGCACGTTTCACTGCAGCCATCATCAACTTGGCAAAGTAGGCAAGCTCAGC TTGTGGTTTCCATTTTGCAGATGGGCCCCCGGGGTCAGTGATGGGAAGAGCAGAAGAGGATGCCAGCCCAGCCTGGCCCAGGAGCAGGTG TCGTTGTGGAGGAAAGCTTAAACTCAACAGCCTCTACCCAACATCTGCCACCTGCATGGCCCCAGATGCCCCCGGTGCAGGCCAGGTCAA GGTGGGAGTAAACTTTCTTTGCCTCCCTCCCCACAGAAGCACCAGACCCACCTGAGCCCCAGAGCCTCATGCCAGCAGCTCCTGGCTGTT CCTCACCTGAGGCTAGAGCAGCAGCTGCCAGCTTATAGATGGGGCGGATGGCAGGTGATAGACTGGGAAGCACTGCTGGGTGGTGGAGGT GGGCAGTGGCACCATGACGGGCCCCTAGCCACACAGCTGTCCTCACTACGGGGCAGGGAGCAGCCTCGGCAACAGGCAAAGGCCAGAGTC AGAGTGGTGGGGAGGACAGGGGCTGCTGCCCCGCTCCTGGAAAGCCACTGCAGAGGGGCAGTGGCTGGCAGTGCCAGGCCTGGGGGAAGT GGAAGCGTCCTGTCTGGGGGCAGATTCCCAAGCAAGGGTGACCCATGGTTAAGGGCCACTGGAAAGCTGGAGAGAGCTTGGGATCCCTTC CACCTGGGGCCAATGGTGTTGATTGCAGACTGGAGGGGTAACCTCCGCTGAGGGTCATTATGTGCCAGGCATGGCATTGGGTACTTTCTG CATTGGGACCAGGCAGCCGGGCCTGCCATTTGAGGCAGCGAGCCTCCGTGACCTGTCCTCCCTCATTTGTAAAGTGGGGTAACAGCTATG TCACTGGCCAGTTGTGGGGATTAAAGTGCTGAGCTTAGTGCCCAGCCCAAAATGCTCAATAAAGCTATTCAGTGATACCTGTTTCCCACA >45705_45705_3_LLGL2-TMEM104_LLGL2_chr17_73567181_ENST00000577200_TMEM104_chr17_72832066_ENST00000335464_length(amino acids)=992AA_BP=740 MGLQVSSGGCRLSKMRRFLRPGHDPVRERLKRDLFQFNKTVEHGFPHQPSALGYSPSLRILAIGTRSGAIKLYGAPGVEFMGLHQENNAV TQIHLLPGQCQLVTLLDDNSLHLWSLKVKGGASELQEDESFTLRGPPGAAPSATQITVVLPHSSCELLYLGTESGNVFVVQLPAFRALED RTISSDAVLQRLPEEARHRRVFEMVEALQEHPRDPNQILIGYSRGLVVIWDLQGSRVLYHFLSSQQLENIWWQRDGRLLVSCHSDGSYCQ WPVSSEAQQPEPLRSLVPYGPFPCKAITRILWLTTRQGLPFTIFQGGMPRASYGDRHCISVIHDGQQTAFDFTSRVIGFTVLTEADPAAT FDDPYALVVLAEEELVVIDLQTAGWPPVQLPYLASLHCSAITCSHHVSNIPLKLWERIIAAGSRQNAHFSTMEWPIDGGTSLTPAPPQRD LLLTGHEDGTVRFWDASGVCLRLLYKLSTVRVFLTDTDPNENFSAQGEDEWPPLRKVGSFDPYSDDPRLGIQKIFLCKYSGYLAVAGTAG QVLVLELNDEAAEQAVEQVEADLLQDQEGYRWKGHERLAARSGPVRFEPGFQPFVLVQCQPPAVVTSLALHSEWRLVAFGTSHGFGLFDH QQRRQVFVKCTLHPSDQLALEGPLSRVKSLKKSLRQSFRRMRRSRVSSRKRHPAGPPGEAQEGSAKAERPGLQNMELAPVQRKIEARSAE DSFTGFVRTLYFADTYLKDTFAVMIVLALIRIGHGQGEGHPPLADFSGVRNLFGVCVYSFMCQHSLPSLITPVSSKRHLTRLVFLDYVLI LAFYGLLSFTAIFCFRGDSLMDMYTLNFARCDVVGLAAVRFFLGLFPVFTISTNFPIIAVTLRNNWKTLFHREGGTYPWVVDRVVFPTIT LVPPVLVAFCTHDLESLVGITGAYAGTGIQYVIPAFLVYHCRRDTQLAFGCGVSNKHRSPFRHTFWVGFVLLWAFSCFIFVTANIILSET -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for LLGL2-TMEM104 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for LLGL2-TMEM104 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for LLGL2-TMEM104 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
