
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:LMBR1L-KCNK6 (FusionGDB2 ID:45737) |
Fusion Gene Summary for LMBR1L-KCNK6 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: LMBR1L-KCNK6 | Fusion gene ID: 45737 | Hgene | Tgene | Gene symbol | LMBR1L | KCNK6 | Gene ID | 55716 | 9424 |
| Gene name | limb development membrane protein 1 like | potassium two pore domain channel subfamily K member 6 | |
| Synonyms | LIMR | K2p6.1|KCNK8|TOSS|TWIK-2|TWIK2 | |
| Cytomap | 12q13.12 | 19q13.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | protein LMBR1Llimb region 1 homolog-likelimb region 1 protein homolog-likelimb region 1-like protein -likelipocalin-1 interacting membrane receptorlipocalin-interacting membrane receptor | potassium channel subfamily K member 6K2P6.1 potassium channelTWIK-originated similarity sequenceTWIK-originated sodium similarity sequenceinward rectifying potassium channel protein TWIK-2potassium channel, two pore domain subfamily K, member 6 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q6UX01 | Q9Y257 | |
| Ensembl transtripts involved in fusion gene | ENST00000267102, ENST00000395141, ENST00000547382, ENST00000553204, | ENST00000588137, ENST00000263372, | |
| Fusion gene scores | * DoF score | 6 X 6 X 4=144 | 3 X 3 X 2=18 |
| # samples | 5 | 3 | |
| ** MAII score | log2(5/144*10)=-1.52606881166759 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(3/18*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: LMBR1L [Title/Abstract] AND KCNK6 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | LMBR1L(49504267)-KCNK6(38817233), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | LMBR1L-KCNK6 seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | LMBR1L | GO:0007165 | signal transduction | 11287427 |
 Fusion gene breakpoints across LMBR1L (5'-gene) Fusion gene breakpoints across LMBR1L (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
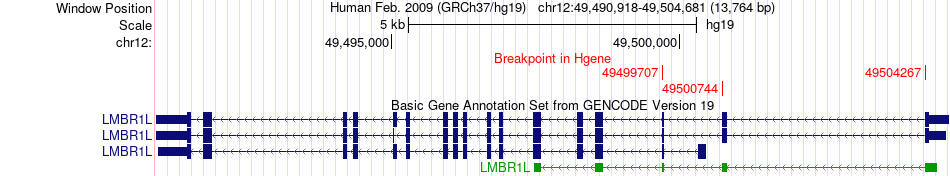 |
 Fusion gene breakpoints across KCNK6 (3'-gene) Fusion gene breakpoints across KCNK6 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
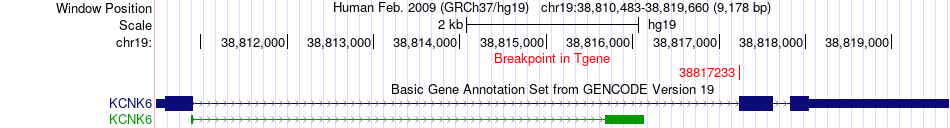 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BLCA | TCGA-CU-A0YN-01A | LMBR1L | chr12 | 49499707 | - | KCNK6 | chr19 | 38817233 | + |
| ChimerDB4 | BLCA | TCGA-CU-A0YN-01A | LMBR1L | chr12 | 49500744 | - | KCNK6 | chr19 | 38817233 | + |
| ChimerDB4 | BLCA | TCGA-CU-A0YN-01A | LMBR1L | chr12 | 49504267 | - | KCNK6 | chr19 | 38817233 | + |
Top |
Fusion Gene ORF analysis for LMBR1L-KCNK6 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000267102 | ENST00000588137 | LMBR1L | chr12 | 49499707 | - | KCNK6 | chr19 | 38817233 | + |
| 5CDS-intron | ENST00000267102 | ENST00000588137 | LMBR1L | chr12 | 49500744 | - | KCNK6 | chr19 | 38817233 | + |
| 5CDS-intron | ENST00000267102 | ENST00000588137 | LMBR1L | chr12 | 49504267 | - | KCNK6 | chr19 | 38817233 | + |
| 5CDS-intron | ENST00000395141 | ENST00000588137 | LMBR1L | chr12 | 49499707 | - | KCNK6 | chr19 | 38817233 | + |
| 5CDS-intron | ENST00000547382 | ENST00000588137 | LMBR1L | chr12 | 49499707 | - | KCNK6 | chr19 | 38817233 | + |
| 5CDS-intron | ENST00000547382 | ENST00000588137 | LMBR1L | chr12 | 49500744 | - | KCNK6 | chr19 | 38817233 | + |
| 5CDS-intron | ENST00000547382 | ENST00000588137 | LMBR1L | chr12 | 49504267 | - | KCNK6 | chr19 | 38817233 | + |
| 5UTR-3CDS | ENST00000553204 | ENST00000263372 | LMBR1L | chr12 | 49499707 | - | KCNK6 | chr19 | 38817233 | + |
| 5UTR-3CDS | ENST00000553204 | ENST00000263372 | LMBR1L | chr12 | 49500744 | - | KCNK6 | chr19 | 38817233 | + |
| 5UTR-3CDS | ENST00000553204 | ENST00000263372 | LMBR1L | chr12 | 49504267 | - | KCNK6 | chr19 | 38817233 | + |
| 5UTR-intron | ENST00000553204 | ENST00000588137 | LMBR1L | chr12 | 49499707 | - | KCNK6 | chr19 | 38817233 | + |
| 5UTR-intron | ENST00000553204 | ENST00000588137 | LMBR1L | chr12 | 49500744 | - | KCNK6 | chr19 | 38817233 | + |
| 5UTR-intron | ENST00000553204 | ENST00000588137 | LMBR1L | chr12 | 49504267 | - | KCNK6 | chr19 | 38817233 | + |
| Frame-shift | ENST00000267102 | ENST00000263372 | LMBR1L | chr12 | 49499707 | - | KCNK6 | chr19 | 38817233 | + |
| Frame-shift | ENST00000267102 | ENST00000263372 | LMBR1L | chr12 | 49504267 | - | KCNK6 | chr19 | 38817233 | + |
| Frame-shift | ENST00000395141 | ENST00000263372 | LMBR1L | chr12 | 49499707 | - | KCNK6 | chr19 | 38817233 | + |
| Frame-shift | ENST00000547382 | ENST00000263372 | LMBR1L | chr12 | 49499707 | - | KCNK6 | chr19 | 38817233 | + |
| Frame-shift | ENST00000547382 | ENST00000263372 | LMBR1L | chr12 | 49504267 | - | KCNK6 | chr19 | 38817233 | + |
| In-frame | ENST00000267102 | ENST00000263372 | LMBR1L | chr12 | 49500744 | - | KCNK6 | chr19 | 38817233 | + |
| In-frame | ENST00000547382 | ENST00000263372 | LMBR1L | chr12 | 49500744 | - | KCNK6 | chr19 | 38817233 | + |
| intron-3CDS | ENST00000395141 | ENST00000263372 | LMBR1L | chr12 | 49500744 | - | KCNK6 | chr19 | 38817233 | + |
| intron-3CDS | ENST00000395141 | ENST00000263372 | LMBR1L | chr12 | 49504267 | - | KCNK6 | chr19 | 38817233 | + |
| intron-intron | ENST00000395141 | ENST00000588137 | LMBR1L | chr12 | 49500744 | - | KCNK6 | chr19 | 38817233 | + |
| intron-intron | ENST00000395141 | ENST00000588137 | LMBR1L | chr12 | 49504267 | - | KCNK6 | chr19 | 38817233 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000267102 | LMBR1L | chr12 | 49500744 | - | ENST00000263372 | KCNK6 | chr19 | 38817233 | + | 2737 | 500 | 190 | 1119 | 309 |
| ENST00000547382 | LMBR1L | chr12 | 49500744 | - | ENST00000263372 | KCNK6 | chr19 | 38817233 | + | 2689 | 452 | 142 | 1071 | 309 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000267102 | ENST00000263372 | LMBR1L | chr12 | 49500744 | - | KCNK6 | chr19 | 38817233 | + | 0.03995088 | 0.96004915 |
| ENST00000547382 | ENST00000263372 | LMBR1L | chr12 | 49500744 | - | KCNK6 | chr19 | 38817233 | + | 0.044108063 | 0.95589197 |
Top |
Fusion Genomic Features for LMBR1L-KCNK6 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| LMBR1L | chr12 | 49500743 | - | KCNK6 | chr19 | 38817232 | + | 3.29E-05 | 0.9999671 |
| LMBR1L | chr12 | 49499706 | - | KCNK6 | chr19 | 38817232 | + | 5.98E-07 | 0.9999994 |
| LMBR1L | chr12 | 49504266 | - | KCNK6 | chr19 | 38817232 | + | 2.08E-09 | 1 |
| LMBR1L | chr12 | 49500743 | - | KCNK6 | chr19 | 38817232 | + | 3.29E-05 | 0.9999671 |
| LMBR1L | chr12 | 49499706 | - | KCNK6 | chr19 | 38817232 | + | 5.98E-07 | 0.9999994 |
| LMBR1L | chr12 | 49504266 | - | KCNK6 | chr19 | 38817232 | + | 2.08E-09 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
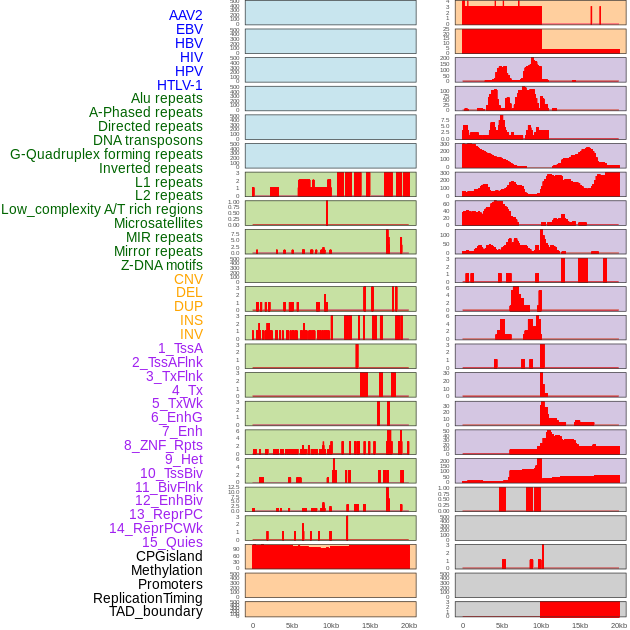 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
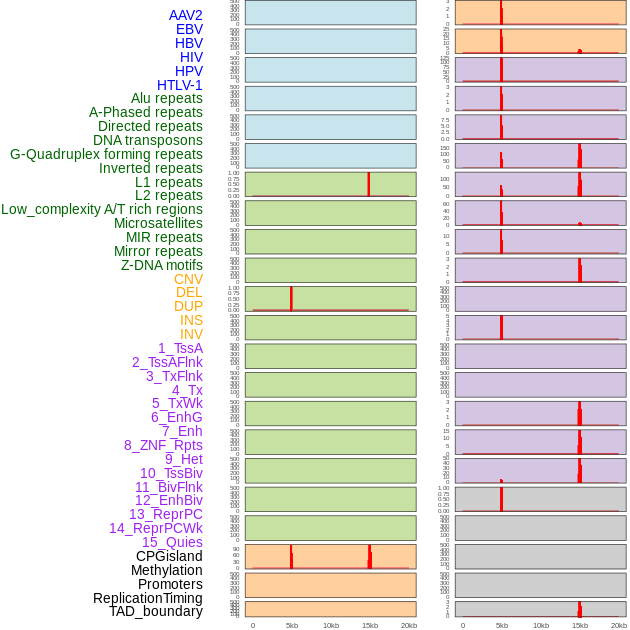 |
Top |
Fusion Protein Features for LMBR1L-KCNK6 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:49504267/chr19:38817233) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| LMBR1L | KCNK6 |
| FUNCTION: Plays an essential role in lymphocyte development by negatively regulating the canonical Wnt signaling pathway (By similarity). In association with UBAC2 and E3 ubiquitin-protein ligase AMFR, promotes the ubiquitin-mediated degradation of CTNNB1 and Wnt receptors FZD6 and LRP6 (By similarity). LMBR1L stabilizes the beta-catenin destruction complex that is required for regulating CTNNB1 levels (By similarity). Acts as a LCN1 receptor and can mediate its endocytosis (PubMed:11287427, PubMed:12591932, PubMed:23964685). {ECO:0000250|UniProtKB:Q9D1E5, ECO:0000269|PubMed:11287427, ECO:0000269|PubMed:12591932, ECO:0000269|PubMed:23964685}. | FUNCTION: Exhibits outward rectification in a physiological K(+) gradient and mild inward rectification in symmetrical K(+) conditions. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000267102 | - | 2 | 17 | 1_21 | 52 | 490.0 | Topological domain | Extracellular |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000547382 | - | 2 | 17 | 1_21 | 52 | 470.0 | Topological domain | Extracellular |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000267102 | - | 2 | 17 | 22_42 | 52 | 490.0 | Transmembrane | Helical |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000547382 | - | 2 | 17 | 22_42 | 52 | 470.0 | Transmembrane | Helical |
| Tgene | KCNK6 | chr12:49500744 | chr19:38817233 | ENST00000263372 | 0 | 3 | 199_223 | 107 | 314.0 | Intramembrane | Pore-forming%3B Name%3DPore-forming 2 | |
| Tgene | KCNK6 | chr12:49500744 | chr19:38817233 | ENST00000263372 | 0 | 3 | 142_172 | 107 | 314.0 | Topological domain | Cytoplasmic | |
| Tgene | KCNK6 | chr12:49500744 | chr19:38817233 | ENST00000263372 | 0 | 3 | 257_313 | 107 | 314.0 | Topological domain | Cytoplasmic | |
| Tgene | KCNK6 | chr12:49500744 | chr19:38817233 | ENST00000263372 | 0 | 3 | 121_141 | 107 | 314.0 | Transmembrane | Helical | |
| Tgene | KCNK6 | chr12:49500744 | chr19:38817233 | ENST00000263372 | 0 | 3 | 173_193 | 107 | 314.0 | Transmembrane | Helical | |
| Tgene | KCNK6 | chr12:49500744 | chr19:38817233 | ENST00000263372 | 0 | 3 | 236_256 | 107 | 314.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000267102 | - | 2 | 17 | 1_76 | 52 | 490.0 | Region | LCN1-binding |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000395141 | - | 1 | 16 | 1_76 | 0 | 485.0 | Region | LCN1-binding |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000547382 | - | 2 | 17 | 1_76 | 52 | 470.0 | Region | LCN1-binding |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000267102 | - | 2 | 17 | 136_154 | 52 | 490.0 | Topological domain | Cytoplasmic |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000267102 | - | 2 | 17 | 176_196 | 52 | 490.0 | Topological domain | Extracellular |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000267102 | - | 2 | 17 | 218_305 | 52 | 490.0 | Topological domain | Cytoplasmic |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000267102 | - | 2 | 17 | 327_350 | 52 | 490.0 | Topological domain | Extracellular |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000267102 | - | 2 | 17 | 372_388 | 52 | 490.0 | Topological domain | Cytoplasmic |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000267102 | - | 2 | 17 | 410_431 | 52 | 490.0 | Topological domain | Extracellular |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000267102 | - | 2 | 17 | 43_66 | 52 | 490.0 | Topological domain | Cytoplasmic |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000267102 | - | 2 | 17 | 453_489 | 52 | 490.0 | Topological domain | Cytoplasmic |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000267102 | - | 2 | 17 | 88_114 | 52 | 490.0 | Topological domain | Extracellular |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000395141 | - | 1 | 16 | 136_154 | 0 | 485.0 | Topological domain | Cytoplasmic |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000395141 | - | 1 | 16 | 176_196 | 0 | 485.0 | Topological domain | Extracellular |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000395141 | - | 1 | 16 | 1_21 | 0 | 485.0 | Topological domain | Extracellular |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000395141 | - | 1 | 16 | 218_305 | 0 | 485.0 | Topological domain | Cytoplasmic |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000395141 | - | 1 | 16 | 327_350 | 0 | 485.0 | Topological domain | Extracellular |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000395141 | - | 1 | 16 | 372_388 | 0 | 485.0 | Topological domain | Cytoplasmic |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000395141 | - | 1 | 16 | 410_431 | 0 | 485.0 | Topological domain | Extracellular |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000395141 | - | 1 | 16 | 43_66 | 0 | 485.0 | Topological domain | Cytoplasmic |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000395141 | - | 1 | 16 | 453_489 | 0 | 485.0 | Topological domain | Cytoplasmic |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000395141 | - | 1 | 16 | 88_114 | 0 | 485.0 | Topological domain | Extracellular |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000547382 | - | 2 | 17 | 136_154 | 52 | 470.0 | Topological domain | Cytoplasmic |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000547382 | - | 2 | 17 | 176_196 | 52 | 470.0 | Topological domain | Extracellular |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000547382 | - | 2 | 17 | 218_305 | 52 | 470.0 | Topological domain | Cytoplasmic |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000547382 | - | 2 | 17 | 327_350 | 52 | 470.0 | Topological domain | Extracellular |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000547382 | - | 2 | 17 | 372_388 | 52 | 470.0 | Topological domain | Cytoplasmic |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000547382 | - | 2 | 17 | 410_431 | 52 | 470.0 | Topological domain | Extracellular |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000547382 | - | 2 | 17 | 43_66 | 52 | 470.0 | Topological domain | Cytoplasmic |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000547382 | - | 2 | 17 | 453_489 | 52 | 470.0 | Topological domain | Cytoplasmic |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000547382 | - | 2 | 17 | 88_114 | 52 | 470.0 | Topological domain | Extracellular |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000267102 | - | 2 | 17 | 115_135 | 52 | 490.0 | Transmembrane | Helical |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000267102 | - | 2 | 17 | 155_175 | 52 | 490.0 | Transmembrane | Helical |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000267102 | - | 2 | 17 | 197_217 | 52 | 490.0 | Transmembrane | Helical |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000267102 | - | 2 | 17 | 306_326 | 52 | 490.0 | Transmembrane | Helical |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000267102 | - | 2 | 17 | 351_371 | 52 | 490.0 | Transmembrane | Helical |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000267102 | - | 2 | 17 | 389_409 | 52 | 490.0 | Transmembrane | Helical |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000267102 | - | 2 | 17 | 432_452 | 52 | 490.0 | Transmembrane | Helical |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000267102 | - | 2 | 17 | 67_87 | 52 | 490.0 | Transmembrane | Helical |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000395141 | - | 1 | 16 | 115_135 | 0 | 485.0 | Transmembrane | Helical |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000395141 | - | 1 | 16 | 155_175 | 0 | 485.0 | Transmembrane | Helical |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000395141 | - | 1 | 16 | 197_217 | 0 | 485.0 | Transmembrane | Helical |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000395141 | - | 1 | 16 | 22_42 | 0 | 485.0 | Transmembrane | Helical |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000395141 | - | 1 | 16 | 306_326 | 0 | 485.0 | Transmembrane | Helical |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000395141 | - | 1 | 16 | 351_371 | 0 | 485.0 | Transmembrane | Helical |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000395141 | - | 1 | 16 | 389_409 | 0 | 485.0 | Transmembrane | Helical |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000395141 | - | 1 | 16 | 432_452 | 0 | 485.0 | Transmembrane | Helical |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000395141 | - | 1 | 16 | 67_87 | 0 | 485.0 | Transmembrane | Helical |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000547382 | - | 2 | 17 | 115_135 | 52 | 470.0 | Transmembrane | Helical |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000547382 | - | 2 | 17 | 155_175 | 52 | 470.0 | Transmembrane | Helical |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000547382 | - | 2 | 17 | 197_217 | 52 | 470.0 | Transmembrane | Helical |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000547382 | - | 2 | 17 | 306_326 | 52 | 470.0 | Transmembrane | Helical |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000547382 | - | 2 | 17 | 351_371 | 52 | 470.0 | Transmembrane | Helical |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000547382 | - | 2 | 17 | 389_409 | 52 | 470.0 | Transmembrane | Helical |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000547382 | - | 2 | 17 | 432_452 | 52 | 470.0 | Transmembrane | Helical |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000547382 | - | 2 | 17 | 67_87 | 52 | 470.0 | Transmembrane | Helical |
| Tgene | KCNK6 | chr12:49500744 | chr19:38817233 | ENST00000263372 | 0 | 3 | 90_115 | 107 | 314.0 | Intramembrane | Pore-forming%3B Name%3DPore-forming 1 | |
| Tgene | KCNK6 | chr12:49500744 | chr19:38817233 | ENST00000263372 | 0 | 3 | 1_4 | 107 | 314.0 | Topological domain | Cytoplasmic | |
| Tgene | KCNK6 | chr12:49500744 | chr19:38817233 | ENST00000263372 | 0 | 3 | 5_25 | 107 | 314.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for LMBR1L-KCNK6 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >45737_45737_1_LMBR1L-KCNK6_LMBR1L_chr12_49500744_ENST00000267102_KCNK6_chr19_38817233_ENST00000263372_length(transcript)=2737nt_BP=500nt TGTGTCGTCACTAGATCTCTTAAAAAGATGGACCCTCAGCGGCGCTTCCTCGTAGCGAGCCTAGTGGCGGGTGTTTGCATTGAAACGTGA GCGCGACCCGACCTTAAAGAGTGGGGAGCAAAGGGAGGACAGAGCCCTTTAAAACGAGGCGGGTGGTGCCTGCCCCTTTAAGGGCGGGGC GTCCGGACGACTGTATCTGAGCCCCAGACTGCCCCGAGTTTCTGTCGCAGGCTGCGAGGAAAGGCCCCTAGGCTGGGTCTGGGTGCTTGG CGGCGGCGGCTTCCTCCCCGCTCGTCCTCCCCGGGCCCAGAGGCACCTCGGCTTCAGTCATGCTGAGCAGAGTATGGAAGCACCTGACTA CGAAGTGCTATCCGTGCGAGAACAGCTATTCCACGAGAGGATCCGCGAGTGTATTATATCAACACTTCTGTTTGCAACACTGTACATCCT CTGCCACATCTTCCTGACCCGCTTCAAGAAGCCTGCTGAGTTCACCACAGGCTATGGGTACACAACGCCACTGACTGATGCGGGCAAGGC CTTCTCCATCGCCTTTGCGCTCCTGGGCGTGCCGACCACCATGCTGCTGCTGACCGCCTCAGCCCAGCGCCTGTCACTGCTGCTGACTCA CGTGCCCCTGTCTTGGCTGAGCATGCGTTGGGGCTGGGACCCCCGGCGGGCGGCCTGCTGGCACTTGGTGGCCCTGTTGGGGGTCGTAGT GACCGTCTGCTTTCTGGTGCCGGCTGTGATCTTTGCCCACCTCGAGGAGGCCTGGAGCTTCTTGGATGCCTTCTACTTCTGCTTTATCTC TCTGTCCACCATCGGCCTGGGCGACTACGTGCCCGGGGAGGCCCCTGGCCAGCCCTACCGGGCCCTCTACAAGGTGCTGGTCACAGTCTA CCTCTTCCTGGGCCTGGTGGCCATGGTGCTGGTGCTGCAGACCTTCCGCCACGTGTCCGACCTCCACGGCCTCACGGAGCTCATCCTGCT GCCCCCTCCGTGCCCTGCCAGTTTCAATGCGGATGAGGACGATCGGGTGGACATCCTGGGCCCCCAGCCGGAGTCGCACCAGCAACTCTC TGCCAGCTCCCACACCGACTACGCTTCCATCCCCAGGTAGCTGGGGCAGCCTCTGCCAGGCTTGGGTGTGCCTGGCCTGGGACTGAGGGG TCCAGGCGACCAGAGCTGGCTGTACAGGAATGTCCACGAGCACAGCAGGTGATCTTGAGGCCTTGCCGTCCACCGTCTCTCCTTTGTTTC CCAGCATCTGGCTGGGATGTGAAGGGCAGCACTCCCTGTCCCCATGTCCCGGGCTCCACTGGGCACCAACATAACCTTGTTCTCTGTCCT TTCTCTCATCCTCTTTACACTGTGTCTCTCTGGCTCTCTGGCATTCTCGCTGCCTCTGTCTTTCCCTCTTGCTGTCTCTGTTTCTCATTC TCTTTCATGTTCCGTCTGTGTCTCTCAATTAACCACTCGTCAACTGCTGATTCTACTGGGCTGTGGGCTCAGACCTCATTTCAGGCACCA GATTGGTCGCTACACCCTGGACAAGTGACTGCCCGTCTCTGAGCCTTGATTTCCTCAGCTGCCAAATGGGAAGAATAGAAGAATTTGCCC CTAAACCCCTCCTGTGTGCTGGCCCTGTGCTAGACAGTGCTGGAGACATAGTTGGGGGTGGAGAACTGCCCTTATGGAGCTTGCAGTCCA GTGAGGTGGACAGACCTGTCCCCAGACAGTGATGGCCCAAAATGGTCAGGACTTTAATGGAGGAGGTGAGGTGTTGAAAGCACAGGCAGA GTGGTCAGGGCTGAAGTCGGAGAAGCATAGGGACTAGGCCCAATCCAGCCTGGAAAGTCAGGGAGGACTTCCTAGAGGAAGGGACATCGA ACTAAGACCTGAACTATGAGAAATAGGCAGGAAGAAGTTGTACCTGACTCATTTTTCTCAGGTGTCTCCAGGGAGCAGGACCCATGGAGG GACCCCTGGTGTAGGCCTGGGCGATAGACTCTTCCTCAGCAGCCTGGCAGGCAGGAAACAGACATAGGACCCCAGCCCAGATCTGAATGG CATGGGAGGTGCTGCCCTTAACCATGACACCATTGTAAGAGCTGTCCACATTTGTATGTTGTGCCCTGGAATCAGCCTGGTTGAGCTCAA ATCCCAACTTAGCCACGTCTGGCCTGTGTCCTTGGGCAGTCACACTACCTCTCTGATTTTGTTTCCTTATCTGTAAAATGGTGATCATCA TAATACAACTTCAAAAGGATTTCAGGCTGAGTGTGGTGGCTCACGCCTATACACCCAGCACTTTGGAAGGCTGAGGAAGGAGGATCGCTT GAGGCCAGGAGTTTGAGACTAGCCTAGGCAACACAGTGAGGCCTTATCTCAACAACAACCACAAAATCTAAAAATTAGCTGGGTGTGGTG GTGCATGCCTGTGATCCTGGCTACTTCAGAGGCTGAGGTGGAAGGATCACTTGAGGCCAGGAGTTTGAGGCTGCAGTGAGTTATGATGGC ACTGCTGCACTCCAGCCTGCGGGACAGAGTGAGACCCTGTCTGAAAGAAAGAGAGAAAGAAAGAAAGAAAGAGAGAGAAAGAAAGAAAGA AAGAAAGGGAAAGATGGAAGGAAGGAAGGAAAAAGAAAGAGAAAGAGAGAGAGAGAGAAAGAAAAAGAAAGAAAAGTAAGAAAGAAAAAA >45737_45737_1_LMBR1L-KCNK6_LMBR1L_chr12_49500744_ENST00000267102_KCNK6_chr19_38817233_ENST00000263372_length(amino acids)=309AA_BP=103 MYLSPRLPRVSVAGCEERPLGWVWVLGGGGFLPARPPRAQRHLGFSHAEQSMEAPDYEVLSVREQLFHERIRECIISTLLFATLYILCHI FLTRFKKPAEFTTGYGYTTPLTDAGKAFSIAFALLGVPTTMLLLTASAQRLSLLLTHVPLSWLSMRWGWDPRRAACWHLVALLGVVVTVC FLVPAVIFAHLEEAWSFLDAFYFCFISLSTIGLGDYVPGEAPGQPYRALYKVLVTVYLFLGLVAMVLVLQTFRHVSDLHGLTELILLPPP -------------------------------------------------------------- >45737_45737_2_LMBR1L-KCNK6_LMBR1L_chr12_49500744_ENST00000547382_KCNK6_chr19_38817233_ENST00000263372_length(transcript)=2689nt_BP=452nt CTCGTAGCGAGCCTAGTGGCGGGTGTTTGCATTGAAACGTGAGCGCGACCCGACCTTAAAGAGTGGGGAGCAAAGGGAGGACAGAGCCCT TTAAAACGAGGCGGGTGGTGCCTGCCCCTTTAAGGGCGGGGCGTCCGGACGACTGTATCTGAGCCCCAGACTGCCCCGAGTTTCTGTCGC AGGCTGCGAGGAAAGGCCCCTAGGCTGGGTCTGGGTGCTTGGCGGCGGCGGCTTCCTCCCCGCTCGTCCTCCCCGGGCCCAGAGGCACCT CGGCTTCAGTCATGCTGAGCAGAGTATGGAAGCACCTGACTACGAAGTGCTATCCGTGCGAGAACAGCTATTCCACGAGAGGATCCGCGA GTGTATTATATCAACACTTCTGTTTGCAACACTGTACATCCTCTGCCACATCTTCCTGACCCGCTTCAAGAAGCCTGCTGAGTTCACCAC AGGCTATGGGTACACAACGCCACTGACTGATGCGGGCAAGGCCTTCTCCATCGCCTTTGCGCTCCTGGGCGTGCCGACCACCATGCTGCT GCTGACCGCCTCAGCCCAGCGCCTGTCACTGCTGCTGACTCACGTGCCCCTGTCTTGGCTGAGCATGCGTTGGGGCTGGGACCCCCGGCG GGCGGCCTGCTGGCACTTGGTGGCCCTGTTGGGGGTCGTAGTGACCGTCTGCTTTCTGGTGCCGGCTGTGATCTTTGCCCACCTCGAGGA GGCCTGGAGCTTCTTGGATGCCTTCTACTTCTGCTTTATCTCTCTGTCCACCATCGGCCTGGGCGACTACGTGCCCGGGGAGGCCCCTGG CCAGCCCTACCGGGCCCTCTACAAGGTGCTGGTCACAGTCTACCTCTTCCTGGGCCTGGTGGCCATGGTGCTGGTGCTGCAGACCTTCCG CCACGTGTCCGACCTCCACGGCCTCACGGAGCTCATCCTGCTGCCCCCTCCGTGCCCTGCCAGTTTCAATGCGGATGAGGACGATCGGGT GGACATCCTGGGCCCCCAGCCGGAGTCGCACCAGCAACTCTCTGCCAGCTCCCACACCGACTACGCTTCCATCCCCAGGTAGCTGGGGCA GCCTCTGCCAGGCTTGGGTGTGCCTGGCCTGGGACTGAGGGGTCCAGGCGACCAGAGCTGGCTGTACAGGAATGTCCACGAGCACAGCAG GTGATCTTGAGGCCTTGCCGTCCACCGTCTCTCCTTTGTTTCCCAGCATCTGGCTGGGATGTGAAGGGCAGCACTCCCTGTCCCCATGTC CCGGGCTCCACTGGGCACCAACATAACCTTGTTCTCTGTCCTTTCTCTCATCCTCTTTACACTGTGTCTCTCTGGCTCTCTGGCATTCTC GCTGCCTCTGTCTTTCCCTCTTGCTGTCTCTGTTTCTCATTCTCTTTCATGTTCCGTCTGTGTCTCTCAATTAACCACTCGTCAACTGCT GATTCTACTGGGCTGTGGGCTCAGACCTCATTTCAGGCACCAGATTGGTCGCTACACCCTGGACAAGTGACTGCCCGTCTCTGAGCCTTG ATTTCCTCAGCTGCCAAATGGGAAGAATAGAAGAATTTGCCCCTAAACCCCTCCTGTGTGCTGGCCCTGTGCTAGACAGTGCTGGAGACA TAGTTGGGGGTGGAGAACTGCCCTTATGGAGCTTGCAGTCCAGTGAGGTGGACAGACCTGTCCCCAGACAGTGATGGCCCAAAATGGTCA GGACTTTAATGGAGGAGGTGAGGTGTTGAAAGCACAGGCAGAGTGGTCAGGGCTGAAGTCGGAGAAGCATAGGGACTAGGCCCAATCCAG CCTGGAAAGTCAGGGAGGACTTCCTAGAGGAAGGGACATCGAACTAAGACCTGAACTATGAGAAATAGGCAGGAAGAAGTTGTACCTGAC TCATTTTTCTCAGGTGTCTCCAGGGAGCAGGACCCATGGAGGGACCCCTGGTGTAGGCCTGGGCGATAGACTCTTCCTCAGCAGCCTGGC AGGCAGGAAACAGACATAGGACCCCAGCCCAGATCTGAATGGCATGGGAGGTGCTGCCCTTAACCATGACACCATTGTAAGAGCTGTCCA CATTTGTATGTTGTGCCCTGGAATCAGCCTGGTTGAGCTCAAATCCCAACTTAGCCACGTCTGGCCTGTGTCCTTGGGCAGTCACACTAC CTCTCTGATTTTGTTTCCTTATCTGTAAAATGGTGATCATCATAATACAACTTCAAAAGGATTTCAGGCTGAGTGTGGTGGCTCACGCCT ATACACCCAGCACTTTGGAAGGCTGAGGAAGGAGGATCGCTTGAGGCCAGGAGTTTGAGACTAGCCTAGGCAACACAGTGAGGCCTTATC TCAACAACAACCACAAAATCTAAAAATTAGCTGGGTGTGGTGGTGCATGCCTGTGATCCTGGCTACTTCAGAGGCTGAGGTGGAAGGATC ACTTGAGGCCAGGAGTTTGAGGCTGCAGTGAGTTATGATGGCACTGCTGCACTCCAGCCTGCGGGACAGAGTGAGACCCTGTCTGAAAGA AAGAGAGAAAGAAAGAAAGAAAGAGAGAGAAAGAAAGAAAGAAAGAAAGGGAAAGATGGAAGGAAGGAAGGAAAAAGAAAGAGAAAGAGA >45737_45737_2_LMBR1L-KCNK6_LMBR1L_chr12_49500744_ENST00000547382_KCNK6_chr19_38817233_ENST00000263372_length(amino acids)=309AA_BP=103 MYLSPRLPRVSVAGCEERPLGWVWVLGGGGFLPARPPRAQRHLGFSHAEQSMEAPDYEVLSVREQLFHERIRECIISTLLFATLYILCHI FLTRFKKPAEFTTGYGYTTPLTDAGKAFSIAFALLGVPTTMLLLTASAQRLSLLLTHVPLSWLSMRWGWDPRRAACWHLVALLGVVVTVC FLVPAVIFAHLEEAWSFLDAFYFCFISLSTIGLGDYVPGEAPGQPYRALYKVLVTVYLFLGLVAMVLVLQTFRHVSDLHGLTELILLPPP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for LMBR1L-KCNK6 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000267102 | - | 2 | 17 | 1_59 | 52.333333333333336 | 490.0 | LGB |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000395141 | - | 1 | 16 | 1_59 | 0.0 | 485.0 | LGB |
| Hgene | LMBR1L | chr12:49500744 | chr19:38817233 | ENST00000547382 | - | 2 | 17 | 1_59 | 52.333333333333336 | 470.0 | LGB |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for LMBR1L-KCNK6 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for LMBR1L-KCNK6 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
