
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:LMBR1-UBE3C (FusionGDB2 ID:45751) |
Fusion Gene Summary for LMBR1-UBE3C |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: LMBR1-UBE3C | Fusion gene ID: 45751 | Hgene | Tgene | Gene symbol | LMBR1 | UBE3C | Gene ID | 64327 | 9690 |
| Gene name | limb development membrane protein 1 | ubiquitin protein ligase E3C | |
| Synonyms | ACHP|C7orf2|DIF14|LSS|PPD2|THYP|TPT|ZRS | HECTH2 | |
| Cytomap | 7q36.3 | 7q36.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | limb region 1 protein homologdifferentiation-related gene 14 proteinlimb region 1 homolog | ubiquitin-protein ligase E3CHECT-type ubiquitin transferase E3Cubiquitin-protein isopeptide ligase (E3) | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q8WVP7 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000353442, ENST00000354505, ENST00000540390, ENST00000359422, ENST00000430825, | ENST00000348165, ENST00000389103, | |
| Fusion gene scores | * DoF score | 14 X 11 X 12=1848 | 11 X 13 X 7=1001 |
| # samples | 20 | 13 | |
| ** MAII score | log2(20/1848*10)=-3.20789285164133 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(13/1001*10)=-2.94485844580754 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: LMBR1 [Title/Abstract] AND UBE3C [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | LMBR1(156685622)-UBE3C(156979528), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | UBE3C | GO:0000209 | protein polyubiquitination | 11278995 |
 Fusion gene breakpoints across LMBR1 (5'-gene) Fusion gene breakpoints across LMBR1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
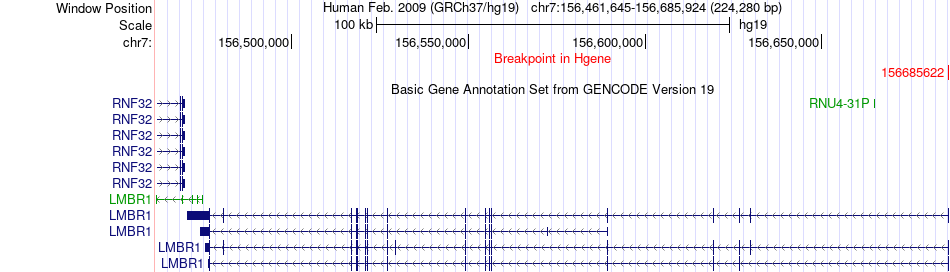 |
 Fusion gene breakpoints across UBE3C (3'-gene) Fusion gene breakpoints across UBE3C (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
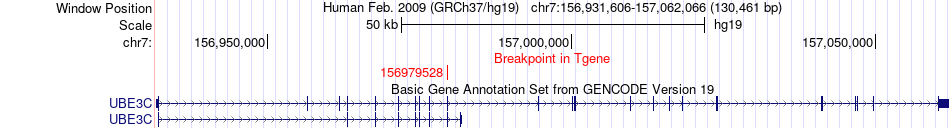 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-DX-A6Z0-01A | LMBR1 | chr7 | 156685622 | - | UBE3C | chr7 | 156979528 | + |
Top |
Fusion Gene ORF analysis for LMBR1-UBE3C |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000353442 | ENST00000348165 | LMBR1 | chr7 | 156685622 | - | UBE3C | chr7 | 156979528 | + |
| In-frame | ENST00000353442 | ENST00000389103 | LMBR1 | chr7 | 156685622 | - | UBE3C | chr7 | 156979528 | + |
| In-frame | ENST00000354505 | ENST00000348165 | LMBR1 | chr7 | 156685622 | - | UBE3C | chr7 | 156979528 | + |
| In-frame | ENST00000354505 | ENST00000389103 | LMBR1 | chr7 | 156685622 | - | UBE3C | chr7 | 156979528 | + |
| In-frame | ENST00000540390 | ENST00000348165 | LMBR1 | chr7 | 156685622 | - | UBE3C | chr7 | 156979528 | + |
| In-frame | ENST00000540390 | ENST00000389103 | LMBR1 | chr7 | 156685622 | - | UBE3C | chr7 | 156979528 | + |
| intron-3CDS | ENST00000359422 | ENST00000348165 | LMBR1 | chr7 | 156685622 | - | UBE3C | chr7 | 156979528 | + |
| intron-3CDS | ENST00000359422 | ENST00000389103 | LMBR1 | chr7 | 156685622 | - | UBE3C | chr7 | 156979528 | + |
| intron-3CDS | ENST00000430825 | ENST00000348165 | LMBR1 | chr7 | 156685622 | - | UBE3C | chr7 | 156979528 | + |
| intron-3CDS | ENST00000430825 | ENST00000389103 | LMBR1 | chr7 | 156685622 | - | UBE3C | chr7 | 156979528 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000353442 | LMBR1 | chr7 | 156685622 | - | ENST00000348165 | UBE3C | chr7 | 156979528 | + | 4029 | 303 | 237 | 2411 | 724 |
| ENST00000353442 | LMBR1 | chr7 | 156685622 | - | ENST00000389103 | UBE3C | chr7 | 156979528 | + | 837 | 303 | 577 | 2 | 192 |
| ENST00000354505 | LMBR1 | chr7 | 156685622 | - | ENST00000348165 | UBE3C | chr7 | 156979528 | + | 3946 | 220 | 154 | 2328 | 724 |
| ENST00000354505 | LMBR1 | chr7 | 156685622 | - | ENST00000389103 | UBE3C | chr7 | 156979528 | + | 754 | 220 | 494 | 0 | 165 |
| ENST00000540390 | LMBR1 | chr7 | 156685622 | - | ENST00000348165 | UBE3C | chr7 | 156979528 | + | 3978 | 252 | 186 | 2360 | 724 |
| ENST00000540390 | LMBR1 | chr7 | 156685622 | - | ENST00000389103 | UBE3C | chr7 | 156979528 | + | 786 | 252 | 526 | 2 | 175 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000353442 | ENST00000348165 | LMBR1 | chr7 | 156685622 | - | UBE3C | chr7 | 156979528 | + | 0.001057617 | 0.99894243 |
| ENST00000353442 | ENST00000389103 | LMBR1 | chr7 | 156685622 | - | UBE3C | chr7 | 156979528 | + | 0.23979849 | 0.7602015 |
| ENST00000354505 | ENST00000348165 | LMBR1 | chr7 | 156685622 | - | UBE3C | chr7 | 156979528 | + | 0.000966268 | 0.9990338 |
| ENST00000354505 | ENST00000389103 | LMBR1 | chr7 | 156685622 | - | UBE3C | chr7 | 156979528 | + | 0.25940228 | 0.7405977 |
| ENST00000540390 | ENST00000348165 | LMBR1 | chr7 | 156685622 | - | UBE3C | chr7 | 156979528 | + | 0.001052068 | 0.9989479 |
| ENST00000540390 | ENST00000389103 | LMBR1 | chr7 | 156685622 | - | UBE3C | chr7 | 156979528 | + | 0.25149974 | 0.7485003 |
Top |
Fusion Genomic Features for LMBR1-UBE3C |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| LMBR1 | chr7 | 156685621 | - | UBE3C | chr7 | 156979527 | + | 2.58E-09 | 1 |
| LMBR1 | chr7 | 156685621 | - | UBE3C | chr7 | 156979527 | + | 2.58E-09 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
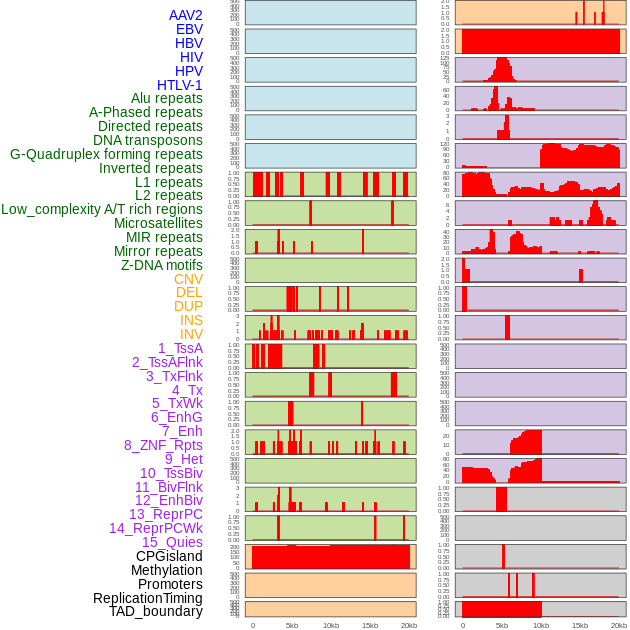 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
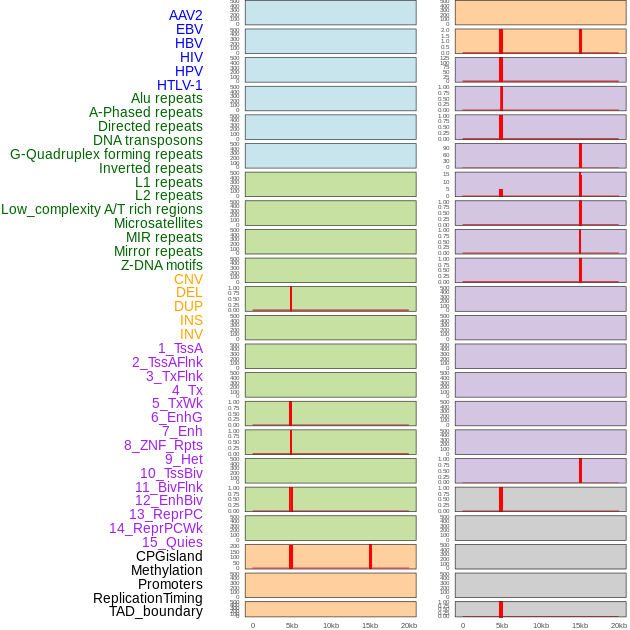 |
Top |
Fusion Protein Features for LMBR1-UBE3C |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr7:156685622/chr7:156979528) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| LMBR1 | . |
| FUNCTION: Putative membrane receptor. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000353442 | - | 1 | 17 | 1_19 | 22 | 491.0 | Topological domain | Extracellular |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000354505 | - | 1 | 18 | 1_19 | 22 | 532.0 | Topological domain | Extracellular |
| Tgene | UBE3C | chr7:156685622 | chr7:156979528 | ENST00000348165 | 8 | 23 | 744_1083 | 381 | 1084.0 | Domain | HECT | |
| Tgene | UBE3C | chr7:156685622 | chr7:156979528 | ENST00000389103 | 6 | 9 | 744_1083 | 338 | 405.0 | Domain | HECT |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000353442 | - | 1 | 17 | 250_287 | 22 | 491.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000354505 | - | 1 | 18 | 250_287 | 22 | 532.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000353442 | - | 1 | 17 | 132_151 | 22 | 491.0 | Topological domain | Cytoplasmic |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000353442 | - | 1 | 17 | 173_187 | 22 | 491.0 | Topological domain | Extracellular |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000353442 | - | 1 | 17 | 209_291 | 22 | 491.0 | Topological domain | Cytoplasmic |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000353442 | - | 1 | 17 | 313_339 | 22 | 491.0 | Topological domain | Extracellular |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000353442 | - | 1 | 17 | 361_383 | 22 | 491.0 | Topological domain | Cytoplasmic |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000353442 | - | 1 | 17 | 405_426 | 22 | 491.0 | Topological domain | Extracellular |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000353442 | - | 1 | 17 | 41_62 | 22 | 491.0 | Topological domain | Cytoplasmic |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000353442 | - | 1 | 17 | 448_490 | 22 | 491.0 | Topological domain | Cytoplasmic |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000353442 | - | 1 | 17 | 84_110 | 22 | 491.0 | Topological domain | Extracellular |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000354505 | - | 1 | 18 | 132_151 | 22 | 532.0 | Topological domain | Cytoplasmic |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000354505 | - | 1 | 18 | 173_187 | 22 | 532.0 | Topological domain | Extracellular |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000354505 | - | 1 | 18 | 209_291 | 22 | 532.0 | Topological domain | Cytoplasmic |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000354505 | - | 1 | 18 | 313_339 | 22 | 532.0 | Topological domain | Extracellular |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000354505 | - | 1 | 18 | 361_383 | 22 | 532.0 | Topological domain | Cytoplasmic |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000354505 | - | 1 | 18 | 405_426 | 22 | 532.0 | Topological domain | Extracellular |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000354505 | - | 1 | 18 | 41_62 | 22 | 532.0 | Topological domain | Cytoplasmic |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000354505 | - | 1 | 18 | 448_490 | 22 | 532.0 | Topological domain | Cytoplasmic |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000354505 | - | 1 | 18 | 84_110 | 22 | 532.0 | Topological domain | Extracellular |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000353442 | - | 1 | 17 | 111_131 | 22 | 491.0 | Transmembrane | Helical |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000353442 | - | 1 | 17 | 152_172 | 22 | 491.0 | Transmembrane | Helical |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000353442 | - | 1 | 17 | 188_208 | 22 | 491.0 | Transmembrane | Helical |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000353442 | - | 1 | 17 | 20_40 | 22 | 491.0 | Transmembrane | Helical |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000353442 | - | 1 | 17 | 292_312 | 22 | 491.0 | Transmembrane | Helical |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000353442 | - | 1 | 17 | 340_360 | 22 | 491.0 | Transmembrane | Helical |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000353442 | - | 1 | 17 | 384_404 | 22 | 491.0 | Transmembrane | Helical |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000353442 | - | 1 | 17 | 427_447 | 22 | 491.0 | Transmembrane | Helical |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000353442 | - | 1 | 17 | 63_83 | 22 | 491.0 | Transmembrane | Helical |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000354505 | - | 1 | 18 | 111_131 | 22 | 532.0 | Transmembrane | Helical |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000354505 | - | 1 | 18 | 152_172 | 22 | 532.0 | Transmembrane | Helical |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000354505 | - | 1 | 18 | 188_208 | 22 | 532.0 | Transmembrane | Helical |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000354505 | - | 1 | 18 | 20_40 | 22 | 532.0 | Transmembrane | Helical |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000354505 | - | 1 | 18 | 292_312 | 22 | 532.0 | Transmembrane | Helical |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000354505 | - | 1 | 18 | 340_360 | 22 | 532.0 | Transmembrane | Helical |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000354505 | - | 1 | 18 | 384_404 | 22 | 532.0 | Transmembrane | Helical |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000354505 | - | 1 | 18 | 427_447 | 22 | 532.0 | Transmembrane | Helical |
| Hgene | LMBR1 | chr7:156685622 | chr7:156979528 | ENST00000354505 | - | 1 | 18 | 63_83 | 22 | 532.0 | Transmembrane | Helical |
| Tgene | UBE3C | chr7:156685622 | chr7:156979528 | ENST00000348165 | 8 | 23 | 45_74 | 381 | 1084.0 | Domain | IQ | |
| Tgene | UBE3C | chr7:156685622 | chr7:156979528 | ENST00000389103 | 6 | 9 | 45_74 | 338 | 405.0 | Domain | IQ | |
| Tgene | UBE3C | chr7:156685622 | chr7:156979528 | ENST00000348165 | 8 | 23 | 1_60 | 381 | 1084.0 | Region | Note=Cis-determinant of acceptor ubiquitin-binding | |
| Tgene | UBE3C | chr7:156685622 | chr7:156979528 | ENST00000389103 | 6 | 9 | 1_60 | 338 | 405.0 | Region | Note=Cis-determinant of acceptor ubiquitin-binding |
Top |
Fusion Gene Sequence for LMBR1-UBE3C |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >45751_45751_1_LMBR1-UBE3C_LMBR1_chr7_156685622_ENST00000353442_UBE3C_chr7_156979528_ENST00000348165_length(transcript)=4029nt_BP=303nt GCCCCGCCCCCGCCCCGCCCCCGACTGCGCGGCGCCCGCCCTCCCTTAGTCAGAGCTGCTCGTCTGAGGCTGCTGAGGCGACGGCCGGTG TCGTGGTCGCGGTACCTGTTCCAACACGGCTCGCGGGCCCGTGCCGGCTCCGGTCCCCGGCGCGGCTGTCCGAGCCCCTGCGGCGGGCGG ACGATGGTGTGGCGGAGCACGCGGACGCGGGCGGCGCGGCGGCGGGCATGAAGGAGGATGGAAGGGCAGGACGAGGTGTCGGCGCGGGAG CAGCACTTCCACAGCCAAGTGCGGGAGTCCACGGAGGATGGCAGACTGTCAGTATCATACATAACAGAGGAATGCCTGAAGAAGCTGGAC ACAAAGCAGCAGACCAACACCCTGCTCAACCTGGTGTGGAGGGACTCTGCGAGCGAGGAGGTCTTCACCACCATGGCCTCCGTCTGCCAC ACGCTGATGGTGCAGCACCGCATGATGGTACCCAAAGTCAGGCTTCTCTACAGTTTAGCCTTTAATGCCAGGTTTCTGAGACATCTTTGG TTTCTAATATCTTCCATGTCAACACGGATGATCACAGGGTCTATGGTACCGTTGCTTCAGGTGATATCCAGGGGTTCTCCTATGTCTTTT GAAGATTCTAGTCGAATCATCCCACTCTTTTATCTTTTTAGCTCCTTGTTTAGTCATTCACTAATTTCCATACATGATAACGAATTCTTC GGTGATCCCATAGAAGTTGTAGGTCAAAGACAATCATCAATGATGCCTTTTACTTTAGAAGAGCTGATAATGTTGTCTCGATGCCTTCGA GATGCATGCCTGGGGATCATCAAGTTGGCTTATCCAGAAACCAAGCCAGAAGTTCGAGAAGAATATATTACAGCATTTCAGAGTATTGGA GTTACTACTAGCTCTGAAATGCAACAATGCATACAGATGGAACAGAAAAGATGGATTCAGTTATTTAAGGTTATCACCAATCTAGTGAAA ATGTTGAAGTCCAGAGACACGAGGAGAAATTTTTGTCCTCCAAACCACTGGCTGTCAGAACAAGAAGATATTAAAGCAGATAAGGTCACT CAGCTCTATGTGCCAGCATCCAGACATGTGTGGAGGTTCCGGCGGATGGGGAGGATAGGCCCGCTGCAGTCCACCCTGGACGTGGGTTTG GAGTCCCCGCCGCTGTCTGTGTCTGAGGAAAGACAGCTTGCTGTCCTGACAGAGTTGCCTTTTGTGGTTCCATTTGAGGAACGAGTAAAG ATCTTTCAGAGGTTGATTTATGCAGATAAGCAAGAAGTTCAAGGAGATGGTCCATTTCTGGATGGAATTAATGTCACAATAAGAAGAAAT TACATTTATGAAGATGCTTATGACAAACTTTCTCCAGAAAATGAGCCTGATTTGAAAAAGCGGATCCGTGTGCACTTGCTCAATGCCCAT GGCCTGGATGAAGCTGGCATTGATGGTGGTGGTATTTTCAGAGAGTTTTTAAATGAACTACTGAAGTCAGGATTTAACCCCAACCAGGGG TTCTTTAAGACTACTAATGAAGGGCTTCTGTACCCCAACCCGGCTGCTCAGATGCTTGTGGGAGATTCTTTTGCCAGACATTACTACTTC CTAGGCAGAATGCTTGGAAAGGCTCTCTATGAGAACATGCTGGTGGAGCTGCCCTTTGCAGGCTTCTTTCTTTCCAAGTTGCTTGGAACC AGTGCCGACGTGGACATTCACCACCTCGCCTCCCTAGACCCTGAGGTGTATAAGAATTTGCTCTTTCTGAAGAGCTACGAAGACGATGTG GAGGAGCTTGGGCTGAACTTCACTGTGGTGAACAATGACCTGGGAGAGGCGCAGGTAGTTGAACTAAAATTCGGTGGGAAAGACATCCCT GTCACCAGCGCCAACCGGATTGCGTACATCCACTTGGTGGCAGACTACAGGCTGAACAGGCAGATCCGCCAGCACTGCCTGGCTTTCCGC CAGGGCCTTGCCAATGTCGTCAGCCTCGAGTGGCTCCGAATGTTTGATCAGCAAGAAATTCAGGTATTAATTTCTGGTGCACAAGTTCCC ATAAGCCTAGAGGACCTAAAATCCTTTACAAACTATTCAGGAGGCTATTCTGCAGACCATCCTGTTATTAAGGTCTTCTGGAGAGTTGTG GAAGGGTTCACTGATGAAGAAAAGCGCAAACTGCTGAAGTTTGTAACAAGCTGCTCTCGACCCCCTCTCTTGGGGTTTAAGGAGTTGTAT CCCGCATTTTGTATTCACAACGGAGGCTCCGACCTTGAGCGGCTCCCCACAGCCAGCACCTGCATGAACCTGCTGAAGCTCCCCGAGTTC TATGACGAGACACTTTTGCGAAGTAAACTTCTCTATGCGATTGAATGTGCCGCTGGCTTTGAGCTGAGCTGAAGCTGATGCTGGGGTCAG ACCCCTACAGAGAACCAGTGCTTCCTTCGTCAGCAGCGCCTCCCCAGACCCACGAGGATACTCACACTGCACGCCTGAGGCTCTCCTAAG CTCCTTCTTTCATTCTGCCATTCCTCCCTCCCTTCCTTTTTTAAATGATTTTTATTACGGTGTGGTCACTTATTTAGATGGACATTGCTT TTCAAATAACTTAAAATAACACGTTATGTGCCATGTGGCTACTTTAGTAATATTGCCAAGAAGAGCACAGTTTTTACACTAGTGGCATCT CAGTGAAATTAACCAAAGATGAAGCTTTGGCTTTGCTGGTGAGATCAGAGCCCTCCTGAGCAGGCAGCGCCACTCCAGGGTTCAGACAGG GCTGCACAGGCGGCAGAGATACAGGGTCTGAGGGCTGAGACGCCATGGGGCCGCTGCTGCTTATGTGGTTGGATTGTTTACAAGCCTCAT TATTAAAACTGAAGGCATTTTTTTTTTCTGCTGCCTTTCCCAAAGTGGTTAGGTTTGGAAAAGAGATGATGATGGTAATATTTTATTTGT GCTTTTTAAGCCATTTCCCCAAATGGGACTAGCATGCTTGTTTTCAGTATACCGTGGCCTGCCTCATGATGGTTTGGAGATACTGTCTGT GGATGTGAGGTGGGGACTTCATTCATTGTCCTATTTCTATCTCCACTTTGTGCCTGGAGAGCTTTCAGGGGAGGTGGAGGAGGAGGGTCT GCCAAGCTACTGCAACATCTGTCACCCACTATACCCAGTTACTTGGGGGAGGACAGACACTGTGGTGTCATTAAAGTTGTTTGAACCAAA GTGGCGGCTGCATCTTTGTCCCGATGCTAGCCGTGCCGGTCTCCCATCATCCGCTCGCCCTCCTTTCCCCTGGGCTGCGCCCACTTGTCT TCCTGGATATTTGGGGGTGACTCGCCATGCTTGGCACCCTCTGCTTCCTGGTGCTGCTCTGACTCGAAGACGGGACAGTCCCTGGTGCAC ATCCAGGGAAGAGGAGTGTCGGTAGTTCTTGCAGTAGGCACTTTATCAGGACCTGACCTGTTGCTGGGTGATTTTAGTCTCTACAAACAG AAAGCGTTTCAAAGCGTCAGCTGTGGGAGCAGAGTGACCCTTTGCTGATGCTGGGGGGAGGGGATCTAAATCCTCATTTATCTCTTCTAT GTCTCGTATTTTACTGTCACTGGAGGCTCTGTGGGCTGTCATAGTTAATTGACCATAATTAGCAATATACTTTTAAAGTGGGAAAGCTGA ATGACACTTTTAAGACAATGAACATTATCAAAACAAAATGTATAATTTCTTAATTTGAATAATAAATTAAGCGTTTAAATGCTATTTGTA GTCTTGATATACAGAAATAAAATAATTAGGGTTGGTCTTTTTTATTTTAGGTTGTTTTATGTTGAATGTTCTATATCTTATTAGTTAATT TGTATATTTTATTAGTATTTTGGAAATAGCATATCTGAGACTGAAGAGAAATTGACAATTCACTTATTTGTGGTTTTTTTCTCAGCTATT >45751_45751_1_LMBR1-UBE3C_LMBR1_chr7_156685622_ENST00000353442_UBE3C_chr7_156979528_ENST00000348165_length(amino acids)=724AA_BP=22 MEGQDEVSAREQHFHSQVRESTEDGRLSVSYITEECLKKLDTKQQTNTLLNLVWRDSASEEVFTTMASVCHTLMVQHRMMVPKVRLLYSL AFNARFLRHLWFLISSMSTRMITGSMVPLLQVISRGSPMSFEDSSRIIPLFYLFSSLFSHSLISIHDNEFFGDPIEVVGQRQSSMMPFTL EELIMLSRCLRDACLGIIKLAYPETKPEVREEYITAFQSIGVTTSSEMQQCIQMEQKRWIQLFKVITNLVKMLKSRDTRRNFCPPNHWLS EQEDIKADKVTQLYVPASRHVWRFRRMGRIGPLQSTLDVGLESPPLSVSEERQLAVLTELPFVVPFEERVKIFQRLIYADKQEVQGDGPF LDGINVTIRRNYIYEDAYDKLSPENEPDLKKRIRVHLLNAHGLDEAGIDGGGIFREFLNELLKSGFNPNQGFFKTTNEGLLYPNPAAQML VGDSFARHYYFLGRMLGKALYENMLVELPFAGFFLSKLLGTSADVDIHHLASLDPEVYKNLLFLKSYEDDVEELGLNFTVVNNDLGEAQV VELKFGGKDIPVTSANRIAYIHLVADYRLNRQIRQHCLAFRQGLANVVSLEWLRMFDQQEIQVLISGAQVPISLEDLKSFTNYSGGYSAD HPVIKVFWRVVEGFTDEEKRKLLKFVTSCSRPPLLGFKELYPAFCIHNGGSDLERLPTASTCMNLLKLPEFYDETLLRSKLLYAIECAAG -------------------------------------------------------------- >45751_45751_2_LMBR1-UBE3C_LMBR1_chr7_156685622_ENST00000353442_UBE3C_chr7_156979528_ENST00000389103_length(transcript)=837nt_BP=303nt GCCCCGCCCCCGCCCCGCCCCCGACTGCGCGGCGCCCGCCCTCCCTTAGTCAGAGCTGCTCGTCTGAGGCTGCTGAGGCGACGGCCGGTG TCGTGGTCGCGGTACCTGTTCCAACACGGCTCGCGGGCCCGTGCCGGCTCCGGTCCCCGGCGCGGCTGTCCGAGCCCCTGCGGCGGGCGG ACGATGGTGTGGCGGAGCACGCGGACGCGGGCGGCGCGGCGGCGGGCATGAAGGAGGATGGAAGGGCAGGACGAGGTGTCGGCGCGGGAG CAGCACTTCCACAGCCAAGTGCGGGAGTCCACGGAGGATGGCAGACTGTCAGTATCATACATAACAGAGGAATGCCTGAAGAAGCTGGAC ACAAAGCAGCAGACCAACACCCTGCTCAACCTGGTGTGGAGGGACTCTGCGAGCGAGGAGGTCTTCACCACCATGGCCTCCGTCTGCCAC ACGCTGATGGTGCAGCACCGCATGATGGTACCCAAAGTCAGGGTGTACAAATGAATGTGGAGACCTTTGTACCCTGACATGGAAAGATTT CCGAGACAGATGCGTACATGAAAACCGGGAAGACACAGGACAGTATGTAGAAGAACCGGCTTTTCCTATAAGAAGGATGGAAAAAATAGG AATATATATTCATATTGCATAAAGAAACTTGGAAGCATATACAAGAAACTCATGAAAGTGAACACTCGTGTCTGAATATGCCGAGGAAGC TGGGGCTGGAGCAGAAGTGCTCTTCTGAGTACACTATTTTTATATCACTTTTAAGTTTTGGATTAGGTGAATGACTTAATATTTTAAAAT >45751_45751_2_LMBR1-UBE3C_LMBR1_chr7_156685622_ENST00000353442_UBE3C_chr7_156979528_ENST00000389103_length(amino acids)=192AA_BP=1 MCLPGFHVRICLGNLSMSGYKGLHIHLYTLTLGTIMRCCTISVWQTEAMVVKTSSLAESLHTRLSRVLVCCFVSSFFRHSSVMYDTDSLP SSVDSRTWLWKCCSRADTSSCPSILLHARRRAARVRVLRHTIVRPPQGLGQPRRGPEPARAREPCWNRYRDHDTGRRLSSLRRAALTKGG -------------------------------------------------------------- >45751_45751_3_LMBR1-UBE3C_LMBR1_chr7_156685622_ENST00000354505_UBE3C_chr7_156979528_ENST00000348165_length(transcript)=3946nt_BP=220nt GCCGGTGTCGTGGTCGCGGTACCTGTTCCAACACGGCTCGCGGGCCCGTGCCGGCTCCGGTCCCCGGCGCGGCTGTCCGAGCCCCTGCGG CGGGCGGACGATGGTGTGGCGGAGCACGCGGACGCGGGCGGCGCGGCGGCGGGCATGAAGGAGGATGGAAGGGCAGGACGAGGTGTCGGC GCGGGAGCAGCACTTCCACAGCCAAGTGCGGGAGTCCACGGAGGATGGCAGACTGTCAGTATCATACATAACAGAGGAATGCCTGAAGAA GCTGGACACAAAGCAGCAGACCAACACCCTGCTCAACCTGGTGTGGAGGGACTCTGCGAGCGAGGAGGTCTTCACCACCATGGCCTCCGT CTGCCACACGCTGATGGTGCAGCACCGCATGATGGTACCCAAAGTCAGGCTTCTCTACAGTTTAGCCTTTAATGCCAGGTTTCTGAGACA TCTTTGGTTTCTAATATCTTCCATGTCAACACGGATGATCACAGGGTCTATGGTACCGTTGCTTCAGGTGATATCCAGGGGTTCTCCTAT GTCTTTTGAAGATTCTAGTCGAATCATCCCACTCTTTTATCTTTTTAGCTCCTTGTTTAGTCATTCACTAATTTCCATACATGATAACGA ATTCTTCGGTGATCCCATAGAAGTTGTAGGTCAAAGACAATCATCAATGATGCCTTTTACTTTAGAAGAGCTGATAATGTTGTCTCGATG CCTTCGAGATGCATGCCTGGGGATCATCAAGTTGGCTTATCCAGAAACCAAGCCAGAAGTTCGAGAAGAATATATTACAGCATTTCAGAG TATTGGAGTTACTACTAGCTCTGAAATGCAACAATGCATACAGATGGAACAGAAAAGATGGATTCAGTTATTTAAGGTTATCACCAATCT AGTGAAAATGTTGAAGTCCAGAGACACGAGGAGAAATTTTTGTCCTCCAAACCACTGGCTGTCAGAACAAGAAGATATTAAAGCAGATAA GGTCACTCAGCTCTATGTGCCAGCATCCAGACATGTGTGGAGGTTCCGGCGGATGGGGAGGATAGGCCCGCTGCAGTCCACCCTGGACGT GGGTTTGGAGTCCCCGCCGCTGTCTGTGTCTGAGGAAAGACAGCTTGCTGTCCTGACAGAGTTGCCTTTTGTGGTTCCATTTGAGGAACG AGTAAAGATCTTTCAGAGGTTGATTTATGCAGATAAGCAAGAAGTTCAAGGAGATGGTCCATTTCTGGATGGAATTAATGTCACAATAAG AAGAAATTACATTTATGAAGATGCTTATGACAAACTTTCTCCAGAAAATGAGCCTGATTTGAAAAAGCGGATCCGTGTGCACTTGCTCAA TGCCCATGGCCTGGATGAAGCTGGCATTGATGGTGGTGGTATTTTCAGAGAGTTTTTAAATGAACTACTGAAGTCAGGATTTAACCCCAA CCAGGGGTTCTTTAAGACTACTAATGAAGGGCTTCTGTACCCCAACCCGGCTGCTCAGATGCTTGTGGGAGATTCTTTTGCCAGACATTA CTACTTCCTAGGCAGAATGCTTGGAAAGGCTCTCTATGAGAACATGCTGGTGGAGCTGCCCTTTGCAGGCTTCTTTCTTTCCAAGTTGCT TGGAACCAGTGCCGACGTGGACATTCACCACCTCGCCTCCCTAGACCCTGAGGTGTATAAGAATTTGCTCTTTCTGAAGAGCTACGAAGA CGATGTGGAGGAGCTTGGGCTGAACTTCACTGTGGTGAACAATGACCTGGGAGAGGCGCAGGTAGTTGAACTAAAATTCGGTGGGAAAGA CATCCCTGTCACCAGCGCCAACCGGATTGCGTACATCCACTTGGTGGCAGACTACAGGCTGAACAGGCAGATCCGCCAGCACTGCCTGGC TTTCCGCCAGGGCCTTGCCAATGTCGTCAGCCTCGAGTGGCTCCGAATGTTTGATCAGCAAGAAATTCAGGTATTAATTTCTGGTGCACA AGTTCCCATAAGCCTAGAGGACCTAAAATCCTTTACAAACTATTCAGGAGGCTATTCTGCAGACCATCCTGTTATTAAGGTCTTCTGGAG AGTTGTGGAAGGGTTCACTGATGAAGAAAAGCGCAAACTGCTGAAGTTTGTAACAAGCTGCTCTCGACCCCCTCTCTTGGGGTTTAAGGA GTTGTATCCCGCATTTTGTATTCACAACGGAGGCTCCGACCTTGAGCGGCTCCCCACAGCCAGCACCTGCATGAACCTGCTGAAGCTCCC CGAGTTCTATGACGAGACACTTTTGCGAAGTAAACTTCTCTATGCGATTGAATGTGCCGCTGGCTTTGAGCTGAGCTGAAGCTGATGCTG GGGTCAGACCCCTACAGAGAACCAGTGCTTCCTTCGTCAGCAGCGCCTCCCCAGACCCACGAGGATACTCACACTGCACGCCTGAGGCTC TCCTAAGCTCCTTCTTTCATTCTGCCATTCCTCCCTCCCTTCCTTTTTTAAATGATTTTTATTACGGTGTGGTCACTTATTTAGATGGAC ATTGCTTTTCAAATAACTTAAAATAACACGTTATGTGCCATGTGGCTACTTTAGTAATATTGCCAAGAAGAGCACAGTTTTTACACTAGT GGCATCTCAGTGAAATTAACCAAAGATGAAGCTTTGGCTTTGCTGGTGAGATCAGAGCCCTCCTGAGCAGGCAGCGCCACTCCAGGGTTC AGACAGGGCTGCACAGGCGGCAGAGATACAGGGTCTGAGGGCTGAGACGCCATGGGGCCGCTGCTGCTTATGTGGTTGGATTGTTTACAA GCCTCATTATTAAAACTGAAGGCATTTTTTTTTTCTGCTGCCTTTCCCAAAGTGGTTAGGTTTGGAAAAGAGATGATGATGGTAATATTT TATTTGTGCTTTTTAAGCCATTTCCCCAAATGGGACTAGCATGCTTGTTTTCAGTATACCGTGGCCTGCCTCATGATGGTTTGGAGATAC TGTCTGTGGATGTGAGGTGGGGACTTCATTCATTGTCCTATTTCTATCTCCACTTTGTGCCTGGAGAGCTTTCAGGGGAGGTGGAGGAGG AGGGTCTGCCAAGCTACTGCAACATCTGTCACCCACTATACCCAGTTACTTGGGGGAGGACAGACACTGTGGTGTCATTAAAGTTGTTTG AACCAAAGTGGCGGCTGCATCTTTGTCCCGATGCTAGCCGTGCCGGTCTCCCATCATCCGCTCGCCCTCCTTTCCCCTGGGCTGCGCCCA CTTGTCTTCCTGGATATTTGGGGGTGACTCGCCATGCTTGGCACCCTCTGCTTCCTGGTGCTGCTCTGACTCGAAGACGGGACAGTCCCT GGTGCACATCCAGGGAAGAGGAGTGTCGGTAGTTCTTGCAGTAGGCACTTTATCAGGACCTGACCTGTTGCTGGGTGATTTTAGTCTCTA CAAACAGAAAGCGTTTCAAAGCGTCAGCTGTGGGAGCAGAGTGACCCTTTGCTGATGCTGGGGGGAGGGGATCTAAATCCTCATTTATCT CTTCTATGTCTCGTATTTTACTGTCACTGGAGGCTCTGTGGGCTGTCATAGTTAATTGACCATAATTAGCAATATACTTTTAAAGTGGGA AAGCTGAATGACACTTTTAAGACAATGAACATTATCAAAACAAAATGTATAATTTCTTAATTTGAATAATAAATTAAGCGTTTAAATGCT ATTTGTAGTCTTGATATACAGAAATAAAATAATTAGGGTTGGTCTTTTTTATTTTAGGTTGTTTTATGTTGAATGTTCTATATCTTATTA GTTAATTTGTATATTTTATTAGTATTTTGGAAATAGCATATCTGAGACTGAAGAGAAATTGACAATTCACTTATTTGTGGTTTTTTTCTC >45751_45751_3_LMBR1-UBE3C_LMBR1_chr7_156685622_ENST00000354505_UBE3C_chr7_156979528_ENST00000348165_length(amino acids)=724AA_BP=22 MEGQDEVSAREQHFHSQVRESTEDGRLSVSYITEECLKKLDTKQQTNTLLNLVWRDSASEEVFTTMASVCHTLMVQHRMMVPKVRLLYSL AFNARFLRHLWFLISSMSTRMITGSMVPLLQVISRGSPMSFEDSSRIIPLFYLFSSLFSHSLISIHDNEFFGDPIEVVGQRQSSMMPFTL EELIMLSRCLRDACLGIIKLAYPETKPEVREEYITAFQSIGVTTSSEMQQCIQMEQKRWIQLFKVITNLVKMLKSRDTRRNFCPPNHWLS EQEDIKADKVTQLYVPASRHVWRFRRMGRIGPLQSTLDVGLESPPLSVSEERQLAVLTELPFVVPFEERVKIFQRLIYADKQEVQGDGPF LDGINVTIRRNYIYEDAYDKLSPENEPDLKKRIRVHLLNAHGLDEAGIDGGGIFREFLNELLKSGFNPNQGFFKTTNEGLLYPNPAAQML VGDSFARHYYFLGRMLGKALYENMLVELPFAGFFLSKLLGTSADVDIHHLASLDPEVYKNLLFLKSYEDDVEELGLNFTVVNNDLGEAQV VELKFGGKDIPVTSANRIAYIHLVADYRLNRQIRQHCLAFRQGLANVVSLEWLRMFDQQEIQVLISGAQVPISLEDLKSFTNYSGGYSAD HPVIKVFWRVVEGFTDEEKRKLLKFVTSCSRPPLLGFKELYPAFCIHNGGSDLERLPTASTCMNLLKLPEFYDETLLRSKLLYAIECAAG -------------------------------------------------------------- >45751_45751_4_LMBR1-UBE3C_LMBR1_chr7_156685622_ENST00000354505_UBE3C_chr7_156979528_ENST00000389103_length(transcript)=754nt_BP=220nt GCCGGTGTCGTGGTCGCGGTACCTGTTCCAACACGGCTCGCGGGCCCGTGCCGGCTCCGGTCCCCGGCGCGGCTGTCCGAGCCCCTGCGG CGGGCGGACGATGGTGTGGCGGAGCACGCGGACGCGGGCGGCGCGGCGGCGGGCATGAAGGAGGATGGAAGGGCAGGACGAGGTGTCGGC GCGGGAGCAGCACTTCCACAGCCAAGTGCGGGAGTCCACGGAGGATGGCAGACTGTCAGTATCATACATAACAGAGGAATGCCTGAAGAA GCTGGACACAAAGCAGCAGACCAACACCCTGCTCAACCTGGTGTGGAGGGACTCTGCGAGCGAGGAGGTCTTCACCACCATGGCCTCCGT CTGCCACACGCTGATGGTGCAGCACCGCATGATGGTACCCAAAGTCAGGGTGTACAAATGAATGTGGAGACCTTTGTACCCTGACATGGA AAGATTTCCGAGACAGATGCGTACATGAAAACCGGGAAGACACAGGACAGTATGTAGAAGAACCGGCTTTTCCTATAAGAAGGATGGAAA AAATAGGAATATATATTCATATTGCATAAAGAAACTTGGAAGCATATACAAGAAACTCATGAAAGTGAACACTCGTGTCTGAATATGCCG AGGAAGCTGGGGCTGGAGCAGAAGTGCTCTTCTGAGTACACTATTTTTATATCACTTTTAAGTTTTGGATTAGGTGAATGACTTAATATT >45751_45751_4_LMBR1-UBE3C_LMBR1_chr7_156685622_ENST00000354505_UBE3C_chr7_156979528_ENST00000389103_length(amino acids)=165AA_BP=1 MCLPGFHVRICLGNLSMSGYKGLHIHLYTLTLGTIMRCCTISVWQTEAMVVKTSSLAESLHTRLSRVLVCCFVSSFFRHSSVMYDTDSLP -------------------------------------------------------------- >45751_45751_5_LMBR1-UBE3C_LMBR1_chr7_156685622_ENST00000540390_UBE3C_chr7_156979528_ENST00000348165_length(transcript)=3978nt_BP=252nt AGAGCTGCTCGTCTGAGGCTGCTGAGGCGACGGCCGGTGTCGTGGTCGCGGTACCTGTTCCAACACGGCTCGCGGGCCCGTGCCGGCTCC GGTCCCCGGCGCGGCTGTCCGAGCCCCTGCGGCGGGCGGACGATGGTGTGGCGGAGCACGCGGACGCGGGCGGCGCGGCGGCGGGCATGA AGGAGGATGGAAGGGCAGGACGAGGTGTCGGCGCGGGAGCAGCACTTCCACAGCCAAGTGCGGGAGTCCACGGAGGATGGCAGACTGTCA GTATCATACATAACAGAGGAATGCCTGAAGAAGCTGGACACAAAGCAGCAGACCAACACCCTGCTCAACCTGGTGTGGAGGGACTCTGCG AGCGAGGAGGTCTTCACCACCATGGCCTCCGTCTGCCACACGCTGATGGTGCAGCACCGCATGATGGTACCCAAAGTCAGGCTTCTCTAC AGTTTAGCCTTTAATGCCAGGTTTCTGAGACATCTTTGGTTTCTAATATCTTCCATGTCAACACGGATGATCACAGGGTCTATGGTACCG TTGCTTCAGGTGATATCCAGGGGTTCTCCTATGTCTTTTGAAGATTCTAGTCGAATCATCCCACTCTTTTATCTTTTTAGCTCCTTGTTT AGTCATTCACTAATTTCCATACATGATAACGAATTCTTCGGTGATCCCATAGAAGTTGTAGGTCAAAGACAATCATCAATGATGCCTTTT ACTTTAGAAGAGCTGATAATGTTGTCTCGATGCCTTCGAGATGCATGCCTGGGGATCATCAAGTTGGCTTATCCAGAAACCAAGCCAGAA GTTCGAGAAGAATATATTACAGCATTTCAGAGTATTGGAGTTACTACTAGCTCTGAAATGCAACAATGCATACAGATGGAACAGAAAAGA TGGATTCAGTTATTTAAGGTTATCACCAATCTAGTGAAAATGTTGAAGTCCAGAGACACGAGGAGAAATTTTTGTCCTCCAAACCACTGG CTGTCAGAACAAGAAGATATTAAAGCAGATAAGGTCACTCAGCTCTATGTGCCAGCATCCAGACATGTGTGGAGGTTCCGGCGGATGGGG AGGATAGGCCCGCTGCAGTCCACCCTGGACGTGGGTTTGGAGTCCCCGCCGCTGTCTGTGTCTGAGGAAAGACAGCTTGCTGTCCTGACA GAGTTGCCTTTTGTGGTTCCATTTGAGGAACGAGTAAAGATCTTTCAGAGGTTGATTTATGCAGATAAGCAAGAAGTTCAAGGAGATGGT CCATTTCTGGATGGAATTAATGTCACAATAAGAAGAAATTACATTTATGAAGATGCTTATGACAAACTTTCTCCAGAAAATGAGCCTGAT TTGAAAAAGCGGATCCGTGTGCACTTGCTCAATGCCCATGGCCTGGATGAAGCTGGCATTGATGGTGGTGGTATTTTCAGAGAGTTTTTA AATGAACTACTGAAGTCAGGATTTAACCCCAACCAGGGGTTCTTTAAGACTACTAATGAAGGGCTTCTGTACCCCAACCCGGCTGCTCAG ATGCTTGTGGGAGATTCTTTTGCCAGACATTACTACTTCCTAGGCAGAATGCTTGGAAAGGCTCTCTATGAGAACATGCTGGTGGAGCTG CCCTTTGCAGGCTTCTTTCTTTCCAAGTTGCTTGGAACCAGTGCCGACGTGGACATTCACCACCTCGCCTCCCTAGACCCTGAGGTGTAT AAGAATTTGCTCTTTCTGAAGAGCTACGAAGACGATGTGGAGGAGCTTGGGCTGAACTTCACTGTGGTGAACAATGACCTGGGAGAGGCG CAGGTAGTTGAACTAAAATTCGGTGGGAAAGACATCCCTGTCACCAGCGCCAACCGGATTGCGTACATCCACTTGGTGGCAGACTACAGG CTGAACAGGCAGATCCGCCAGCACTGCCTGGCTTTCCGCCAGGGCCTTGCCAATGTCGTCAGCCTCGAGTGGCTCCGAATGTTTGATCAG CAAGAAATTCAGGTATTAATTTCTGGTGCACAAGTTCCCATAAGCCTAGAGGACCTAAAATCCTTTACAAACTATTCAGGAGGCTATTCT GCAGACCATCCTGTTATTAAGGTCTTCTGGAGAGTTGTGGAAGGGTTCACTGATGAAGAAAAGCGCAAACTGCTGAAGTTTGTAACAAGC TGCTCTCGACCCCCTCTCTTGGGGTTTAAGGAGTTGTATCCCGCATTTTGTATTCACAACGGAGGCTCCGACCTTGAGCGGCTCCCCACA GCCAGCACCTGCATGAACCTGCTGAAGCTCCCCGAGTTCTATGACGAGACACTTTTGCGAAGTAAACTTCTCTATGCGATTGAATGTGCC GCTGGCTTTGAGCTGAGCTGAAGCTGATGCTGGGGTCAGACCCCTACAGAGAACCAGTGCTTCCTTCGTCAGCAGCGCCTCCCCAGACCC ACGAGGATACTCACACTGCACGCCTGAGGCTCTCCTAAGCTCCTTCTTTCATTCTGCCATTCCTCCCTCCCTTCCTTTTTTAAATGATTT TTATTACGGTGTGGTCACTTATTTAGATGGACATTGCTTTTCAAATAACTTAAAATAACACGTTATGTGCCATGTGGCTACTTTAGTAAT ATTGCCAAGAAGAGCACAGTTTTTACACTAGTGGCATCTCAGTGAAATTAACCAAAGATGAAGCTTTGGCTTTGCTGGTGAGATCAGAGC CCTCCTGAGCAGGCAGCGCCACTCCAGGGTTCAGACAGGGCTGCACAGGCGGCAGAGATACAGGGTCTGAGGGCTGAGACGCCATGGGGC CGCTGCTGCTTATGTGGTTGGATTGTTTACAAGCCTCATTATTAAAACTGAAGGCATTTTTTTTTTCTGCTGCCTTTCCCAAAGTGGTTA GGTTTGGAAAAGAGATGATGATGGTAATATTTTATTTGTGCTTTTTAAGCCATTTCCCCAAATGGGACTAGCATGCTTGTTTTCAGTATA CCGTGGCCTGCCTCATGATGGTTTGGAGATACTGTCTGTGGATGTGAGGTGGGGACTTCATTCATTGTCCTATTTCTATCTCCACTTTGT GCCTGGAGAGCTTTCAGGGGAGGTGGAGGAGGAGGGTCTGCCAAGCTACTGCAACATCTGTCACCCACTATACCCAGTTACTTGGGGGAG GACAGACACTGTGGTGTCATTAAAGTTGTTTGAACCAAAGTGGCGGCTGCATCTTTGTCCCGATGCTAGCCGTGCCGGTCTCCCATCATC CGCTCGCCCTCCTTTCCCCTGGGCTGCGCCCACTTGTCTTCCTGGATATTTGGGGGTGACTCGCCATGCTTGGCACCCTCTGCTTCCTGG TGCTGCTCTGACTCGAAGACGGGACAGTCCCTGGTGCACATCCAGGGAAGAGGAGTGTCGGTAGTTCTTGCAGTAGGCACTTTATCAGGA CCTGACCTGTTGCTGGGTGATTTTAGTCTCTACAAACAGAAAGCGTTTCAAAGCGTCAGCTGTGGGAGCAGAGTGACCCTTTGCTGATGC TGGGGGGAGGGGATCTAAATCCTCATTTATCTCTTCTATGTCTCGTATTTTACTGTCACTGGAGGCTCTGTGGGCTGTCATAGTTAATTG ACCATAATTAGCAATATACTTTTAAAGTGGGAAAGCTGAATGACACTTTTAAGACAATGAACATTATCAAAACAAAATGTATAATTTCTT AATTTGAATAATAAATTAAGCGTTTAAATGCTATTTGTAGTCTTGATATACAGAAATAAAATAATTAGGGTTGGTCTTTTTTATTTTAGG TTGTTTTATGTTGAATGTTCTATATCTTATTAGTTAATTTGTATATTTTATTAGTATTTTGGAAATAGCATATCTGAGACTGAAGAGAAA TTGACAATTCACTTATTTGTGGTTTTTTTCTCAGCTATTCTGAGCTTATTTATTTATTTGTATGTTCTAATGGCTAAACATTTACATTAA >45751_45751_5_LMBR1-UBE3C_LMBR1_chr7_156685622_ENST00000540390_UBE3C_chr7_156979528_ENST00000348165_length(amino acids)=724AA_BP=22 MEGQDEVSAREQHFHSQVRESTEDGRLSVSYITEECLKKLDTKQQTNTLLNLVWRDSASEEVFTTMASVCHTLMVQHRMMVPKVRLLYSL AFNARFLRHLWFLISSMSTRMITGSMVPLLQVISRGSPMSFEDSSRIIPLFYLFSSLFSHSLISIHDNEFFGDPIEVVGQRQSSMMPFTL EELIMLSRCLRDACLGIIKLAYPETKPEVREEYITAFQSIGVTTSSEMQQCIQMEQKRWIQLFKVITNLVKMLKSRDTRRNFCPPNHWLS EQEDIKADKVTQLYVPASRHVWRFRRMGRIGPLQSTLDVGLESPPLSVSEERQLAVLTELPFVVPFEERVKIFQRLIYADKQEVQGDGPF LDGINVTIRRNYIYEDAYDKLSPENEPDLKKRIRVHLLNAHGLDEAGIDGGGIFREFLNELLKSGFNPNQGFFKTTNEGLLYPNPAAQML VGDSFARHYYFLGRMLGKALYENMLVELPFAGFFLSKLLGTSADVDIHHLASLDPEVYKNLLFLKSYEDDVEELGLNFTVVNNDLGEAQV VELKFGGKDIPVTSANRIAYIHLVADYRLNRQIRQHCLAFRQGLANVVSLEWLRMFDQQEIQVLISGAQVPISLEDLKSFTNYSGGYSAD HPVIKVFWRVVEGFTDEEKRKLLKFVTSCSRPPLLGFKELYPAFCIHNGGSDLERLPTASTCMNLLKLPEFYDETLLRSKLLYAIECAAG -------------------------------------------------------------- >45751_45751_6_LMBR1-UBE3C_LMBR1_chr7_156685622_ENST00000540390_UBE3C_chr7_156979528_ENST00000389103_length(transcript)=786nt_BP=252nt AGAGCTGCTCGTCTGAGGCTGCTGAGGCGACGGCCGGTGTCGTGGTCGCGGTACCTGTTCCAACACGGCTCGCGGGCCCGTGCCGGCTCC GGTCCCCGGCGCGGCTGTCCGAGCCCCTGCGGCGGGCGGACGATGGTGTGGCGGAGCACGCGGACGCGGGCGGCGCGGCGGCGGGCATGA AGGAGGATGGAAGGGCAGGACGAGGTGTCGGCGCGGGAGCAGCACTTCCACAGCCAAGTGCGGGAGTCCACGGAGGATGGCAGACTGTCA GTATCATACATAACAGAGGAATGCCTGAAGAAGCTGGACACAAAGCAGCAGACCAACACCCTGCTCAACCTGGTGTGGAGGGACTCTGCG AGCGAGGAGGTCTTCACCACCATGGCCTCCGTCTGCCACACGCTGATGGTGCAGCACCGCATGATGGTACCCAAAGTCAGGGTGTACAAA TGAATGTGGAGACCTTTGTACCCTGACATGGAAAGATTTCCGAGACAGATGCGTACATGAAAACCGGGAAGACACAGGACAGTATGTAGA AGAACCGGCTTTTCCTATAAGAAGGATGGAAAAAATAGGAATATATATTCATATTGCATAAAGAAACTTGGAAGCATATACAAGAAACTC ATGAAAGTGAACACTCGTGTCTGAATATGCCGAGGAAGCTGGGGCTGGAGCAGAAGTGCTCTTCTGAGTACACTATTTTTATATCACTTT >45751_45751_6_LMBR1-UBE3C_LMBR1_chr7_156685622_ENST00000540390_UBE3C_chr7_156979528_ENST00000389103_length(amino acids)=175AA_BP=1 MCLPGFHVRICLGNLSMSGYKGLHIHLYTLTLGTIMRCCTISVWQTEAMVVKTSSLAESLHTRLSRVLVCCFVSSFFRHSSVMYDTDSLP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for LMBR1-UBE3C |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for LMBR1-UBE3C |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for LMBR1-UBE3C |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
