
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ANKRD44-NFATC3 (FusionGDB2 ID:4742) |
Fusion Gene Summary for ANKRD44-NFATC3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ANKRD44-NFATC3 | Fusion gene ID: 4742 | Hgene | Tgene | Gene symbol | ANKRD44 | NFATC3 | Gene ID | 91526 | 4775 |
| Gene name | ankyrin repeat domain 44 | nuclear factor of activated T cells 3 | |
| Synonyms | PP6-ARS-B | NF-AT4c|NFAT4|NFATX | |
| Cytomap | 2q33.1 | 16q22.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit Bankyrin repeat domain-containing protein 44serine/threonine-protein phosphatase 6 regulatory subunit ARS-B | nuclear factor of activated T-cells, cytoplasmic 3NF-ATc3T cell transcription factor NFAT4nuclear factor of activated T-cells c3 isoform IE-Xanuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | Q12968 | |
| Ensembl transtripts involved in fusion gene | ENST00000282272, ENST00000328737, ENST00000409153, ENST00000409919, ENST00000450567, ENST00000539527, ENST00000337207, ENST00000477852, | ENST00000535127, ENST00000329524, ENST00000346183, ENST00000349223, ENST00000575270, | |
| Fusion gene scores | * DoF score | 11 X 6 X 6=396 | 15 X 17 X 5=1275 |
| # samples | 11 | 17 | |
| ** MAII score | log2(11/396*10)=-1.84799690655495 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(17/1275*10)=-2.90689059560852 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ANKRD44 [Title/Abstract] AND NFATC3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ANKRD44(198051747)-NFATC3(68200746), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | ANKRD44-NFATC3 seems lost the major protein functional domain in Tgene partner, which is a transcription factor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | NFATC3 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 18815128 |
| Tgene | NFATC3 | GO:1905064 | negative regulation of vascular smooth muscle cell differentiation | 23853098 |
 Fusion gene breakpoints across ANKRD44 (5'-gene) Fusion gene breakpoints across ANKRD44 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
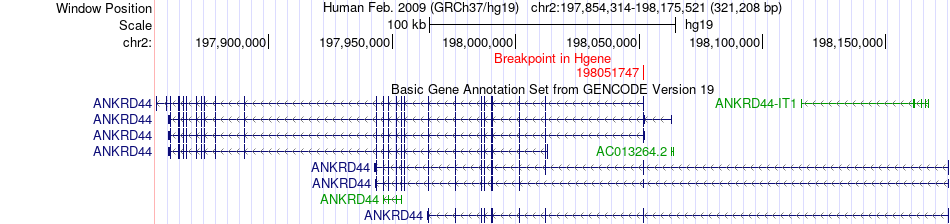 |
 Fusion gene breakpoints across NFATC3 (3'-gene) Fusion gene breakpoints across NFATC3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
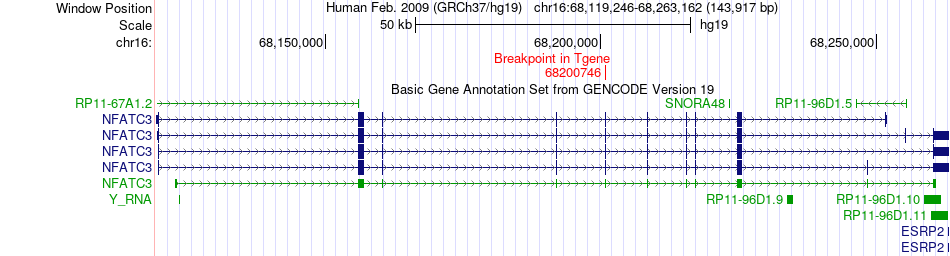 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-HU-A4GJ-01A | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
Top |
Fusion Gene ORF analysis for ANKRD44-NFATC3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000282272 | ENST00000535127 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| 5CDS-3UTR | ENST00000328737 | ENST00000535127 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| 5CDS-3UTR | ENST00000409153 | ENST00000535127 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| 5CDS-3UTR | ENST00000409919 | ENST00000535127 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| 5CDS-3UTR | ENST00000450567 | ENST00000535127 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| 5UTR-3CDS | ENST00000539527 | ENST00000329524 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| 5UTR-3CDS | ENST00000539527 | ENST00000346183 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| 5UTR-3CDS | ENST00000539527 | ENST00000349223 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| 5UTR-3CDS | ENST00000539527 | ENST00000575270 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| 5UTR-3UTR | ENST00000539527 | ENST00000535127 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| Frame-shift | ENST00000282272 | ENST00000329524 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| Frame-shift | ENST00000282272 | ENST00000346183 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| Frame-shift | ENST00000282272 | ENST00000349223 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| Frame-shift | ENST00000282272 | ENST00000575270 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| Frame-shift | ENST00000409153 | ENST00000329524 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| Frame-shift | ENST00000409153 | ENST00000346183 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| Frame-shift | ENST00000409153 | ENST00000349223 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| Frame-shift | ENST00000409153 | ENST00000575270 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| Frame-shift | ENST00000409919 | ENST00000329524 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| Frame-shift | ENST00000409919 | ENST00000346183 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| Frame-shift | ENST00000409919 | ENST00000349223 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| Frame-shift | ENST00000409919 | ENST00000575270 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| Frame-shift | ENST00000450567 | ENST00000329524 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| Frame-shift | ENST00000450567 | ENST00000346183 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| Frame-shift | ENST00000450567 | ENST00000349223 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| Frame-shift | ENST00000450567 | ENST00000575270 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| In-frame | ENST00000328737 | ENST00000329524 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| In-frame | ENST00000328737 | ENST00000346183 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| In-frame | ENST00000328737 | ENST00000349223 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| In-frame | ENST00000328737 | ENST00000575270 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| intron-3CDS | ENST00000337207 | ENST00000329524 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| intron-3CDS | ENST00000337207 | ENST00000346183 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| intron-3CDS | ENST00000337207 | ENST00000349223 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| intron-3CDS | ENST00000337207 | ENST00000575270 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| intron-3CDS | ENST00000477852 | ENST00000329524 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| intron-3CDS | ENST00000477852 | ENST00000346183 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| intron-3CDS | ENST00000477852 | ENST00000349223 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| intron-3CDS | ENST00000477852 | ENST00000575270 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| intron-3UTR | ENST00000337207 | ENST00000535127 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
| intron-3UTR | ENST00000477852 | ENST00000535127 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000328737 | ANKRD44 | chr2 | 198051747 | - | ENST00000575270 | NFATC3 | chr16 | 68200746 | + | 1998 | 113 | 132 | 1655 | 507 |
| ENST00000328737 | ANKRD44 | chr2 | 198051747 | - | ENST00000349223 | NFATC3 | chr16 | 68200746 | + | 4616 | 113 | 132 | 1709 | 525 |
| ENST00000328737 | ANKRD44 | chr2 | 198051747 | - | ENST00000329524 | NFATC3 | chr16 | 68200746 | + | 4632 | 113 | 132 | 1718 | 528 |
| ENST00000328737 | ANKRD44 | chr2 | 198051747 | - | ENST00000346183 | NFATC3 | chr16 | 68200746 | + | 4528 | 113 | 132 | 1739 | 535 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000328737 | ENST00000575270 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + | 0.01334272 | 0.9866573 |
| ENST00000328737 | ENST00000349223 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + | 0.007446815 | 0.9925532 |
| ENST00000328737 | ENST00000329524 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + | 0.009067248 | 0.9909327 |
| ENST00000328737 | ENST00000346183 | ANKRD44 | chr2 | 198051747 | - | NFATC3 | chr16 | 68200746 | + | 0.004823918 | 0.9951761 |
Top |
Fusion Genomic Features for ANKRD44-NFATC3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| ANKRD44 | chr2 | 198051746 | - | NFATC3 | chr16 | 68200745 | + | 1.48E-08 | 1 |
| ANKRD44 | chr2 | 198051746 | - | NFATC3 | chr16 | 68200745 | + | 1.48E-08 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
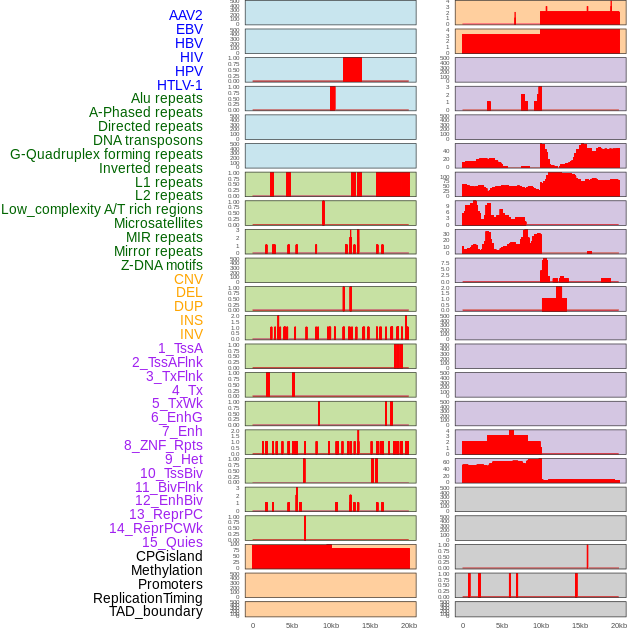 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
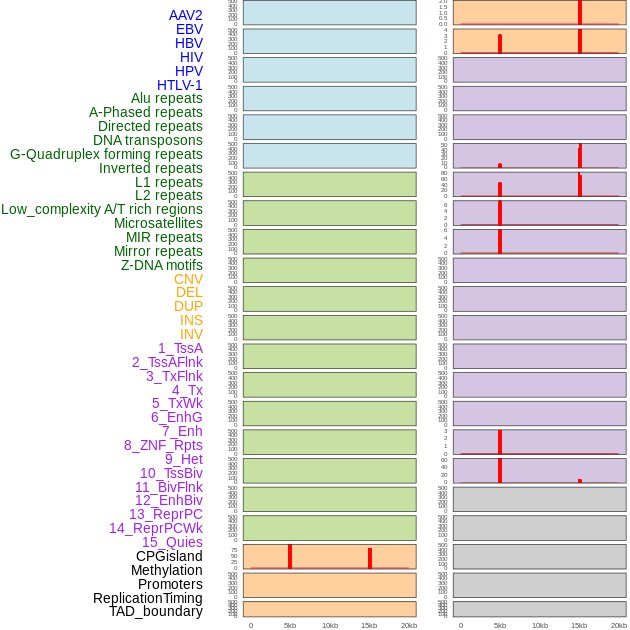 |
Top |
Fusion Protein Features for ANKRD44-NFATC3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:198051747/chr16:68200746) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | NFATC3 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Acts as a regulator of transcriptional activation. Plays a role in the inducible expression of cytokine genes in T-cells, especially in the induction of the IL-2 (PubMed:18815128). Along with NFATC4, involved in embryonic heart development (By similarity). {ECO:0000250|UniProtKB:P97305, ECO:0000269|PubMed:18815128}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 7_36 | 37 | 580.0 | Repeat | Note=ANK 1 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 7_36 | 37 | 368.0 | Repeat | Note=ANK 1 |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000329524 | 3 | 11 | 1032_1041 | 533 | 1996.6666666666667 | Motif | Note=Nuclear export signal | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000329524 | 3 | 11 | 686_688 | 533 | 1996.6666666666667 | Motif | Note=Nuclear localization signal | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000346183 | 3 | 10 | 1032_1041 | 533 | 1076.0 | Motif | Note=Nuclear export signal | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000346183 | 3 | 10 | 686_688 | 533 | 1076.0 | Motif | Note=Nuclear localization signal | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000349223 | 3 | 11 | 1032_1041 | 533 | 1066.0 | Motif | Note=Nuclear export signal | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000349223 | 3 | 11 | 686_688 | 533 | 1066.0 | Motif | Note=Nuclear localization signal |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 106_135 | 12 | 920.0 | Repeat | Note=ANK 4 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 139_168 | 12 | 920.0 | Repeat | Note=ANK 5 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 172_201 | 12 | 920.0 | Repeat | Note=ANK 6 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 205_234 | 12 | 920.0 | Repeat | Note=ANK 7 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 238_267 | 12 | 920.0 | Repeat | Note=ANK 8 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 271_301 | 12 | 920.0 | Repeat | Note=ANK 9 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 305_334 | 12 | 920.0 | Repeat | Note=ANK 10 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 338_367 | 12 | 920.0 | Repeat | Note=ANK 11 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 371_400 | 12 | 920.0 | Repeat | Note=ANK 12 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 404_433 | 12 | 920.0 | Repeat | Note=ANK 13 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 40_69 | 12 | 920.0 | Repeat | Note=ANK 2 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 437_466 | 12 | 920.0 | Repeat | Note=ANK 14 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 470_498 | 12 | 920.0 | Repeat | Note=ANK 15 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 531_561 | 12 | 920.0 | Repeat | Note=ANK 16 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 566_595 | 12 | 920.0 | Repeat | Note=ANK 17 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 599_628 | 12 | 920.0 | Repeat | Note=ANK 18 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 633_662 | 12 | 920.0 | Repeat | Note=ANK 19 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 669_698 | 12 | 920.0 | Repeat | Note=ANK 20 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 702_731 | 12 | 920.0 | Repeat | Note=ANK 21 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 735_764 | 12 | 920.0 | Repeat | Note=ANK 22 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 73_102 | 12 | 920.0 | Repeat | Note=ANK 3 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 771_800 | 12 | 920.0 | Repeat | Note=ANK 23 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 7_36 | 12 | 920.0 | Repeat | Note=ANK 1 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 803_832 | 12 | 920.0 | Repeat | Note=ANK 24 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 838_867 | 12 | 920.0 | Repeat | Note=ANK 25 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 871_901 | 12 | 920.0 | Repeat | Note=ANK 26 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 905_934 | 12 | 920.0 | Repeat | Note=ANK 27 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000328737 | - | 2 | 26 | 941_970 | 12 | 920.0 | Repeat | Note=ANK 28 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 106_135 | 37 | 580.0 | Repeat | Note=ANK 4 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 139_168 | 37 | 580.0 | Repeat | Note=ANK 5 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 172_201 | 37 | 580.0 | Repeat | Note=ANK 6 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 205_234 | 37 | 580.0 | Repeat | Note=ANK 7 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 238_267 | 37 | 580.0 | Repeat | Note=ANK 8 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 271_301 | 37 | 580.0 | Repeat | Note=ANK 9 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 305_334 | 37 | 580.0 | Repeat | Note=ANK 10 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 338_367 | 37 | 580.0 | Repeat | Note=ANK 11 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 371_400 | 37 | 580.0 | Repeat | Note=ANK 12 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 404_433 | 37 | 580.0 | Repeat | Note=ANK 13 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 40_69 | 37 | 580.0 | Repeat | Note=ANK 2 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 437_466 | 37 | 580.0 | Repeat | Note=ANK 14 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 470_498 | 37 | 580.0 | Repeat | Note=ANK 15 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 531_561 | 37 | 580.0 | Repeat | Note=ANK 16 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 566_595 | 37 | 580.0 | Repeat | Note=ANK 17 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 599_628 | 37 | 580.0 | Repeat | Note=ANK 18 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 633_662 | 37 | 580.0 | Repeat | Note=ANK 19 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 669_698 | 37 | 580.0 | Repeat | Note=ANK 20 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 702_731 | 37 | 580.0 | Repeat | Note=ANK 21 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 735_764 | 37 | 580.0 | Repeat | Note=ANK 22 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 73_102 | 37 | 580.0 | Repeat | Note=ANK 3 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 771_800 | 37 | 580.0 | Repeat | Note=ANK 23 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 803_832 | 37 | 580.0 | Repeat | Note=ANK 24 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 838_867 | 37 | 580.0 | Repeat | Note=ANK 25 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 871_901 | 37 | 580.0 | Repeat | Note=ANK 26 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 905_934 | 37 | 580.0 | Repeat | Note=ANK 27 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409153 | - | 2 | 16 | 941_970 | 37 | 580.0 | Repeat | Note=ANK 28 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 106_135 | 37 | 368.0 | Repeat | Note=ANK 4 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 139_168 | 37 | 368.0 | Repeat | Note=ANK 5 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 172_201 | 37 | 368.0 | Repeat | Note=ANK 6 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 205_234 | 37 | 368.0 | Repeat | Note=ANK 7 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 238_267 | 37 | 368.0 | Repeat | Note=ANK 8 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 271_301 | 37 | 368.0 | Repeat | Note=ANK 9 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 305_334 | 37 | 368.0 | Repeat | Note=ANK 10 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 338_367 | 37 | 368.0 | Repeat | Note=ANK 11 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 371_400 | 37 | 368.0 | Repeat | Note=ANK 12 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 404_433 | 37 | 368.0 | Repeat | Note=ANK 13 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 40_69 | 37 | 368.0 | Repeat | Note=ANK 2 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 437_466 | 37 | 368.0 | Repeat | Note=ANK 14 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 470_498 | 37 | 368.0 | Repeat | Note=ANK 15 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 531_561 | 37 | 368.0 | Repeat | Note=ANK 16 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 566_595 | 37 | 368.0 | Repeat | Note=ANK 17 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 599_628 | 37 | 368.0 | Repeat | Note=ANK 18 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 633_662 | 37 | 368.0 | Repeat | Note=ANK 19 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 669_698 | 37 | 368.0 | Repeat | Note=ANK 20 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 702_731 | 37 | 368.0 | Repeat | Note=ANK 21 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 735_764 | 37 | 368.0 | Repeat | Note=ANK 22 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 73_102 | 37 | 368.0 | Repeat | Note=ANK 3 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 771_800 | 37 | 368.0 | Repeat | Note=ANK 23 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 803_832 | 37 | 368.0 | Repeat | Note=ANK 24 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 838_867 | 37 | 368.0 | Repeat | Note=ANK 25 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 871_901 | 37 | 368.0 | Repeat | Note=ANK 26 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 905_934 | 37 | 368.0 | Repeat | Note=ANK 27 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000409919 | - | 2 | 10 | 941_970 | 37 | 368.0 | Repeat | Note=ANK 28 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 106_135 | 12 | 920.0 | Repeat | Note=ANK 4 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 139_168 | 12 | 920.0 | Repeat | Note=ANK 5 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 172_201 | 12 | 920.0 | Repeat | Note=ANK 6 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 205_234 | 12 | 920.0 | Repeat | Note=ANK 7 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 238_267 | 12 | 920.0 | Repeat | Note=ANK 8 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 271_301 | 12 | 920.0 | Repeat | Note=ANK 9 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 305_334 | 12 | 920.0 | Repeat | Note=ANK 10 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 338_367 | 12 | 920.0 | Repeat | Note=ANK 11 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 371_400 | 12 | 920.0 | Repeat | Note=ANK 12 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 404_433 | 12 | 920.0 | Repeat | Note=ANK 13 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 40_69 | 12 | 920.0 | Repeat | Note=ANK 2 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 437_466 | 12 | 920.0 | Repeat | Note=ANK 14 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 470_498 | 12 | 920.0 | Repeat | Note=ANK 15 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 531_561 | 12 | 920.0 | Repeat | Note=ANK 16 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 566_595 | 12 | 920.0 | Repeat | Note=ANK 17 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 599_628 | 12 | 920.0 | Repeat | Note=ANK 18 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 633_662 | 12 | 920.0 | Repeat | Note=ANK 19 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 669_698 | 12 | 920.0 | Repeat | Note=ANK 20 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 702_731 | 12 | 920.0 | Repeat | Note=ANK 21 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 735_764 | 12 | 920.0 | Repeat | Note=ANK 22 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 73_102 | 12 | 920.0 | Repeat | Note=ANK 3 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 771_800 | 12 | 920.0 | Repeat | Note=ANK 23 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 7_36 | 12 | 920.0 | Repeat | Note=ANK 1 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 803_832 | 12 | 920.0 | Repeat | Note=ANK 24 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 838_867 | 12 | 920.0 | Repeat | Note=ANK 25 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 871_901 | 12 | 920.0 | Repeat | Note=ANK 26 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 905_934 | 12 | 920.0 | Repeat | Note=ANK 27 |
| Hgene | ANKRD44 | chr2:198051747 | chr16:68200746 | ENST00000450567 | - | 1 | 25 | 941_970 | 12 | 920.0 | Repeat | Note=ANK 28 |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000329524 | 3 | 11 | 24_29 | 533 | 1996.6666666666667 | Compositional bias | Note=Poly-Pro | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000346183 | 3 | 10 | 24_29 | 533 | 1076.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000349223 | 3 | 11 | 24_29 | 533 | 1066.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000329524 | 3 | 11 | 444_451 | 533 | 1996.6666666666667 | DNA binding | . | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000346183 | 3 | 10 | 444_451 | 533 | 1076.0 | DNA binding | . | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000349223 | 3 | 11 | 444_451 | 533 | 1066.0 | DNA binding | . | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000329524 | 3 | 11 | 415_596 | 533 | 1996.6666666666667 | Domain | RHD | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000346183 | 3 | 10 | 415_596 | 533 | 1076.0 | Domain | RHD | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000349223 | 3 | 11 | 415_596 | 533 | 1066.0 | Domain | RHD | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000329524 | 3 | 11 | 273_275 | 533 | 1996.6666666666667 | Motif | Note=Nuclear localization signal | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000346183 | 3 | 10 | 273_275 | 533 | 1076.0 | Motif | Note=Nuclear localization signal | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000349223 | 3 | 11 | 273_275 | 533 | 1066.0 | Motif | Note=Nuclear localization signal | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000329524 | 3 | 11 | 109_114 | 533 | 1996.6666666666667 | Region | Note=Calcineurin-binding | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000329524 | 3 | 11 | 207_308 | 533 | 1996.6666666666667 | Region | Note=3 X SP repeats | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000346183 | 3 | 10 | 109_114 | 533 | 1076.0 | Region | Note=Calcineurin-binding | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000346183 | 3 | 10 | 207_308 | 533 | 1076.0 | Region | Note=3 X SP repeats | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000349223 | 3 | 11 | 109_114 | 533 | 1066.0 | Region | Note=Calcineurin-binding | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000349223 | 3 | 11 | 207_308 | 533 | 1066.0 | Region | Note=3 X SP repeats | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000329524 | 3 | 11 | 207_223 | 533 | 1996.6666666666667 | Repeat | Note=1 | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000329524 | 3 | 11 | 236_252 | 533 | 1996.6666666666667 | Repeat | Note=2 | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000329524 | 3 | 11 | 292_308 | 533 | 1996.6666666666667 | Repeat | Note=3 | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000346183 | 3 | 10 | 207_223 | 533 | 1076.0 | Repeat | Note=1 | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000346183 | 3 | 10 | 236_252 | 533 | 1076.0 | Repeat | Note=2 | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000346183 | 3 | 10 | 292_308 | 533 | 1076.0 | Repeat | Note=3 | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000349223 | 3 | 11 | 207_223 | 533 | 1066.0 | Repeat | Note=1 | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000349223 | 3 | 11 | 236_252 | 533 | 1066.0 | Repeat | Note=2 | |
| Tgene | NFATC3 | chr2:198051747 | chr16:68200746 | ENST00000349223 | 3 | 11 | 292_308 | 533 | 1066.0 | Repeat | Note=3 |
Top |
Fusion Gene Sequence for ANKRD44-NFATC3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >4742_4742_1_ANKRD44-NFATC3_ANKRD44_chr2_198051747_ENST00000328737_NFATC3_chr16_68200746_ENST00000329524_length(transcript)=4632nt_BP=113nt TTCAGAGGCTGCGTCTTTCAGGAGGCCAGCCACCATTGGTTCAGGCAATCTTCAGCGGTGATCCAGAGGAGATCCGGATGCTCATCCATA AAACTGAAGATGTGAATACTCTGTATTGATTGTGCAGGTATTTTGAAACTCCGCAATTCAGATATAGAACTTCGAAAAGGAGAAACTGAT ATTGGCAGAAAGAATACTAGAGTACGACTTGTGTTTCGTGTACACATCCCACAGCCCAGTGGAAAAGTCCTTTCTCTGCAGATAGCCTCT ATACCCGTTGAGTGCTCCCAGCGGTCTGCTCAAGAACTTCCTCATATTGAGAAGTACAGTATCAACAGTTGTTCTGTAAATGGAGGTCAT GAAATGGTTGTGACTGGATCTAATTTTCTTCCAGAATCCAAAATCATTTTTCTTGAAAAAGGACAAGATGGACGACCTCAGTGGGAGGTA GAAGGGAAGATAATCAGGGAAAAATGTCAAGGGGCTCACATTGTCCTTGAAGTTCCTCCATATCATAACCCAGCAGTTACAGCTGCAGTG CAGGTGCACTTTTATCTTTGCAATGGCAAGAGGAAAAAAAGCCAGTCTCAACGTTTTACTTATACACCAGTTTTGATGAAGCAAGAACAC AGAGAAGAGATTGATTTGTCTTCAGTTCCATCTTTGCCTGTGCCTCATCCTGCTCAGACCCAGAGGCCTTCCTCTGATTCAGGGTGTTCA CATGACAGTGTACTGTCAGGACAGAGAAGTTTGATTTGCTCCATCCCACAAACATATGCATCCATGGTGACCTCATCCCATCTGCCACAG TTGCAGTGTAGAGATGAGAGTGTTAGTAAAGAACAGCATATGATTCCTTCTCCAATTGTACACCAGCCTTTTCAAGTCACACCAACACCT CCTGTGGGGTCTTCCTATCAGCCTATGCAAACTAATGTTGTGTACAATGGACCAACTTGTCTTCCTATTAATGCTGCCTCTAGTCAAGAA TTTGATTCAGTTTTGTTTCAGCAGGATGCAACTCTTTCTGGTTTAGTGAATCTTGGCTGTCAACCACTGTCATCCATACCATTTCATTCT TCAAATTCAGGCTCAACAGGACATCTCTTAGCCCATACACCTCATTCTGTGCATACCCTGCCTCATCTGCAATCAATGGGATATCATTGT TCAAATACAGGACAAAGATCTCTTTCTTCTCCAGTGGCTGACCAGATTACAGGTCAGCCTTCGTCTCAGTTACAACCTATTACATATGGT CCTTCACATTCAGGGTCTGCTACAACAGCTTCCCCAGCAGCTTCTCATCCCTTGGCTAGTTCACCGCTTTCTGGGCCACCATCTCCTCAG CTTCAGCCTATGCCTTACCAATCTCCTAGCTCAGGAACTGCCTCATCACCGTCTCCAGCCACCAGAATGCATTCTGGACAGCACTCAACT CAAGCACAAAGTACGGGCCAGGGGGGTCTTTCTGCACCTTCATCCTTAATATGTCACAGTTTGTGTGATCCAGCGTCATTTCCACCTGAT GGGGCAACTGTGAGCATTAAACCTGAACCAGAAGATCGAGAGCCTAACTTTGCAACCATTGGTCTGCAGGACATCACTTTAGATGATGAC CAATTTATATCTGACTTGGAACACCAGCCATCAGGTTCAGCAGAGAAATGGCCTAACCACAGTGTGCTCTCATGTCCAGCTCCTTTCTGG AGAATCTAGAGGTGAACGAGATAATTGGGAGAGACATGTCCCAGATTTCTGTTTCCCAAGGAGCAGGGGTGAGCAGGCAGGCTCCCCTCC CGAGTCCTGAGTCCCTGGATTTAGGAAGATCTGATGGGCTCTAACAGTGCTTACTGCAGCCTTGTGTCCACCACCAACTTCTCAGCATGT TTCTCTCCTTGGACCTTGGGTTTCCAACTCTGCAGCCTTCAGGTCTGGGGCCAGGAGTGGGACCCACCATTTGTGGGGAAAGTAGCATTC CTCCACCTCAGGCCTTGGGTAGATTTGGCAAAAGAACAGGAGCAGCATAGGCTGTTTGAGCTTTGGGGAAATGAACTTTGCTTTTTATAT TTAACTAGGATACTTTTATATGATGGGTGCTTTGAGTGTGAATGCAGCAGGCTCTCTTGTTTCCGAGGTGCTGCTTTTGCAGGTGACCTG GTTACTTAGCTAGGATTGGTGATTTGTACTGCTTTATGGTCATTTGAAGGGCCCTTTAGTTTTTATGATAATTTTTAAAATAGGAACTTT TGATAAGACCTTCTAGAAGCAAAAAAAAAAAAAAAAAAAAAAAAAAGAAAAAAAAAGAAAAGAAAAAAAGAAAAAGAAAAAAATATCCCA AGCCCACACCTATGCCTCAGAAAGTCAGCATGTGTCCTGAAGACGTTCATAGATTTATAGGAAATAAAGCCAAATGTAGCTCATACCTCT TTATAAAACTAAACTGTTGTTAGATAAAATGAGAAGAGACCATTCCATCATTGAAGTGTTGGAATATAAAAAGACATTCCAGAGCATAGC CTTTTGGTAACTTTCTAGGTAATCTTGCAATCTCTCCATACTGGCTCCATGTTGAAATTGCTTTTTCCCTCAGGACCCTGGGTAAGATGG CTACATTTAAGGTGACTCTGAATGGTGGGGTTCTAGCTTGGTCACTTTCATTTCTTGCCATCATAAAAACTAGTAGGCTGTGGAGTGCGC TTCCTGTTTGTGTCATCTGGTCGTAGAACCTTTTCCAAACAGAAGCATCTGCCTTTGGCTGTTCTGTGACCTTTGAAGCCAGTCTCAGTG GTTTAGGCAGCTTTGCAGTTCTAACTCTAGCTAGAGGCCAGGCTGTGTGCAGAGCTAAAGCACAAATGAGCAGTACATCAGCATGGGCAG AGAAAATACAGGACTCCAGATGGGAAGGTTTTAAGAGTCACACTGGCATGGAAGGCTTATCTGCCATTCTATTTATAGCCTGAAATGATT CTGGCCTACCCTGTCCTGTCTGTATGATCTAAAAACCAGCCTTTTCCCCAGAAGGCTCTGAGCACATCCAGAAGTGATAACCTGACCCTC CACACCAAAGACACCCTGGTGCTGCTGCTGGTGAGACTGGTGTCCAAGGCAGGGCCCAGCAGAAACTGCAGCCTGCTGTCTACCAAAGGA GGTACCCGAGTTGGGTACTTTAAAAAAAAAAAAAACCTGCCCTTACCCCTCTCCCTAATGGGATTTTTTTTACCTGTTCCTGGGCCCATT ACAGAGCCTCTGAAGTTTGCAGTATGCTTTCAAATGAAATAATATACTTCCATTGATTCAGGAAGCAGATTTTTTGAGTGCCTATATGTG AGACAGTGTGCTGGTTGAAGGCCTGATGCTGTTATTCCAGGAGACTGCAGGCCCTTGATGCCAGGACTCTCTCTACTAAGGCCAGTTCAG GTCTGGGAAAGGGAGCTGGCCTATTGCCCACCACCTGTTGTCAGTAGGTTAGGAGCTGTTGACTCAGGATGAGGAAGTATGGTCTTTAAA AAAAACAAAATATATTTTAGATTTCTCCCCTAAAGTTAGTTAGAGAAAAAGGTCTATTCTTCAGCAGATCACTGTGGCCCCTCTTGGAGC TGGGTGTTTTCAAATTTTCTTGAAAAGTGAGTTGTGTCACAGAATGTGGATAGTGAGATTTGCTTCTGGGCCTGTGGTTGGACGCCCACA GCTTCCTTTGGAGGGGCAAGAAGTTGGCCTCTCGGTTGACATTCCAGAGAAATGGCCAGACTTGCCCCTACCCTTTTGCCCCTGGTGTAG CTGCCTCACATGAGGTTATTGCAAGAGTTCCCCTTGCAATACCCTTTGTGCCCTCAAGAATTAAAAGCCTCTATGATCCAGAGTTGCTAT GCACCAAATTTTGATGCCTTCTAAATACCTTGATGCCTGTAGCACTAGAAATGCTTTCCTATTTAAAATAAACATTAAATTTTAAAATAG AGCTTTGTATACTATGTAAGCAGTTTTATATCAGCTTTGTAAAAGCTTTGGTGTTAAGAGTCAATATTTTTTGGCTTTGTAGATAAAGAC TTAAATTTTTGTGCAACATAGTGGTGTTAAAGCACACATCCATTGGACTATGCAAATCAATTTCTACTAGCAGCACTGAGCAACTGGGCA CGTGCTAGTACGCACCCCCTCCTACTCTAACTGTGCTAGCTCTTGGGTTGAGGAATGGAATGGGTAGCCTTCTGCTTACTGGAGATGCAC AGAGAAACATGGCTGAAAACAGCTTTAAAAAGAACATGTCTTTGGGTTCATGTGTGGTGCTGCTGCCCGGGTGTAAACTATCCCCACCCA GGGCCAGCCATTATCACCAGACCTCTTCTGGGCCTGGCTGTGAAGCCCTGTTTTTGGTATTCAGAATGGATCTTTAAGGCTCAGATGGGC TCAAATTGAAACCTTGTGTTTTATAAAATGGATGTTTAGTAAAGGAACTGGTTCTCAGTTATGTTTACAGCACTTGGAATTGTGTGTTCT TGTACATTTTGTATTTTAAAACCTTTTATGGGAGTGCAGTGCTCCTTACACAAATACAAAGGGAAGAAGAGCCAGCAGTTGAGGCTCCTC >4742_4742_1_ANKRD44-NFATC3_ANKRD44_chr2_198051747_ENST00000328737_NFATC3_chr16_68200746_ENST00000329524_length(amino acids)=528AA_BP= MKLRNSDIELRKGETDIGRKNTRVRLVFRVHIPQPSGKVLSLQIASIPVECSQRSAQELPHIEKYSINSCSVNGGHEMVVTGSNFLPESK IIFLEKGQDGRPQWEVEGKIIREKCQGAHIVLEVPPYHNPAVTAAVQVHFYLCNGKRKKSQSQRFTYTPVLMKQEHREEIDLSSVPSLPV PHPAQTQRPSSDSGCSHDSVLSGQRSLICSIPQTYASMVTSSHLPQLQCRDESVSKEQHMIPSPIVHQPFQVTPTPPVGSSYQPMQTNVV YNGPTCLPINAASSQEFDSVLFQQDATLSGLVNLGCQPLSSIPFHSSNSGSTGHLLAHTPHSVHTLPHLQSMGYHCSNTGQRSLSSPVAD QITGQPSSQLQPITYGPSHSGSATTASPAASHPLASSPLSGPPSPQLQPMPYQSPSSGTASSPSPATRMHSGQHSTQAQSTGQGGLSAPS -------------------------------------------------------------- >4742_4742_2_ANKRD44-NFATC3_ANKRD44_chr2_198051747_ENST00000328737_NFATC3_chr16_68200746_ENST00000346183_length(transcript)=4528nt_BP=113nt TTCAGAGGCTGCGTCTTTCAGGAGGCCAGCCACCATTGGTTCAGGCAATCTTCAGCGGTGATCCAGAGGAGATCCGGATGCTCATCCATA AAACTGAAGATGTGAATACTCTGTATTGATTGTGCAGGTATTTTGAAACTCCGCAATTCAGATATAGAACTTCGAAAAGGAGAAACTGAT ATTGGCAGAAAGAATACTAGAGTACGACTTGTGTTTCGTGTACACATCCCACAGCCCAGTGGAAAAGTCCTTTCTCTGCAGATAGCCTCT ATACCCGTTGAGTGCTCCCAGCGGTCTGCTCAAGAACTTCCTCATATTGAGAAGTACAGTATCAACAGTTGTTCTGTAAATGGAGGTCAT GAAATGGTTGTGACTGGATCTAATTTTCTTCCAGAATCCAAAATCATTTTTCTTGAAAAAGGACAAGATGGACGACCTCAGTGGGAGGTA GAAGGGAAGATAATCAGGGAAAAATGTCAAGGGGCTCACATTGTCCTTGAAGTTCCTCCATATCATAACCCAGCAGTTACAGCTGCAGTG CAGGTGCACTTTTATCTTTGCAATGGCAAGAGGAAAAAAAGCCAGTCTCAACGTTTTACTTATACACCAGTTTTGATGAAGCAAGAACAC AGAGAAGAGATTGATTTGTCTTCAGTTCCATCTTTGCCTGTGCCTCATCCTGCTCAGACCCAGAGGCCTTCCTCTGATTCAGGGTGTTCA CATGACAGTGTACTGTCAGGACAGAGAAGTTTGATTTGCTCCATCCCACAAACATATGCATCCATGGTGACCTCATCCCATCTGCCACAG TTGCAGTGTAGAGATGAGAGTGTTAGTAAAGAACAGCATATGATTCCTTCTCCAATTGTACACCAGCCTTTTCAAGTCACACCAACACCT CCTGTGGGGTCTTCCTATCAGCCTATGCAAACTAATGTTGTGTACAATGGACCAACTTGTCTTCCTATTAATGCTGCCTCTAGTCAAGAA TTTGATTCAGTTTTGTTTCAGCAGGATGCAACTCTTTCTGGTTTAGTGAATCTTGGCTGTCAACCACTGTCATCCATACCATTTCATTCT TCAAATTCAGGCTCAACAGGACATCTCTTAGCCCATACACCTCATTCTGTGCATACCCTGCCTCATCTGCAATCAATGGGATATCATTGT TCAAATACAGGACAAAGATCTCTTTCTTCTCCAGTGGCTGACCAGATTACAGGTCAGCCTTCGTCTCAGTTACAACCTATTACATATGGT CCTTCACATTCAGGGTCTGCTACAACAGCTTCCCCAGCAGCTTCTCATCCCTTGGCTAGTTCACCGCTTTCTGGGCCACCATCTCCTCAG CTTCAGCCTATGCCTTACCAATCTCCTAGCTCAGGAACTGCCTCATCACCGTCTCCAGCCACCAGAATGCATTCTGGACAGCACTCAACT CAAGCACAAAGTACGGGCCAGGGGGGTCTTTCTGCACCTTCATCCTTAATATGTCACAGTTTGTGTGATCCAGCGTCATTTCCACCTGAT GGGGCAACTGTGAGCATTAAACCTGAACCAGAAGATCGAGAGCCTAACTTTGCAACCATTGGTCTGCAGGACATCACTTTAGATGATGTG AACGAGATAATTGGGAGAGACATGTCCCAGATTTCTGTTTCCCAAGGAGCAGGGGTGAGCAGGCAGGCTCCCCTCCCGAGTCCTGAGTCC CTGGATTTAGGAAGATCTGATGGGCTCTAACAGTGCTTACTGCAGCCTTGTGTCCACCACCAACTTCTCAGCATGTTTCTCTCCTTGGAC CTTGGGTTTCCAACTCTGCAGCCTTCAGGTCTGGGGCCAGGAGTGGGACCCACCATTTGTGGGGAAAGTAGCATTCCTCCACCTCAGGCC TTGGGTAGATTTGGCAAAAGAACAGGAGCAGCATAGGCTGTTTGAGCTTTGGGGAAATGAACTTTGCTTTTTATATTTAACTAGGATACT TTTATATGATGGGTGCTTTGAGTGTGAATGCAGCAGGCTCTCTTGTTTCCGAGGTGCTGCTTTTGCAGGTGACCTGGTTACTTAGCTAGG ATTGGTGATTTGTACTGCTTTATGGTCATTTGAAGGGCCCTTTAGTTTTTATGATAATTTTTAAAATAGGAACTTTTGATAAGACCTTCT AGAAGCAAAAAAAAAAAAAAAAAAAAAAAAAAGAAAAAAAAAGAAAAGAAAAAAAGAAAAAGAAAAAAATATCCCAAGCCCACACCTATG CCTCAGAAAGTCAGCATGTGTCCTGAAGACGTTCATAGATTTATAGGAAATAAAGCCAAATGTAGCTCATACCTCTTTATAAAACTAAAC TGTTGTTAGATAAAATGAGAAGAGACCATTCCATCATTGAAGTGTTGGAATATAAAAAGACATTCCAGAGCATAGCCTTTTGGTAACTTT CTAGGTAATCTTGCAATCTCTCCATACTGGCTCCATGTTGAAATTGCTTTTTCCCTCAGGACCCTGGGTAAGATGGCTACATTTAAGGTG ACTCTGAATGGTGGGGTTCTAGCTTGGTCACTTTCATTTCTTGCCATCATAAAAACTAGTAGGCTGTGGAGTGCGCTTCCTGTTTGTGTC ATCTGGTCGTAGAACCTTTTCCAAACAGAAGCATCTGCCTTTGGCTGTTCTGTGACCTTTGAAGCCAGTCTCAGTGGTTTAGGCAGCTTT GCAGTTCTAACTCTAGCTAGAGGCCAGGCTGTGTGCAGAGCTAAAGCACAAATGAGCAGTACATCAGCATGGGCAGAGAAAATACAGGAC TCCAGATGGGAAGGTTTTAAGAGTCACACTGGCATGGAAGGCTTATCTGCCATTCTATTTATAGCCTGAAATGATTCTGGCCTACCCTGT CCTGTCTGTATGATCTAAAAACCAGCCTTTTCCCCAGAAGGCTCTGAGCACATCCAGAAGTGATAACCTGACCCTCCACACCAAAGACAC CCTGGTGCTGCTGCTGGTGAGACTGGTGTCCAAGGCAGGGCCCAGCAGAAACTGCAGCCTGCTGTCTACCAAAGGAGGTACCCGAGTTGG GTACTTTAAAAAAAAAAAAAACCTGCCCTTACCCCTCTCCCTAATGGGATTTTTTTTACCTGTTCCTGGGCCCATTACAGAGCCTCTGAA GTTTGCAGTATGCTTTCAAATGAAATAATATACTTCCATTGATTCAGGAAGCAGATTTTTTGAGTGCCTATATGTGAGACAGTGTGCTGG TTGAAGGCCTGATGCTGTTATTCCAGGAGACTGCAGGCCCTTGATGCCAGGACTCTCTCTACTAAGGCCAGTTCAGGTCTGGGAAAGGGA GCTGGCCTATTGCCCACCACCTGTTGTCAGTAGGTTAGGAGCTGTTGACTCAGGATGAGGAAGTATGGTCTTTAAAAAAAACAAAATATA TTTTAGATTTCTCCCCTAAAGTTAGTTAGAGAAAAAGGTCTATTCTTCAGCAGATCACTGTGGCCCCTCTTGGAGCTGGGTGTTTTCAAA TTTTCTTGAAAAGTGAGTTGTGTCACAGAATGTGGATAGTGAGATTTGCTTCTGGGCCTGTGGTTGGACGCCCACAGCTTCCTTTGGAGG GGCAAGAAGTTGGCCTCTCGGTTGACATTCCAGAGAAATGGCCAGACTTGCCCCTACCCTTTTGCCCCTGGTGTAGCTGCCTCACATGAG GTTATTGCAAGAGTTCCCCTTGCAATACCCTTTGTGCCCTCAAGAATTAAAAGCCTCTATGATCCAGAGTTGCTATGCACCAAATTTTGA TGCCTTCTAAATACCTTGATGCCTGTAGCACTAGAAATGCTTTCCTATTTAAAATAAACATTAAATTTTAAAATAGAGCTTTGTATACTA TGTAAGCAGTTTTATATCAGCTTTGTAAAAGCTTTGGTGTTAAGAGTCAATATTTTTTGGCTTTGTAGATAAAGACTTAAATTTTTGTGC AACATAGTGGTGTTAAAGCACACATCCATTGGACTATGCAAATCAATTTCTACTAGCAGCACTGAGCAACTGGGCACGTGCTAGTACGCA CCCCCTCCTACTCTAACTGTGCTAGCTCTTGGGTTGAGGAATGGAATGGGTAGCCTTCTGCTTACTGGAGATGCACAGAGAAACATGGCT GAAAACAGCTTTAAAAAGAACATGTCTTTGGGTTCATGTGTGGTGCTGCTGCCCGGGTGTAAACTATCCCCACCCAGGGCCAGCCATTAT CACCAGACCTCTTCTGGGCCTGGCTGTGAAGCCCTGTTTTTGGTATTCAGAATGGATCTTTAAGGCTCAGATGGGCTCAAATTGAAACCT TGTGTTTTATAAAATGGATGTTTAGTAAAGGAACTGGTTCTCAGTTATGTTTACAGCACTTGGAATTGTGTGTTCTTGTACATTTTGTAT TTTAAAACCTTTTATGGGAGTGCAGTGCTCCTTACACAAATACAAAGGGAAGAAGAGCCAGCAGTTGAGGCTCCTCAGTTTTAGTGCTGA >4742_4742_2_ANKRD44-NFATC3_ANKRD44_chr2_198051747_ENST00000328737_NFATC3_chr16_68200746_ENST00000346183_length(amino acids)=535AA_BP= MKLRNSDIELRKGETDIGRKNTRVRLVFRVHIPQPSGKVLSLQIASIPVECSQRSAQELPHIEKYSINSCSVNGGHEMVVTGSNFLPESK IIFLEKGQDGRPQWEVEGKIIREKCQGAHIVLEVPPYHNPAVTAAVQVHFYLCNGKRKKSQSQRFTYTPVLMKQEHREEIDLSSVPSLPV PHPAQTQRPSSDSGCSHDSVLSGQRSLICSIPQTYASMVTSSHLPQLQCRDESVSKEQHMIPSPIVHQPFQVTPTPPVGSSYQPMQTNVV YNGPTCLPINAASSQEFDSVLFQQDATLSGLVNLGCQPLSSIPFHSSNSGSTGHLLAHTPHSVHTLPHLQSMGYHCSNTGQRSLSSPVAD QITGQPSSQLQPITYGPSHSGSATTASPAASHPLASSPLSGPPSPQLQPMPYQSPSSGTASSPSPATRMHSGQHSTQAQSTGQGGLSAPS -------------------------------------------------------------- >4742_4742_3_ANKRD44-NFATC3_ANKRD44_chr2_198051747_ENST00000328737_NFATC3_chr16_68200746_ENST00000349223_length(transcript)=4616nt_BP=113nt TTCAGAGGCTGCGTCTTTCAGGAGGCCAGCCACCATTGGTTCAGGCAATCTTCAGCGGTGATCCAGAGGAGATCCGGATGCTCATCCATA AAACTGAAGATGTGAATACTCTGTATTGATTGTGCAGGTATTTTGAAACTCCGCAATTCAGATATAGAACTTCGAAAAGGAGAAACTGAT ATTGGCAGAAAGAATACTAGAGTACGACTTGTGTTTCGTGTACACATCCCACAGCCCAGTGGAAAAGTCCTTTCTCTGCAGATAGCCTCT ATACCCGTTGAGTGCTCCCAGCGGTCTGCTCAAGAACTTCCTCATATTGAGAAGTACAGTATCAACAGTTGTTCTGTAAATGGAGGTCAT GAAATGGTTGTGACTGGATCTAATTTTCTTCCAGAATCCAAAATCATTTTTCTTGAAAAAGGACAAGATGGACGACCTCAGTGGGAGGTA GAAGGGAAGATAATCAGGGAAAAATGTCAAGGGGCTCACATTGTCCTTGAAGTTCCTCCATATCATAACCCAGCAGTTACAGCTGCAGTG CAGGTGCACTTTTATCTTTGCAATGGCAAGAGGAAAAAAAGCCAGTCTCAACGTTTTACTTATACACCAGTTTTGATGAAGCAAGAACAC AGAGAAGAGATTGATTTGTCTTCAGTTCCATCTTTGCCTGTGCCTCATCCTGCTCAGACCCAGAGGCCTTCCTCTGATTCAGGGTGTTCA CATGACAGTGTACTGTCAGGACAGAGAAGTTTGATTTGCTCCATCCCACAAACATATGCATCCATGGTGACCTCATCCCATCTGCCACAG TTGCAGTGTAGAGATGAGAGTGTTAGTAAAGAACAGCATATGATTCCTTCTCCAATTGTACACCAGCCTTTTCAAGTCACACCAACACCT CCTGTGGGGTCTTCCTATCAGCCTATGCAAACTAATGTTGTGTACAATGGACCAACTTGTCTTCCTATTAATGCTGCCTCTAGTCAAGAA TTTGATTCAGTTTTGTTTCAGCAGGATGCAACTCTTTCTGGTTTAGTGAATCTTGGCTGTCAACCACTGTCATCCATACCATTTCATTCT TCAAATTCAGGCTCAACAGGACATCTCTTAGCCCATACACCTCATTCTGTGCATACCCTGCCTCATCTGCAATCAATGGGATATCATTGT TCAAATACAGGACAAAGATCTCTTTCTTCTCCAGTGGCTGACCAGATTACAGGTCAGCCTTCGTCTCAGTTACAACCTATTACATATGGT CCTTCACATTCAGGGTCTGCTACAACAGCTTCCCCAGCAGCTTCTCATCCCTTGGCTAGTTCACCGCTTTCTGGGCCACCATCTCCTCAG CTTCAGCCTATGCCTTACCAATCTCCTAGCTCAGGAACTGCCTCATCACCGTCTCCAGCCACCAGAATGCATTCTGGACAGCACTCAACT CAAGCACAAAGTACGGGCCAGGGGGGTCTTTCTGCACCTTCATCCTTAATATGTCACAGTTTGTGTGATCCAGCGTCATTTCCACCTGAT GGGGCAACTGTGAGCATTAAACCTGAACCAGAAGATCGAGAGCCTAACTTTGCAACCATTGGTCTGCAGGACATCACTTTAGATGATGAT TTGTTTACCAGTAATAATTTTGACTTGCTTCAGTTGAGACCTACGTTTTGGCCAGTCCCAGCAGGAAGATATCTGAGGAATCTAGAGTGA ACGAGATAATTGGGAGAGACATGTCCCAGATTTCTGTTTCCCAAGGAGCAGGGGTGAGCAGGCAGGCTCCCCTCCCGAGTCCTGAGTCCC TGGATTTAGGAAGATCTGATGGGCTCTAACAGTGCTTACTGCAGCCTTGTGTCCACCACCAACTTCTCAGCATGTTTCTCTCCTTGGACC TTGGGTTTCCAACTCTGCAGCCTTCAGGTCTGGGGCCAGGAGTGGGACCCACCATTTGTGGGGAAAGTAGCATTCCTCCACCTCAGGCCT TGGGTAGATTTGGCAAAAGAACAGGAGCAGCATAGGCTGTTTGAGCTTTGGGGAAATGAACTTTGCTTTTTATATTTAACTAGGATACTT TTATATGATGGGTGCTTTGAGTGTGAATGCAGCAGGCTCTCTTGTTTCCGAGGTGCTGCTTTTGCAGGTGACCTGGTTACTTAGCTAGGA TTGGTGATTTGTACTGCTTTATGGTCATTTGAAGGGCCCTTTAGTTTTTATGATAATTTTTAAAATAGGAACTTTTGATAAGACCTTCTA GAAGCAAAAAAAAAAAAAAAAAAAAAAAAAAGAAAAAAAAAGAAAAGAAAAAAAGAAAAAGAAAAAAATATCCCAAGCCCACACCTATGC CTCAGAAAGTCAGCATGTGTCCTGAAGACGTTCATAGATTTATAGGAAATAAAGCCAAATGTAGCTCATACCTCTTTATAAAACTAAACT GTTGTTAGATAAAATGAGAAGAGACCATTCCATCATTGAAGTGTTGGAATATAAAAAGACATTCCAGAGCATAGCCTTTTGGTAACTTTC TAGGTAATCTTGCAATCTCTCCATACTGGCTCCATGTTGAAATTGCTTTTTCCCTCAGGACCCTGGGTAAGATGGCTACATTTAAGGTGA CTCTGAATGGTGGGGTTCTAGCTTGGTCACTTTCATTTCTTGCCATCATAAAAACTAGTAGGCTGTGGAGTGCGCTTCCTGTTTGTGTCA TCTGGTCGTAGAACCTTTTCCAAACAGAAGCATCTGCCTTTGGCTGTTCTGTGACCTTTGAAGCCAGTCTCAGTGGTTTAGGCAGCTTTG CAGTTCTAACTCTAGCTAGAGGCCAGGCTGTGTGCAGAGCTAAAGCACAAATGAGCAGTACATCAGCATGGGCAGAGAAAATACAGGACT CCAGATGGGAAGGTTTTAAGAGTCACACTGGCATGGAAGGCTTATCTGCCATTCTATTTATAGCCTGAAATGATTCTGGCCTACCCTGTC CTGTCTGTATGATCTAAAAACCAGCCTTTTCCCCAGAAGGCTCTGAGCACATCCAGAAGTGATAACCTGACCCTCCACACCAAAGACACC CTGGTGCTGCTGCTGGTGAGACTGGTGTCCAAGGCAGGGCCCAGCAGAAACTGCAGCCTGCTGTCTACCAAAGGAGGTACCCGAGTTGGG TACTTTAAAAAAAAAAAAAACCTGCCCTTACCCCTCTCCCTAATGGGATTTTTTTTACCTGTTCCTGGGCCCATTACAGAGCCTCTGAAG TTTGCAGTATGCTTTCAAATGAAATAATATACTTCCATTGATTCAGGAAGCAGATTTTTTGAGTGCCTATATGTGAGACAGTGTGCTGGT TGAAGGCCTGATGCTGTTATTCCAGGAGACTGCAGGCCCTTGATGCCAGGACTCTCTCTACTAAGGCCAGTTCAGGTCTGGGAAAGGGAG CTGGCCTATTGCCCACCACCTGTTGTCAGTAGGTTAGGAGCTGTTGACTCAGGATGAGGAAGTATGGTCTTTAAAAAAAACAAAATATAT TTTAGATTTCTCCCCTAAAGTTAGTTAGAGAAAAAGGTCTATTCTTCAGCAGATCACTGTGGCCCCTCTTGGAGCTGGGTGTTTTCAAAT TTTCTTGAAAAGTGAGTTGTGTCACAGAATGTGGATAGTGAGATTTGCTTCTGGGCCTGTGGTTGGACGCCCACAGCTTCCTTTGGAGGG GCAAGAAGTTGGCCTCTCGGTTGACATTCCAGAGAAATGGCCAGACTTGCCCCTACCCTTTTGCCCCTGGTGTAGCTGCCTCACATGAGG TTATTGCAAGAGTTCCCCTTGCAATACCCTTTGTGCCCTCAAGAATTAAAAGCCTCTATGATCCAGAGTTGCTATGCACCAAATTTTGAT GCCTTCTAAATACCTTGATGCCTGTAGCACTAGAAATGCTTTCCTATTTAAAATAAACATTAAATTTTAAAATAGAGCTTTGTATACTAT GTAAGCAGTTTTATATCAGCTTTGTAAAAGCTTTGGTGTTAAGAGTCAATATTTTTTGGCTTTGTAGATAAAGACTTAAATTTTTGTGCA ACATAGTGGTGTTAAAGCACACATCCATTGGACTATGCAAATCAATTTCTACTAGCAGCACTGAGCAACTGGGCACGTGCTAGTACGCAC CCCCTCCTACTCTAACTGTGCTAGCTCTTGGGTTGAGGAATGGAATGGGTAGCCTTCTGCTTACTGGAGATGCACAGAGAAACATGGCTG AAAACAGCTTTAAAAAGAACATGTCTTTGGGTTCATGTGTGGTGCTGCTGCCCGGGTGTAAACTATCCCCACCCAGGGCCAGCCATTATC ACCAGACCTCTTCTGGGCCTGGCTGTGAAGCCCTGTTTTTGGTATTCAGAATGGATCTTTAAGGCTCAGATGGGCTCAAATTGAAACCTT GTGTTTTATAAAATGGATGTTTAGTAAAGGAACTGGTTCTCAGTTATGTTTACAGCACTTGGAATTGTGTGTTCTTGTACATTTTGTATT TTAAAACCTTTTATGGGAGTGCAGTGCTCCTTACACAAATACAAAGGGAAGAAGAGCCAGCAGTTGAGGCTCCTCAGTTTTAGTGCTGAA >4742_4742_3_ANKRD44-NFATC3_ANKRD44_chr2_198051747_ENST00000328737_NFATC3_chr16_68200746_ENST00000349223_length(amino acids)=525AA_BP= MKLRNSDIELRKGETDIGRKNTRVRLVFRVHIPQPSGKVLSLQIASIPVECSQRSAQELPHIEKYSINSCSVNGGHEMVVTGSNFLPESK IIFLEKGQDGRPQWEVEGKIIREKCQGAHIVLEVPPYHNPAVTAAVQVHFYLCNGKRKKSQSQRFTYTPVLMKQEHREEIDLSSVPSLPV PHPAQTQRPSSDSGCSHDSVLSGQRSLICSIPQTYASMVTSSHLPQLQCRDESVSKEQHMIPSPIVHQPFQVTPTPPVGSSYQPMQTNVV YNGPTCLPINAASSQEFDSVLFQQDATLSGLVNLGCQPLSSIPFHSSNSGSTGHLLAHTPHSVHTLPHLQSMGYHCSNTGQRSLSSPVAD QITGQPSSQLQPITYGPSHSGSATTASPAASHPLASSPLSGPPSPQLQPMPYQSPSSGTASSPSPATRMHSGQHSTQAQSTGQGGLSAPS -------------------------------------------------------------- >4742_4742_4_ANKRD44-NFATC3_ANKRD44_chr2_198051747_ENST00000328737_NFATC3_chr16_68200746_ENST00000575270_length(transcript)=1998nt_BP=113nt TTCAGAGGCTGCGTCTTTCAGGAGGCCAGCCACCATTGGTTCAGGCAATCTTCAGCGGTGATCCAGAGGAGATCCGGATGCTCATCCATA AAACTGAAGATGTGAATACTCTGTATTGATTGTGCAGGTATTTTGAAACTCCGCAATTCAGATATAGAACTTCGAAAAGGAGAAACTGAT ATTGGCAGAAAGAATACTAGAGTACGACTTGTGTTTCGTGTACACATCCCACAGCCCAGTGGAAAAGTCCTTTCTCTGCAGATAGCCTCT ATACCCGTTGAGTGCTCCCAGCGGTCTGCTCAAGAACTTCCTCATATTGAGAAGTACAGTATCAACAGTTGTTCTGTAAATGGAGGTCAT GAAATGGTTGTGACTGGATCTAATTTTCTTCCAGAATCCAAAATCATTTTTCTTGAAAAAGGACAAGATGGACGACCTCAGTGGGAGGTA GAAGGGAAGATAATCAGGGAAAAATGTCAAGGGGCTCACATTGTCCTTGAAGTTCCTCCATATCATAACCCAGCAGTTACAGCTGCAGTG CAGGTGCACTTTTATCTTTGCAATGGCAAGAGGAAAAAAAGCCAGTCTCAACGTTTTACTTATACACCAGTTTTGATGAAGCAAGAACAC AGAGAAGAGATTGATTTGTCTTCAGTTCCATCTTTGCCTGTGCCTCATCCTGCTCAGACCCAGAGGCCTTCCTCTGATTCAGGGTGTTCA CATGACAGTGTACTGTCAGGACAGAGAAGTTTGATTTGCTCCATCCCACAAACATATGCATCCATGGTGACCTCATCCCATCTGCCACAG TTGCAGTGTAGAGATGAGAGTGTTAGTAAAGAACAGCATATGATTCCTTCTCCAATTGTACACCAGCCTTTTCAAGTCACACCAACACCT CCTGTGGGGTCTTCCTATCAGCCTATGCAAACTAATGTTGTGTACAATGGACCAACTTGTCTTCCTATTAATGCTGCCTCTAGTCAAGAA TTTGATTCAGTTTTGTTTCAGCAGGATGCAACTCTTTCTGGTTTAGTGAATCTTGGCTGTCAACCACTGTCATCCATACCATTTCATTCT TCAAATTCAGGCTCAACAGGACATCTCTTAGCCCATACACCTCATTCTGTGCATACCCTGCCTCATCTGCAATCAATGGGATATCATTGT TCAAATACAGGACAAAGATCTCTTTCTTCTCCAGTGGCTGACCAGATTACAGGTCAGCCTTCGTCTCAGTTACAACCTATTACATATGGT CCTTCACATTCAGGGTCTGCTACAACAGCTTCCCCAGCAGCTTCTCATCCCTTGGCTAGTTCACCGCTTTCTGGGCCACCATCTCCTCAG CTTCAGCCTATGCCTTACCAATCTCCTAGCTCAGGAACTGCCTCATCACCGTCTCCAGCCACCAGAATGCATTCTGGACAGCACTCAACT CAAGCACAAAGTACGGGCCAGGGGGGTCTTTCTGCACCTTCATCCTTAATATGTCACAGTTTGTGTGATCCAGCGTCATTTCCACCTGAT GGGGCAACTGTGAGCATTAAACCTGAACCAGAAGATCGAGAGCCTAACTTTGCAACCATTGGTCTGCAGGACATCACTTTAGATGATGGT TTTGTCAAAGCAGAGAAACCACTTAGTCATGACTGAGTCTTAGCCCGAGGAAGGGATGGGAAAAAGGGAGGAAGAAGGAGGCCAAGCAAG TGATGAGGAAGATACGTACATCGCTTTTGCTCACATTTCATTGAAGAAAACGAATTGCATGGGCCATACCTAGCTACACTGGGGAAGCCA GGAAATATACTCTGTAGCTGGGTGTCAGTGTGACTAACTAAACCTCGATTACTGTGGAAGAATGAATTAAGATAATTAGCAGCCTACCAT AGATACCTTCTAAATTTTCAGGAACTTATTTCTGTCTGTAGCTTGAAAAATCAAATGAATATTTTTTGGTTTTTCCCTTTTTTTTGTTTT >4742_4742_4_ANKRD44-NFATC3_ANKRD44_chr2_198051747_ENST00000328737_NFATC3_chr16_68200746_ENST00000575270_length(amino acids)=507AA_BP= MKLRNSDIELRKGETDIGRKNTRVRLVFRVHIPQPSGKVLSLQIASIPVECSQRSAQELPHIEKYSINSCSVNGGHEMVVTGSNFLPESK IIFLEKGQDGRPQWEVEGKIIREKCQGAHIVLEVPPYHNPAVTAAVQVHFYLCNGKRKKSQSQRFTYTPVLMKQEHREEIDLSSVPSLPV PHPAQTQRPSSDSGCSHDSVLSGQRSLICSIPQTYASMVTSSHLPQLQCRDESVSKEQHMIPSPIVHQPFQVTPTPPVGSSYQPMQTNVV YNGPTCLPINAASSQEFDSVLFQQDATLSGLVNLGCQPLSSIPFHSSNSGSTGHLLAHTPHSVHTLPHLQSMGYHCSNTGQRSLSSPVAD QITGQPSSQLQPITYGPSHSGSATTASPAASHPLASSPLSGPPSPQLQPMPYQSPSSGTASSPSPATRMHSGQHSTQAQSTGQGGLSAPS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ANKRD44-NFATC3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ANKRD44-NFATC3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ANKRD44-NFATC3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
