
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:LONP1-SAFB2 (FusionGDB2 ID:49426) |
Fusion Gene Summary for LONP1-SAFB2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: LONP1-SAFB2 | Fusion gene ID: 49426 | Hgene | Tgene | Gene symbol | LONP1 | SAFB2 | Gene ID | 9361 | 9667 |
| Gene name | lon peptidase 1, mitochondrial | scaffold attachment factor B2 | |
| Synonyms | CODASS|LON|LONP|LonHS|PIM1|PRSS15|hLON | - | |
| Cytomap | 19p13.3 | 19p13.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | lon protease homolog, mitochondrialhLON ATP-dependent proteaselon protease-like proteinmitochondrial ATP-dependent protease Lonmitochondrial lon protease-like proteinserine protease 15 | scaffold attachment factor B2 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | P36776 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000360614, ENST00000540670, ENST00000585374, ENST00000590729, ENST00000593119, ENST00000590511, | ENST00000591310, ENST00000252542, | |
| Fusion gene scores | * DoF score | 18 X 14 X 12=3024 | 8 X 8 X 5=320 |
| # samples | 23 | 9 | |
| ** MAII score | log2(23/3024*10)=-3.7167523732767 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/320*10)=-1.83007499855769 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: LONP1 [Title/Abstract] AND SAFB2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | SAFB2(5598804)-LONP1(5708414), # samples:3 LONP1(5705783)-SAFB2(5600271), # samples:1 LONP1(5705782)-SAFB2(5604947), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | SAFB2-LONP1 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. SAFB2-LONP1 seems lost the major protein functional domain in Hgene partner, which is a transcription factor due to the frame-shifted ORF. SAFB2-LONP1 seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. SAFB2-LONP1 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. LONP1-SAFB2 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. LONP1-SAFB2 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. LONP1-SAFB2 seems lost the major protein functional domain in Tgene partner, which is a transcription factor due to the frame-shifted ORF. LONP1-SAFB2 seems lost the major protein functional domain in Tgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | LONP1 | GO:0034599 | cellular response to oxidative stress | 17420247 |
| Hgene | LONP1 | GO:0051603 | proteolysis involved in cellular protein catabolic process | 8248235|17420247 |
 Fusion gene breakpoints across LONP1 (5'-gene) Fusion gene breakpoints across LONP1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
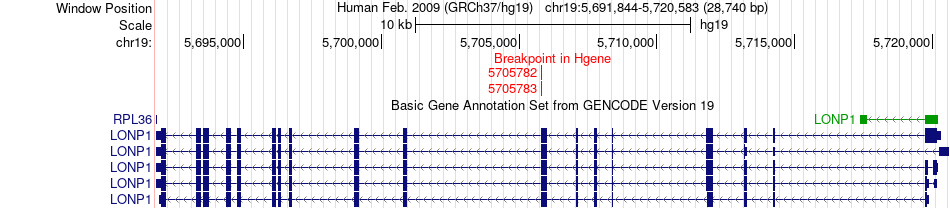 |
 Fusion gene breakpoints across SAFB2 (3'-gene) Fusion gene breakpoints across SAFB2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
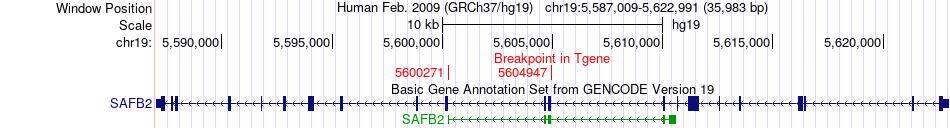 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-AR-A251-01A | LONP1 | chr19 | 5705783 | - | SAFB2 | chr19 | 5600271 | - |
| ChimerDB4 | BRCA | TCGA-AR-A251 | LONP1 | chr19 | 5705782 | - | SAFB2 | chr19 | 5604947 | - |
Top |
Fusion Gene ORF analysis for LONP1-SAFB2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000360614 | ENST00000591310 | LONP1 | chr19 | 5705783 | - | SAFB2 | chr19 | 5600271 | - |
| 5CDS-5UTR | ENST00000360614 | ENST00000591310 | LONP1 | chr19 | 5705782 | - | SAFB2 | chr19 | 5604947 | - |
| 5CDS-5UTR | ENST00000540670 | ENST00000591310 | LONP1 | chr19 | 5705783 | - | SAFB2 | chr19 | 5600271 | - |
| 5CDS-5UTR | ENST00000540670 | ENST00000591310 | LONP1 | chr19 | 5705782 | - | SAFB2 | chr19 | 5604947 | - |
| 5CDS-5UTR | ENST00000585374 | ENST00000591310 | LONP1 | chr19 | 5705783 | - | SAFB2 | chr19 | 5600271 | - |
| 5CDS-5UTR | ENST00000585374 | ENST00000591310 | LONP1 | chr19 | 5705782 | - | SAFB2 | chr19 | 5604947 | - |
| 5CDS-5UTR | ENST00000590729 | ENST00000591310 | LONP1 | chr19 | 5705783 | - | SAFB2 | chr19 | 5600271 | - |
| 5CDS-5UTR | ENST00000590729 | ENST00000591310 | LONP1 | chr19 | 5705782 | - | SAFB2 | chr19 | 5604947 | - |
| 5CDS-5UTR | ENST00000593119 | ENST00000591310 | LONP1 | chr19 | 5705783 | - | SAFB2 | chr19 | 5600271 | - |
| 5CDS-5UTR | ENST00000593119 | ENST00000591310 | LONP1 | chr19 | 5705782 | - | SAFB2 | chr19 | 5604947 | - |
| Frame-shift | ENST00000360614 | ENST00000252542 | LONP1 | chr19 | 5705782 | - | SAFB2 | chr19 | 5604947 | - |
| Frame-shift | ENST00000540670 | ENST00000252542 | LONP1 | chr19 | 5705782 | - | SAFB2 | chr19 | 5604947 | - |
| Frame-shift | ENST00000585374 | ENST00000252542 | LONP1 | chr19 | 5705783 | - | SAFB2 | chr19 | 5600271 | - |
| Frame-shift | ENST00000590729 | ENST00000252542 | LONP1 | chr19 | 5705782 | - | SAFB2 | chr19 | 5604947 | - |
| Frame-shift | ENST00000593119 | ENST00000252542 | LONP1 | chr19 | 5705782 | - | SAFB2 | chr19 | 5604947 | - |
| In-frame | ENST00000360614 | ENST00000252542 | LONP1 | chr19 | 5705783 | - | SAFB2 | chr19 | 5600271 | - |
| In-frame | ENST00000540670 | ENST00000252542 | LONP1 | chr19 | 5705783 | - | SAFB2 | chr19 | 5600271 | - |
| In-frame | ENST00000585374 | ENST00000252542 | LONP1 | chr19 | 5705782 | - | SAFB2 | chr19 | 5604947 | - |
| In-frame | ENST00000590729 | ENST00000252542 | LONP1 | chr19 | 5705783 | - | SAFB2 | chr19 | 5600271 | - |
| In-frame | ENST00000593119 | ENST00000252542 | LONP1 | chr19 | 5705783 | - | SAFB2 | chr19 | 5600271 | - |
| intron-3CDS | ENST00000590511 | ENST00000252542 | LONP1 | chr19 | 5705783 | - | SAFB2 | chr19 | 5600271 | - |
| intron-3CDS | ENST00000590511 | ENST00000252542 | LONP1 | chr19 | 5705782 | - | SAFB2 | chr19 | 5604947 | - |
| intron-5UTR | ENST00000590511 | ENST00000591310 | LONP1 | chr19 | 5705783 | - | SAFB2 | chr19 | 5600271 | - |
| intron-5UTR | ENST00000590511 | ENST00000591310 | LONP1 | chr19 | 5705782 | - | SAFB2 | chr19 | 5604947 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000360614 | LONP1 | chr19 | 5705783 | - | ENST00000252542 | SAFB2 | chr19 | 5600271 | - | 3072 | 1525 | 38 | 2827 | 929 |
| ENST00000593119 | LONP1 | chr19 | 5705783 | - | ENST00000252542 | SAFB2 | chr19 | 5600271 | - | 2755 | 1208 | 33 | 2510 | 825 |
| ENST00000540670 | LONP1 | chr19 | 5705783 | - | ENST00000252542 | SAFB2 | chr19 | 5600271 | - | 2839 | 1292 | 369 | 2594 | 741 |
| ENST00000590729 | LONP1 | chr19 | 5705783 | - | ENST00000252542 | SAFB2 | chr19 | 5600271 | - | 2583 | 1036 | 59 | 2338 | 759 |
| ENST00000585374 | LONP1 | chr19 | 5705782 | - | ENST00000252542 | SAFB2 | chr19 | 5604947 | - | 2982 | 1172 | 1238 | 2737 | 499 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000360614 | ENST00000252542 | LONP1 | chr19 | 5705783 | - | SAFB2 | chr19 | 5600271 | - | 0.005275876 | 0.9947241 |
| ENST00000593119 | ENST00000252542 | LONP1 | chr19 | 5705783 | - | SAFB2 | chr19 | 5600271 | - | 0.009283451 | 0.9907165 |
| ENST00000540670 | ENST00000252542 | LONP1 | chr19 | 5705783 | - | SAFB2 | chr19 | 5600271 | - | 0.016348477 | 0.9836516 |
| ENST00000590729 | ENST00000252542 | LONP1 | chr19 | 5705783 | - | SAFB2 | chr19 | 5600271 | - | 0.00664826 | 0.99335176 |
| ENST00000585374 | ENST00000252542 | LONP1 | chr19 | 5705782 | - | SAFB2 | chr19 | 5604947 | - | 0.033374153 | 0.9666258 |
Top |
Fusion Genomic Features for LONP1-SAFB2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
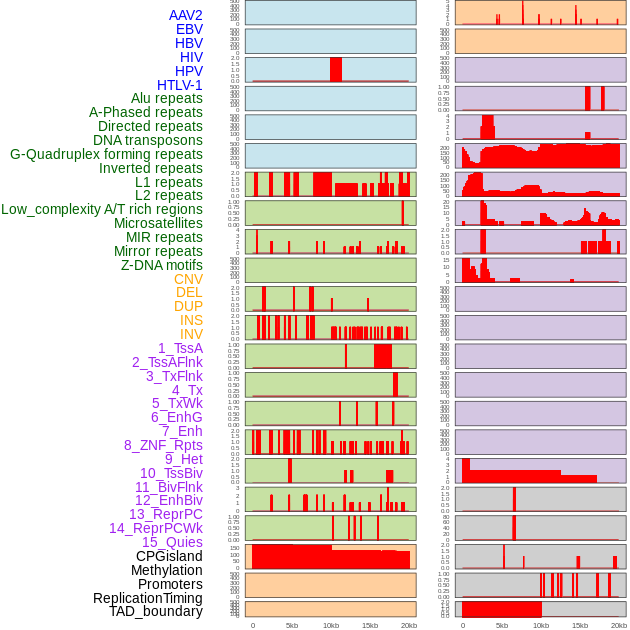 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for LONP1-SAFB2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:5598804/chr19:5708414) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| LONP1 | . |
| FUNCTION: ATP-dependent serine protease that mediates the selective degradation of misfolded, unassembled or oxidatively damaged polypeptides as well as certain short-lived regulatory proteins in the mitochondrial matrix. May also have a chaperone function in the assembly of inner membrane protein complexes. Participates in the regulation of mitochondrial gene expression and in the maintenance of the integrity of the mitochondrial genome. Binds to mitochondrial promoters and RNA in a single-stranded, site-specific, and strand-specific manner. May regulate mitochondrial DNA replication and/or gene expression using site-specific, single-stranded DNA binding to target the degradation of regulatory proteins binding to adjacent sites in mitochondrial promoters (PubMed:12198491, PubMed:15870080, PubMed:17420247, PubMed:8248235). Endogenous substrates include mitochondrial steroidogenic acute regulatory (StAR) protein, helicase Twinkle (TWNK) and the large ribosomal subunit protein bL32m. bL32m is protected from degradation by LONP1 when it is bound to a nucleic acid (RNA), but TWNK is not (PubMed:17579211, PubMed:28377575). {ECO:0000255|HAMAP-Rule:MF_03120, ECO:0000269|PubMed:12198491, ECO:0000269|PubMed:15870080, ECO:0000269|PubMed:17420247, ECO:0000269|PubMed:17579211, ECO:0000269|PubMed:28377575, ECO:0000269|PubMed:8248235}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LONP1 | chr19:5705782 | chr19:5604947 | ENST00000360614 | - | 8 | 18 | 124_368 | 455 | 960.0 | Domain | Lon N-terminal |
| Hgene | LONP1 | chr19:5705783 | chr19:5600271 | ENST00000360614 | - | 8 | 18 | 124_368 | 455 | 960.0 | Domain | Lon N-terminal |
| Tgene | SAFB2 | chr19:5705782 | chr19:5604947 | ENST00000252542 | 8 | 21 | 482_545 | 432 | 954.0 | Compositional bias | Note=Lys-rich | |
| Tgene | SAFB2 | chr19:5705782 | chr19:5604947 | ENST00000252542 | 8 | 21 | 619_724 | 432 | 954.0 | Compositional bias | Note=Glu-rich | |
| Tgene | SAFB2 | chr19:5705782 | chr19:5604947 | ENST00000252542 | 8 | 21 | 621_788 | 432 | 954.0 | Compositional bias | Note=Arg-rich | |
| Tgene | SAFB2 | chr19:5705782 | chr19:5604947 | ENST00000252542 | 8 | 21 | 792_926 | 432 | 954.0 | Compositional bias | Note=Gly-rich | |
| Tgene | SAFB2 | chr19:5705783 | chr19:5600271 | ENST00000252542 | 10 | 21 | 619_724 | 519 | 954.0 | Compositional bias | Note=Glu-rich | |
| Tgene | SAFB2 | chr19:5705783 | chr19:5600271 | ENST00000252542 | 10 | 21 | 621_788 | 519 | 954.0 | Compositional bias | Note=Arg-rich | |
| Tgene | SAFB2 | chr19:5705783 | chr19:5600271 | ENST00000252542 | 10 | 21 | 792_926 | 519 | 954.0 | Compositional bias | Note=Gly-rich | |
| Tgene | SAFB2 | chr19:5705782 | chr19:5604947 | ENST00000252542 | 8 | 21 | 713_730 | 432 | 954.0 | Motif | Nuclear localization signal | |
| Tgene | SAFB2 | chr19:5705783 | chr19:5600271 | ENST00000252542 | 10 | 21 | 713_730 | 519 | 954.0 | Motif | Nuclear localization signal |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LONP1 | chr19:5705782 | chr19:5604947 | ENST00000360614 | - | 8 | 18 | 759_949 | 455 | 960.0 | Domain | Lon proteolytic |
| Hgene | LONP1 | chr19:5705783 | chr19:5600271 | ENST00000360614 | - | 8 | 18 | 759_949 | 455 | 960.0 | Domain | Lon proteolytic |
| Hgene | LONP1 | chr19:5705782 | chr19:5604947 | ENST00000360614 | - | 8 | 18 | 523_530 | 455 | 960.0 | Nucleotide binding | ATP |
| Hgene | LONP1 | chr19:5705783 | chr19:5600271 | ENST00000360614 | - | 8 | 18 | 523_530 | 455 | 960.0 | Nucleotide binding | ATP |
| Tgene | SAFB2 | chr19:5705783 | chr19:5600271 | ENST00000252542 | 10 | 21 | 482_545 | 519 | 954.0 | Compositional bias | Note=Lys-rich | |
| Tgene | SAFB2 | chr19:5705782 | chr19:5604947 | ENST00000252542 | 8 | 21 | 30_64 | 432 | 954.0 | Domain | SAP | |
| Tgene | SAFB2 | chr19:5705782 | chr19:5604947 | ENST00000252542 | 8 | 21 | 407_485 | 432 | 954.0 | Domain | RRM | |
| Tgene | SAFB2 | chr19:5705783 | chr19:5600271 | ENST00000252542 | 10 | 21 | 30_64 | 519 | 954.0 | Domain | SAP | |
| Tgene | SAFB2 | chr19:5705783 | chr19:5600271 | ENST00000252542 | 10 | 21 | 407_485 | 519 | 954.0 | Domain | RRM |
Top |
Fusion Gene Sequence for LONP1-SAFB2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >49426_49426_1_LONP1-SAFB2_LONP1_chr19_5705782_ENST00000585374_SAFB2_chr19_5604947_ENST00000252542_length(transcript)=2982nt_BP=1172nt AGTATGGCCGGGCTATGGCGGCGAGCACTGGCTACGTGCGACTGTGGGGAGCGGCGCGGTGCTGGGTGCTGCGGCGGCCGATGCTGGCCG CCGCCGGGGGGCGGCGCGGGCGCCGGGGAAGGCCCGGTCATAACGGCGCTCACGCCCATGACGATCCCCGATGTGTTTCCGCACCTGCCG CTCATCGCCATCACCCGCAACCCGGTGTTCCCGCGCTTTATCAAGATTATCGAGGTTAAAAATAAGAAGTTGGTTGAGCTGCTGAGAAGG AAAGTTCGTCTCGCCCAGCCTTATGTCGGCGTCTTTCTAAAGAGAGATGACAGCAATGAGTCGGATGTGGTCGAGAGCCTGGATGAAATC TACCACACGGGGACGTTTGCCCAGATCCATGAGATGCAGGACCTTGGGGACAAGCTGCGCATGATCGTCATGGGACACAGAAGAGTCCAT ATCAGCAGACAGCTGGAGGTGGAGCCCGAGGAGCCGGAGGCGGAGAACAAGCACAAGCCCCGCAGGAAGTCAAAGCGGGGCAAGAAGGAG GCGGAGGACGAGCTGAGCGCCAGGCACCCGGCGGAGCTGGCGATGGAGCCCACCCCTGAGCTCCCGGCTGAGGTGCTCATGGTGGAGGTA GAGAACGTTGTCCACGAGGACTTCCAGGTCACGGAGGAGGTGAAAGCCCTGACTGCAGAGATCGTGAAGACCATCCGGGACATCATTGCC TTGAACCCTCTCTACAGGGAGTCAGTGCTGCAGATGATGCAGGCTGGCCAGCGGGTGGTGGACAACCCCATCTACCTGAGCGACATGGGC GCCGCGCTCACCGGGGCCGAGTCCCATGAGCTGCAGGACGTCCTGGAAGAGACCAATATTCCTAAGCGGCTGTACAAGGCCCTCTCCCTG CTGAAGAAGGAATTTGAACTGAGCAAGCTGCAGCAGCGCCTGGGGCGGGAGGTGGAGGAGAAGATCAAGCAGACCCACCGTAAGTACCTG CTGCAGGAGCAGCTAAAGATCATCAAGAAGGAGCTGGGCCTGGAGAAGGACGACAAGGATGCCATCGAGGAGAAGTTCCGGGAGCGCCTG AAGGAGCTCGTGGTCCCCAAGCACGTCATGGATGTTGTGGACGAGGAGCTGAGCAAGCTGGGCCTGCTGGACAACCACTCCTCGGAGTTC AAGTTGTCGGGGCCAAAGTGGTAACGAACGCCCGCAGCCCGGGGGCTCGATGCTATGGATTCGTCACCATGTCGACATCTGACGAGGCGA CCAAGTGCATCAGCCATCTCCACAGAACTGAGCTGCATGGACGAATGATCTCCGTAGAGAAGGCCAAAAATGAGCCTGCTGGGAAAAAGC TTTCCGACAGAAAAGAGTGCGAAGTGAAGAAGGAAAAATTATCGAGTGTCGACAGACATCATTCTGTGGAGATCAAAATTGAAAAAACTG TAATTAAGAAGGAAGAGAAGATTGAGAAGAAGGAGGAAAAAAAGCCTGAAGACATTAAGAAGGAAGAAAAAGACCAGGATGAGCTGAAAC CCGGACCTACAAATCGGTCTAGAGTCACCAAATCAGGAAGCAGAGGAATGGAGCGGACGGTCGTGATGGATAAATCGAAAGGAGAGCCCG TCATTAGCGTGAAAACCACAAGCAGGTCCAAAGAGAGAAGCTCCAAGAGTCAGGATCGCAAGTCAGAAAGCAAAGAAAAGAGAGACATCT TGTCGTTTGATAAAATCAAAGAACAAAGGGAGAGAGAGCGCCAGAGGCAGCGGGAACGGGAGATCCGCGAAACGGAGAGGCGGCGGGAGC GCGAGCAGCGGGAGCGGGAGCAACGCCTCGAGGCCTTCCATGAGCGGAAGGAGAAGGCCCGGCTACAGCGGGAACGCCTGCAGCTCGAGT GCCAGCGCCAGCGGCTGGAGCGGGAGCGCATGGAGCGGGAGCGGCTGGAGCGCGAGCGCATGCGCGTGGAGCGTGAGCGCAGGAAGGAGC AGGAGCGCATCCACCGCGAGCGCGAGGAGCTGCGGCGCCAGCAGGAGCAGCTGCGTTACGAGCAGGAGCGGCGGCCCGGGCGGAGGCCCT ACGACCTGGACCGACGAGATGATGCCTATTGGCCAGAAGGAAAGCGTGTGGCAATGGAGGACCGATATCGTGCAGACTTTCCCCGGCCAG ACCACCGCTTTCACGACTTCGATCATCGAGACCGGGGCCAGTACCAGGACCACGCCATCGACAGGCGGGAGGGTTCGAGGCCAATGATGG GAGACCACCGGGATGGGCAGCACTATGGAGATGACCGCCATGGCCACGGAGGACCCCCAGAGCGCCACGGCCGGGACTCCCGTGATGGCT GGGGGGGCTACGGCTCCGACAAGAGGCTGAGTGAAGGCCGGGGGCTGCCCCCTCCCCCCAGGGGTGGCCGTGACTGGGGAGAGCACAACC AGCGGCTAGAGGAGCACCAGGCACGCGCCTGGCAGGGTGCCATGGACGCAGGCGCGGCTAGCCGGGAGCACGCCAGGTGGCAAGGTGGCG AGAGGGGCCTGTCTGGGCCCTCGGGGCCGGGGCACATGGCAAGCCGCGGTGGAGTGGCGGGGCGAGGCGGCTTTGCACAAGGTGGACATT CCCAGGGCCACGTGGTGCCAGGTGGCGGACTGGAAGGTGGCGGAGTGGCCAGCCAGGACCGGGGCAGCAGAGTCCCTCACCCACACCCTC ATCCCCCCCCGTACCCCCACTTCACCCGCCGCTACTAAGTCCCACTCGCTGCGAGTTTTCGGGTGGGCAGACGCACTGTTGAATCTGGTA GCCAGGGTTCCCTCGAACTTGGGGGATCTTTTTAAAAGCAAAGTAAATCCTGCCACCATGTTGTAGCTCAATACAATGTGAACTCACTTT TTTTTTTTTTTTAAATAAATGTGTTCTTGTTCTGCCATTTTTAAATCAAGGTTTCTGTTAACGAGGCATTCCATTTTCCATTAATAAAGT >49426_49426_1_LONP1-SAFB2_LONP1_chr19_5705782_ENST00000585374_SAFB2_chr19_5604947_ENST00000252542_length(amino acids)=499AA_BP=163 MSTSDEATKCISHLHRTELHGRMISVEKAKNEPAGKKLSDRKECEVKKEKLSSVDRHHSVEIKIEKTVIKKEEKIEKKEEKKPEDIKKEE KDQDELKPGPTNRSRVTKSGSRGMERTVVMDKSKGEPVISVKTTSRSKERSSKSQDRKSESKEKRDILSFDKIKEQRERERQRQREREIR ETERRREREQREREQRLEAFHERKEKARLQRERLQLECQRQRLERERMERERLERERMRVERERRKEQERIHREREELRRQQEQLRYEQE RRPGRRPYDLDRRDDAYWPEGKRVAMEDRYRADFPRPDHRFHDFDHRDRGQYQDHAIDRREGSRPMMGDHRDGQHYGDDRHGHGGPPERH GRDSRDGWGGYGSDKRLSEGRGLPPPPRGGRDWGEHNQRLEEHQARAWQGAMDAGAASREHARWQGGERGLSGPSGPGHMASRGGVAGRG -------------------------------------------------------------- >49426_49426_2_LONP1-SAFB2_LONP1_chr19_5705783_ENST00000360614_SAFB2_chr19_5600271_ENST00000252542_length(transcript)=3072nt_BP=1525nt CGAACTCCGCCCCCTCTTCTCCGCGTAGGCCCAGCTCCCTGAAGCGGCTGTTTCGAGCCACGCGCCCATCGGGTACCGAGGCACGCGCCG GGCGTCACGTGCGTTTCGCGGCGAGCGGAAATGACGCGAGTTGTGTGAGCCGCCAGTATGGCCGGGCTATGGCGGCGAGCACTGGCTACG TGCGACTGTGGGGAGCGGCGCGGTGCTGGGTGCTGCGGCGGCCGATGCTGGCCGCCGCCGGGGGGCGGGTTCCCACTGCAGCAGGAGCGT GGTTGCTCCGAGGCCAGCGGACCTGCGACGCCTCTCCTCCTTGGGCACTGTGGGGCCGAGGCCCGGCAATTGGGGGCCAATGGCGGGGGT TTTGGGAAGCGAGCAGCCGCGGCGGAGGCGCATTCTCGGGGGGCGAGGACGCCTCCGAGGGCGGCGCGGAGGAAGGAGCCGGCGGCGCGG GGGGCAGCGCGGGCGCCGGGGAAGGCCCGGTCATAACGGCGCTCACGCCCATGACGATCCCCGATGTGTTTCCGCACCTGCCGCTCATCG CCATCACCCGCAACCCGGTGTTCCCGCGCTTTATCAAGATTATCGAGGTTAAAAATAAGAAGTTGGTTGAGCTGCTGAGAAGGAAAGTTC GTCTCGCCCAGCCTTATGTCGGCGTCTTTCTAAAGAGAGATGACAGCAATGAGTCGGATGTGGTCGAGAGCCTGGATGAAATCTACCACA CGGGGACGTTTGCCCAGATCCATGAGATGCAGGACCTTGGGGACAAGCTGCGCATGATCGTCATGGGACACAGAAGAGTCCATATCAGCA GACAGCTGGAGGTGGAGCCCGAGGAGCCGGAGGCGGAGAACAAGCACAAGCCCCGCAGGAAGTCAAAGCGGGGCAAGAAGGAGGCGGAGG ACGAGCTGAGCGCCAGGCACCCGGCGGAGCTGGCGATGGAGCCCACCCCTGAGCTCCCGGCTGAGGTGCTCATGGTGGAGGTAGAGAACG TTGTCCACGAGGACTTCCAGGTCACGGAGGAGGTGAAAGCCCTGACTGCAGAGATCGTGAAGACCATCCGGGACATCATTGCCTTGAACC CTCTCTACAGGGAGTCAGTGCTGCAGATGATGCAGGCTGGCCAGCGGGTGGTGGACAACCCCATCTACCTGAGCGACATGGGCGCCGCGC TCACCGGGGCCGAGTCCCATGAGCTGCAGGACGTCCTGGAAGAGACCAATATTCCTAAGCGGCTGTACAAGGCCCTCTCCCTGCTGAAGA AGGAATTTGAACTGAGCAAGCTGCAGCAGCGCCTGGGGCGGGAGGTGGAGGAGAAGATCAAGCAGACCCACCGTAAGTACCTGCTGCAGG AGCAGCTAAAGATCATCAAGAAGGAGCTGGGCCTGGAGAAGGACGACAAGGATGCCATCGAGGAGAAGTTCCGGGAGCGCCTGAAGGAGC TCGTGGTCCCCAAGCACGTCATGGATGTTGTGGACGAGGAGCTGAGCAAGCTGGGCCTGCTGGACAACCACTCCTCGGAGTTCAAAACTG TAATTAAGAAGGAAGAGAAGATTGAGAAGAAGGAGGAAAAAAAGCCTGAAGACATTAAGAAGGAAGAAAAAGACCAGGATGAGCTGAAAC CCGGACCTACAAATCGGTCTAGAGTCACCAAATCAGGAAGCAGAGGAATGGAGCGGACGGTCGTGATGGATAAATCGAAAGGAGAGCCCG TCATTAGCGTGAAAACCACAAGCAGGTCCAAAGAGAGAAGCTCCAAGAGTCAGGATCGCAAGTCAGAAAGCAAAGAAAAGAGAGACATCT TGTCGTTTGATAAAATCAAAGAACAAAGGGAGAGAGAGCGCCAGAGGCAGCGGGAACGGGAGATCCGCGAAACGGAGAGGCGGCGGGAGC GCGAGCAGCGGGAGCGGGAGCAACGCCTCGAGGCCTTCCATGAGCGGAAGGAGAAGGCCCGGCTACAGCGGGAACGCCTGCAGCTCGAGT GCCAGCGCCAGCGGCTGGAGCGGGAGCGCATGGAGCGGGAGCGGCTGGAGCGCGAGCGCATGCGCGTGGAGCGTGAGCGCAGGAAGGAGC AGGAGCGCATCCACCGCGAGCGCGAGGAGCTGCGGCGCCAGCAGGAGCAGCTGCGTTACGAGCAGGAGCGGCGGCCCGGGCGGAGGCCCT ACGACCTGGACCGACGAGATGATGCCTATTGGCCAGAAGGAAAGCGTGTGGCAATGGAGGACCGATATCGTGCAGACTTTCCCCGGCCAG ACCACCGCTTTCACGACTTCGATCATCGAGACCGGGGCCAGTACCAGGACCACGCCATCGACAGGCGGGAGGGTTCGAGGCCAATGATGG GAGACCACCGGGATGGGCAGCACTATGGAGATGACCGCCATGGCCACGGAGGACCCCCAGAGCGCCACGGCCGGGACTCCCGTGATGGCT GGGGGGGCTACGGCTCCGACAAGAGGCTGAGTGAAGGCCGGGGGCTGCCCCCTCCCCCCAGGGGTGGCCGTGACTGGGGAGAGCACAACC AGCGGCTAGAGGAGCACCAGGCACGCGCCTGGCAGGGTGCCATGGACGCAGGCGCGGCTAGCCGGGAGCACGCCAGGTGGCAAGGTGGCG AGAGGGGCCTGTCTGGGCCCTCGGGGCCGGGGCACATGGCAAGCCGCGGTGGAGTGGCGGGGCGAGGCGGCTTTGCACAAGGTGGACATT CCCAGGGCCACGTGGTGCCAGGTGGCGGACTGGAAGGTGGCGGAGTGGCCAGCCAGGACCGGGGCAGCAGAGTCCCTCACCCACACCCTC ATCCCCCCCCGTACCCCCACTTCACCCGCCGCTACTAAGTCCCACTCGCTGCGAGTTTTCGGGTGGGCAGACGCACTGTTGAATCTGGTA GCCAGGGTTCCCTCGAACTTGGGGGATCTTTTTAAAAGCAAAGTAAATCCTGCCACCATGTTGTAGCTCAATACAATGTGAACTCACTTT TTTTTTTTTTTTAAATAAATGTGTTCTTGTTCTGCCATTTTTAAATCAAGGTTTCTGTTAACGAGGCATTCCATTTTCCATTAATAAAGT >49426_49426_2_LONP1-SAFB2_LONP1_chr19_5705783_ENST00000360614_SAFB2_chr19_5600271_ENST00000252542_length(amino acids)=929AA_BP=1 MKRLFRATRPSGTEARAGRHVRFAASGNDASCVSRQYGRAMAASTGYVRLWGAARCWVLRRPMLAAAGGRVPTAAGAWLLRGQRTCDASP PWALWGRGPAIGGQWRGFWEASSRGGGAFSGGEDASEGGAEEGAGGAGGSAGAGEGPVITALTPMTIPDVFPHLPLIAITRNPVFPRFIK IIEVKNKKLVELLRRKVRLAQPYVGVFLKRDDSNESDVVESLDEIYHTGTFAQIHEMQDLGDKLRMIVMGHRRVHISRQLEVEPEEPEAE NKHKPRRKSKRGKKEAEDELSARHPAELAMEPTPELPAEVLMVEVENVVHEDFQVTEEVKALTAEIVKTIRDIIALNPLYRESVLQMMQA GQRVVDNPIYLSDMGAALTGAESHELQDVLEETNIPKRLYKALSLLKKEFELSKLQQRLGREVEEKIKQTHRKYLLQEQLKIIKKELGLE KDDKDAIEEKFRERLKELVVPKHVMDVVDEELSKLGLLDNHSSEFKTVIKKEEKIEKKEEKKPEDIKKEEKDQDELKPGPTNRSRVTKSG SRGMERTVVMDKSKGEPVISVKTTSRSKERSSKSQDRKSESKEKRDILSFDKIKEQRERERQRQREREIRETERRREREQREREQRLEAF HERKEKARLQRERLQLECQRQRLERERMERERLERERMRVERERRKEQERIHREREELRRQQEQLRYEQERRPGRRPYDLDRRDDAYWPE GKRVAMEDRYRADFPRPDHRFHDFDHRDRGQYQDHAIDRREGSRPMMGDHRDGQHYGDDRHGHGGPPERHGRDSRDGWGGYGSDKRLSEG RGLPPPPRGGRDWGEHNQRLEEHQARAWQGAMDAGAASREHARWQGGERGLSGPSGPGHMASRGGVAGRGGFAQGGHSQGHVVPGGGLEG -------------------------------------------------------------- >49426_49426_3_LONP1-SAFB2_LONP1_chr19_5705783_ENST00000540670_SAFB2_chr19_5600271_ENST00000252542_length(transcript)=2839nt_BP=1292nt AGACGAATCCCGGGAAGCCCGAATCCGCGCACCCCAAAGTGCGCTCAGAGCCTGAGTATGCGCACCCGGCCACCCGTGGCCAGTCTAGAG TCTGAGCATGCGCATGTTGTCCGCGCGCCTAAGCATGTGCACAGGCTGCTCATGAGTTCCTAAACGTTGCGTTTGCAACACTACCAGTGT TTTGTACGGGCGCTGGCGCCTGCTCACAGCCTTCCACGTGACCGGAAAAGGCCTTCAGCGCATAAAGTGGGAGACCGGAAGAGGCGGAGA CGATCCGATGCTCGAACTCCGCCCCCTCTTCTCCGCGTAGGCCCAGCTCCCTGAAGCGGCTGTTTCGAGCCACGCGCCCATCGGGTTAAA AATAAGAAGTTGGTTGAGCTGCTGAGAAGGAAAGTTCGTCTCGCCCAGCCTTATGTCGGCGTCTTTCTAAAGAGAGATGACAGCAATGAG TCGGATGTGGTCGAGAGCCTGGATGAAATCTACCACACGGGGACGTTTGCCCAGATCCATGAGATGCAGGACCTTGGGGACAAGCTGCGC ATGATCGTCATGGGACACAGAAGAGTCCATATCAGCAGACAGCTGGAGGTGGAGCCCGAGGAGCCGGAGGCGGAGAACAAGCACAAGCCC CGCAGGAAGTCAAAGCGGGGCAAGAAGGAGGCGGAGGACGAGCTGAGCGCCAGGCACCCGGCGGAGCTGGCGATGGAGCCCACCCCTGAG CTCCCGGCTGAGGTGCTCATGGTGGAGGTAGAGAACGTTGTCCACGAGGACTTCCAGGTCACGGAGGAGGTGAAAGCCCTGACTGCAGAG ATCGTGAAGACCATCCGGGACATCATTGCCTTGAACCCTCTCTACAGGGAGTCAGTGCTGCAGATGATGCAGGCTGGCCAGCGGGTGGTG GACAACCCCATCTACCTGAGCGACATGGGCGCCGCGCTCACCGGGGCCGAGTCCCATGAGCTGCAGGACGTCCTGGAAGAGACCAATATT CCTAAGCGGCTGTACAAGGCCCTCTCCCTGCTGAAGAAGGAATTTGAACTGAGCAAGCTGCAGCAGCGCCTGGGGCGGGAGGTGGAGGAG AAGATCAAGCAGACCCACCGTAAGTACCTGCTGCAGGAGCAGCTAAAGATCATCAAGAAGGAGCTGGGCCTGGAGAAGGACGACAAGGAT GCCATCGAGGAGAAGTTCCGGGAGCGCCTGAAGGAGCTCGTGGTCCCCAAGCACGTCATGGATGTTGTGGACGAGGAGCTGAGCAAGCTG GGCCTGCTGGACAACCACTCCTCGGAGTTCAAAACTGTAATTAAGAAGGAAGAGAAGATTGAGAAGAAGGAGGAAAAAAAGCCTGAAGAC ATTAAGAAGGAAGAAAAAGACCAGGATGAGCTGAAACCCGGACCTACAAATCGGTCTAGAGTCACCAAATCAGGAAGCAGAGGAATGGAG CGGACGGTCGTGATGGATAAATCGAAAGGAGAGCCCGTCATTAGCGTGAAAACCACAAGCAGGTCCAAAGAGAGAAGCTCCAAGAGTCAG GATCGCAAGTCAGAAAGCAAAGAAAAGAGAGACATCTTGTCGTTTGATAAAATCAAAGAACAAAGGGAGAGAGAGCGCCAGAGGCAGCGG GAACGGGAGATCCGCGAAACGGAGAGGCGGCGGGAGCGCGAGCAGCGGGAGCGGGAGCAACGCCTCGAGGCCTTCCATGAGCGGAAGGAG AAGGCCCGGCTACAGCGGGAACGCCTGCAGCTCGAGTGCCAGCGCCAGCGGCTGGAGCGGGAGCGCATGGAGCGGGAGCGGCTGGAGCGC GAGCGCATGCGCGTGGAGCGTGAGCGCAGGAAGGAGCAGGAGCGCATCCACCGCGAGCGCGAGGAGCTGCGGCGCCAGCAGGAGCAGCTG CGTTACGAGCAGGAGCGGCGGCCCGGGCGGAGGCCCTACGACCTGGACCGACGAGATGATGCCTATTGGCCAGAAGGAAAGCGTGTGGCA ATGGAGGACCGATATCGTGCAGACTTTCCCCGGCCAGACCACCGCTTTCACGACTTCGATCATCGAGACCGGGGCCAGTACCAGGACCAC GCCATCGACAGGCGGGAGGGTTCGAGGCCAATGATGGGAGACCACCGGGATGGGCAGCACTATGGAGATGACCGCCATGGCCACGGAGGA CCCCCAGAGCGCCACGGCCGGGACTCCCGTGATGGCTGGGGGGGCTACGGCTCCGACAAGAGGCTGAGTGAAGGCCGGGGGCTGCCCCCT CCCCCCAGGGGTGGCCGTGACTGGGGAGAGCACAACCAGCGGCTAGAGGAGCACCAGGCACGCGCCTGGCAGGGTGCCATGGACGCAGGC GCGGCTAGCCGGGAGCACGCCAGGTGGCAAGGTGGCGAGAGGGGCCTGTCTGGGCCCTCGGGGCCGGGGCACATGGCAAGCCGCGGTGGA GTGGCGGGGCGAGGCGGCTTTGCACAAGGTGGACATTCCCAGGGCCACGTGGTGCCAGGTGGCGGACTGGAAGGTGGCGGAGTGGCCAGC CAGGACCGGGGCAGCAGAGTCCCTCACCCACACCCTCATCCCCCCCCGTACCCCCACTTCACCCGCCGCTACTAAGTCCCACTCGCTGCG AGTTTTCGGGTGGGCAGACGCACTGTTGAATCTGGTAGCCAGGGTTCCCTCGAACTTGGGGGATCTTTTTAAAAGCAAAGTAAATCCTGC CACCATGTTGTAGCTCAATACAATGTGAACTCACTTTTTTTTTTTTTTTAAATAAATGTGTTCTTGTTCTGCCATTTTTAAATCAAGGTT >49426_49426_3_LONP1-SAFB2_LONP1_chr19_5705783_ENST00000540670_SAFB2_chr19_5600271_ENST00000252542_length(amino acids)=741AA_BP=1 MVELLRRKVRLAQPYVGVFLKRDDSNESDVVESLDEIYHTGTFAQIHEMQDLGDKLRMIVMGHRRVHISRQLEVEPEEPEAENKHKPRRK SKRGKKEAEDELSARHPAELAMEPTPELPAEVLMVEVENVVHEDFQVTEEVKALTAEIVKTIRDIIALNPLYRESVLQMMQAGQRVVDNP IYLSDMGAALTGAESHELQDVLEETNIPKRLYKALSLLKKEFELSKLQQRLGREVEEKIKQTHRKYLLQEQLKIIKKELGLEKDDKDAIE EKFRERLKELVVPKHVMDVVDEELSKLGLLDNHSSEFKTVIKKEEKIEKKEEKKPEDIKKEEKDQDELKPGPTNRSRVTKSGSRGMERTV VMDKSKGEPVISVKTTSRSKERSSKSQDRKSESKEKRDILSFDKIKEQRERERQRQREREIRETERRREREQREREQRLEAFHERKEKAR LQRERLQLECQRQRLERERMERERLERERMRVERERRKEQERIHREREELRRQQEQLRYEQERRPGRRPYDLDRRDDAYWPEGKRVAMED RYRADFPRPDHRFHDFDHRDRGQYQDHAIDRREGSRPMMGDHRDGQHYGDDRHGHGGPPERHGRDSRDGWGGYGSDKRLSEGRGLPPPPR GGRDWGEHNQRLEEHQARAWQGAMDAGAASREHARWQGGERGLSGPSGPGHMASRGGVAGRGGFAQGGHSQGHVVPGGGLEGGGVASQDR -------------------------------------------------------------- >49426_49426_4_LONP1-SAFB2_LONP1_chr19_5705783_ENST00000590729_SAFB2_chr19_5600271_ENST00000252542_length(transcript)=2583nt_BP=1036nt GCGGCGCGGGGGGCAGCGCGGGCGCCGGGGAAGGCCCGGTCATAACGGCGCTCACGCCCATGACGATCCCCGATGTGTTTCCGCACCTGC CGCTCATCGCCATCACCCGCAACCCGGTGTTCCCGCGCTTTATCAAGATTATCGAGGTTAAAAATAAGAAGTTGGTTGAGCTGCTGAGAA GGAAAGTTCGTCTCGCCCAGCCTTATGTCGGCGTCTTTCTAAAGAGAGATGACAGCAATGAGTCGGATGTGGTCGAGAGCCTGGATGAAA TCTACCACACGGGGACGTTTGCCCAGATCCATGAGATGCAGGACCTTGGGGACAAGCTGCGCATGATCGTCATGGGACACAGAAGAGTCC ATATCAGCAGACAGCTGGAGGTGGAGCCCGAGGAGCCGGAGGCGGAGAACAAGCACAAGCCCCGCAGGAAGTCAAAGCGGGGCAAGAAGG AGGCGGAGGACGAGCTGAGCGCCAGGCACCCGGCGGAGCTGGCGATGGAGCCCACCCCTGAGCTCCCGGCTGAGGTGCTCATGGTGGAGG CCCTGACTGCAGAGATCGTGAAGACCATCCGGGACATCATTGCCTTGAACCCTCTCTACAGGGAGTCAGTGCTGCAGATGATGCAGGCTG GCCAGCGGGTGGTGGACAACCCCATCTACCTGAGCGACATGGGCGCCGCGCTCACCGGGGCCGAGTCCCATGAGCTGCAGGACGTCCTGG AAGAGACCAATATTCCTAAGCGGCTGTACAAGGCCCTCTCCCTGCTGAAGAAGGAATTTGAACTGAGCAAGCTGCAGCAGCGCCTGGGGC GGGAGGTGGAGGAGAAGATCAAGCAGACCCACCGTAAGTACCTGCTGCAGGAGCAGCTAAAGATCATCAAGAAGGAGCTGGGCCTGGAGA AGGACGACAAGGATGCCATCGAGGAGAAGTTCCGGGAGCGCCTGAAGGAGCTCGTGGTCCCCAAGCACGTCATGGATGTTGTGGACGAGG AGCTGAGCAAGCTGGGCCTGCTGGACAACCACTCCTCGGAGTTCAAAACTGTAATTAAGAAGGAAGAGAAGATTGAGAAGAAGGAGGAAA AAAAGCCTGAAGACATTAAGAAGGAAGAAAAAGACCAGGATGAGCTGAAACCCGGACCTACAAATCGGTCTAGAGTCACCAAATCAGGAA GCAGAGGAATGGAGCGGACGGTCGTGATGGATAAATCGAAAGGAGAGCCCGTCATTAGCGTGAAAACCACAAGCAGGTCCAAAGAGAGAA GCTCCAAGAGTCAGGATCGCAAGTCAGAAAGCAAAGAAAAGAGAGACATCTTGTCGTTTGATAAAATCAAAGAACAAAGGGAGAGAGAGC GCCAGAGGCAGCGGGAACGGGAGATCCGCGAAACGGAGAGGCGGCGGGAGCGCGAGCAGCGGGAGCGGGAGCAACGCCTCGAGGCCTTCC ATGAGCGGAAGGAGAAGGCCCGGCTACAGCGGGAACGCCTGCAGCTCGAGTGCCAGCGCCAGCGGCTGGAGCGGGAGCGCATGGAGCGGG AGCGGCTGGAGCGCGAGCGCATGCGCGTGGAGCGTGAGCGCAGGAAGGAGCAGGAGCGCATCCACCGCGAGCGCGAGGAGCTGCGGCGCC AGCAGGAGCAGCTGCGTTACGAGCAGGAGCGGCGGCCCGGGCGGAGGCCCTACGACCTGGACCGACGAGATGATGCCTATTGGCCAGAAG GAAAGCGTGTGGCAATGGAGGACCGATATCGTGCAGACTTTCCCCGGCCAGACCACCGCTTTCACGACTTCGATCATCGAGACCGGGGCC AGTACCAGGACCACGCCATCGACAGGCGGGAGGGTTCGAGGCCAATGATGGGAGACCACCGGGATGGGCAGCACTATGGAGATGACCGCC ATGGCCACGGAGGACCCCCAGAGCGCCACGGCCGGGACTCCCGTGATGGCTGGGGGGGCTACGGCTCCGACAAGAGGCTGAGTGAAGGCC GGGGGCTGCCCCCTCCCCCCAGGGGTGGCCGTGACTGGGGAGAGCACAACCAGCGGCTAGAGGAGCACCAGGCACGCGCCTGGCAGGGTG CCATGGACGCAGGCGCGGCTAGCCGGGAGCACGCCAGGTGGCAAGGTGGCGAGAGGGGCCTGTCTGGGCCCTCGGGGCCGGGGCACATGG CAAGCCGCGGTGGAGTGGCGGGGCGAGGCGGCTTTGCACAAGGTGGACATTCCCAGGGCCACGTGGTGCCAGGTGGCGGACTGGAAGGTG GCGGAGTGGCCAGCCAGGACCGGGGCAGCAGAGTCCCTCACCCACACCCTCATCCCCCCCCGTACCCCCACTTCACCCGCCGCTACTAAG TCCCACTCGCTGCGAGTTTTCGGGTGGGCAGACGCACTGTTGAATCTGGTAGCCAGGGTTCCCTCGAACTTGGGGGATCTTTTTAAAAGC AAAGTAAATCCTGCCACCATGTTGTAGCTCAATACAATGTGAACTCACTTTTTTTTTTTTTTTAAATAAATGTGTTCTTGTTCTGCCATT >49426_49426_4_LONP1-SAFB2_LONP1_chr19_5705783_ENST00000590729_SAFB2_chr19_5600271_ENST00000252542_length(amino acids)=759AA_BP=0 MTIPDVFPHLPLIAITRNPVFPRFIKIIEVKNKKLVELLRRKVRLAQPYVGVFLKRDDSNESDVVESLDEIYHTGTFAQIHEMQDLGDKL RMIVMGHRRVHISRQLEVEPEEPEAENKHKPRRKSKRGKKEAEDELSARHPAELAMEPTPELPAEVLMVEALTAEIVKTIRDIIALNPLY RESVLQMMQAGQRVVDNPIYLSDMGAALTGAESHELQDVLEETNIPKRLYKALSLLKKEFELSKLQQRLGREVEEKIKQTHRKYLLQEQL KIIKKELGLEKDDKDAIEEKFRERLKELVVPKHVMDVVDEELSKLGLLDNHSSEFKTVIKKEEKIEKKEEKKPEDIKKEEKDQDELKPGP TNRSRVTKSGSRGMERTVVMDKSKGEPVISVKTTSRSKERSSKSQDRKSESKEKRDILSFDKIKEQRERERQRQREREIRETERRREREQ REREQRLEAFHERKEKARLQRERLQLECQRQRLERERMERERLERERMRVERERRKEQERIHREREELRRQQEQLRYEQERRPGRRPYDL DRRDDAYWPEGKRVAMEDRYRADFPRPDHRFHDFDHRDRGQYQDHAIDRREGSRPMMGDHRDGQHYGDDRHGHGGPPERHGRDSRDGWGG YGSDKRLSEGRGLPPPPRGGRDWGEHNQRLEEHQARAWQGAMDAGAASREHARWQGGERGLSGPSGPGHMASRGGVAGRGGFAQGGHSQG -------------------------------------------------------------- >49426_49426_5_LONP1-SAFB2_LONP1_chr19_5705783_ENST00000593119_SAFB2_chr19_5600271_ENST00000252542_length(transcript)=2755nt_BP=1208nt GCGAGTTGTGTGAGCCGCCAGTATGGCCGGGCTATGGCGGCGAGCACTGGCTACGTGCGACTGTGGGGAGCGGCGCGGTGCTGGGTGCTG CGGCGGCCGATGCTGGCCGCCGCCGGGGGGCGGGTTCCCACTGCAGCAGGAGCGTGGTTGCTCCGAGGCCCGGTCATAACGGCGCTCACG CCCATGACGATCCCCGATGTGTTTCCGCACCTGCCGCTCATCGCCATCACCCGCAACCCGGTGTTCCCGCGCTTTATCAAGATTATCGAG GTTAAAAATAAGAAGTTGGTTGAGCTGCTGAGAAGGAAAGTTCGTCTCGCCCAGCCTTATGTCGGCGTCTTTCTAAAGAGAGATGACAGC AATGAGTCGGATGTGGTCGAGAGCCTGGATGAAATCTACCACACGGGGACGTTTGCCCAGATCCATGAGATGCAGGACCTTGGGGACAAG CTGCGCATGATCGTCATGGGACACAGAAGAGTCCATATCAGCAGACAGCTGGAGGTGGAGCCCGAGGAGCCGGAGGCGGAGAACAAGCAC AAGCCCCGCAGGAAGTCAAAGCGGGGCAAGAAGGAGGCGGAGGACGAGCTGAGCGCCAGGCACCCGGCGGAGCTGGCGATGGAGCCCACC CCTGAGCTCCCGGCTGAGGTGCTCATGGTGGAGGTAGAGAACGTTGTCCACGAGGACTTCCAGGTCACGGAGGAGGTGAAAGCCCTGACT GCAGAGATCGTGAAGACCATCCGGGACATCATTGCCTTGAACCCTCTCTACAGGGAGTCAGTGCTGCAGATGATGCAGGCTGGCCAGCGG GTGGTGGACAACCCCATCTACCTGAGCGACATGGGCGCCGCGCTCACCGGGGCCGAGTCCCATGAGCTGCAGGACGTCCTGGAAGAGACC AATATTCCTAAGCGGCTGTACAAGGCCCTCTCCCTGCTGAAGAAGGAATTTGAACTGAGCAAGCTGCAGCAGCGCCTGGGGCGGGAGGTG GAGGAGAAGATCAAGCAGACCCACCGTAAGTACCTGCTGCAGGAGCAGCTAAAGATCATCAAGAAGGAGCTGGGCCTGGAGAAGGACGAC AAGGATGCCATCGAGGAGAAGTTCCGGGAGCGCCTGAAGGAGCTCGTGGTCCCCAAGCACGTCATGGATGTTGTGGACGAGGAGCTGAGC AAGCTGGGCCTGCTGGACAACCACTCCTCGGAGTTCAAAACTGTAATTAAGAAGGAAGAGAAGATTGAGAAGAAGGAGGAAAAAAAGCCT GAAGACATTAAGAAGGAAGAAAAAGACCAGGATGAGCTGAAACCCGGACCTACAAATCGGTCTAGAGTCACCAAATCAGGAAGCAGAGGA ATGGAGCGGACGGTCGTGATGGATAAATCGAAAGGAGAGCCCGTCATTAGCGTGAAAACCACAAGCAGGTCCAAAGAGAGAAGCTCCAAG AGTCAGGATCGCAAGTCAGAAAGCAAAGAAAAGAGAGACATCTTGTCGTTTGATAAAATCAAAGAACAAAGGGAGAGAGAGCGCCAGAGG CAGCGGGAACGGGAGATCCGCGAAACGGAGAGGCGGCGGGAGCGCGAGCAGCGGGAGCGGGAGCAACGCCTCGAGGCCTTCCATGAGCGG AAGGAGAAGGCCCGGCTACAGCGGGAACGCCTGCAGCTCGAGTGCCAGCGCCAGCGGCTGGAGCGGGAGCGCATGGAGCGGGAGCGGCTG GAGCGCGAGCGCATGCGCGTGGAGCGTGAGCGCAGGAAGGAGCAGGAGCGCATCCACCGCGAGCGCGAGGAGCTGCGGCGCCAGCAGGAG CAGCTGCGTTACGAGCAGGAGCGGCGGCCCGGGCGGAGGCCCTACGACCTGGACCGACGAGATGATGCCTATTGGCCAGAAGGAAAGCGT GTGGCAATGGAGGACCGATATCGTGCAGACTTTCCCCGGCCAGACCACCGCTTTCACGACTTCGATCATCGAGACCGGGGCCAGTACCAG GACCACGCCATCGACAGGCGGGAGGGTTCGAGGCCAATGATGGGAGACCACCGGGATGGGCAGCACTATGGAGATGACCGCCATGGCCAC GGAGGACCCCCAGAGCGCCACGGCCGGGACTCCCGTGATGGCTGGGGGGGCTACGGCTCCGACAAGAGGCTGAGTGAAGGCCGGGGGCTG CCCCCTCCCCCCAGGGGTGGCCGTGACTGGGGAGAGCACAACCAGCGGCTAGAGGAGCACCAGGCACGCGCCTGGCAGGGTGCCATGGAC GCAGGCGCGGCTAGCCGGGAGCACGCCAGGTGGCAAGGTGGCGAGAGGGGCCTGTCTGGGCCCTCGGGGCCGGGGCACATGGCAAGCCGC GGTGGAGTGGCGGGGCGAGGCGGCTTTGCACAAGGTGGACATTCCCAGGGCCACGTGGTGCCAGGTGGCGGACTGGAAGGTGGCGGAGTG GCCAGCCAGGACCGGGGCAGCAGAGTCCCTCACCCACACCCTCATCCCCCCCCGTACCCCCACTTCACCCGCCGCTACTAAGTCCCACTC GCTGCGAGTTTTCGGGTGGGCAGACGCACTGTTGAATCTGGTAGCCAGGGTTCCCTCGAACTTGGGGGATCTTTTTAAAAGCAAAGTAAA TCCTGCCACCATGTTGTAGCTCAATACAATGTGAACTCACTTTTTTTTTTTTTTTAAATAAATGTGTTCTTGTTCTGCCATTTTTAAATC >49426_49426_5_LONP1-SAFB2_LONP1_chr19_5705783_ENST00000593119_SAFB2_chr19_5600271_ENST00000252542_length(amino acids)=825AA_BP=0 MAASTGYVRLWGAARCWVLRRPMLAAAGGRVPTAAGAWLLRGPVITALTPMTIPDVFPHLPLIAITRNPVFPRFIKIIEVKNKKLVELLR RKVRLAQPYVGVFLKRDDSNESDVVESLDEIYHTGTFAQIHEMQDLGDKLRMIVMGHRRVHISRQLEVEPEEPEAENKHKPRRKSKRGKK EAEDELSARHPAELAMEPTPELPAEVLMVEVENVVHEDFQVTEEVKALTAEIVKTIRDIIALNPLYRESVLQMMQAGQRVVDNPIYLSDM GAALTGAESHELQDVLEETNIPKRLYKALSLLKKEFELSKLQQRLGREVEEKIKQTHRKYLLQEQLKIIKKELGLEKDDKDAIEEKFRER LKELVVPKHVMDVVDEELSKLGLLDNHSSEFKTVIKKEEKIEKKEEKKPEDIKKEEKDQDELKPGPTNRSRVTKSGSRGMERTVVMDKSK GEPVISVKTTSRSKERSSKSQDRKSESKEKRDILSFDKIKEQRERERQRQREREIRETERRREREQREREQRLEAFHERKEKARLQRERL QLECQRQRLERERMERERLERERMRVERERRKEQERIHREREELRRQQEQLRYEQERRPGRRPYDLDRRDDAYWPEGKRVAMEDRYRADF PRPDHRFHDFDHRDRGQYQDHAIDRREGSRPMMGDHRDGQHYGDDRHGHGGPPERHGRDSRDGWGGYGSDKRLSEGRGLPPPPRGGRDWG EHNQRLEEHQARAWQGAMDAGAASREHARWQGGERGLSGPSGPGHMASRGGVAGRGGFAQGGHSQGHVVPGGGLEGGGVASQDRGSRVPH -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for LONP1-SAFB2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | SAFB2 | chr19:5705782 | chr19:5604947 | ENST00000252542 | 8 | 21 | 600_953 | 432.0 | 954.0 | SAFB1 | |
| Tgene | SAFB2 | chr19:5705783 | chr19:5600271 | ENST00000252542 | 10 | 21 | 600_953 | 519.6666666666666 | 954.0 | SAFB1 |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for LONP1-SAFB2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for LONP1-SAFB2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
