
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:LRCH1-ATP13A3 (FusionGDB2 ID:49633) |
Fusion Gene Summary for LRCH1-ATP13A3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: LRCH1-ATP13A3 | Fusion gene ID: 49633 | Hgene | Tgene | Gene symbol | LRCH1 | ATP13A3 | Gene ID | 23143 | 79572 |
| Gene name | leucine rich repeats and calponin homology domain containing 1 | ATPase 13A3 | |
| Synonyms | CHDC1|NP81 | AFURS1 | |
| Cytomap | 13q14.13-q14.2 | 3q29 | |
| Type of gene | protein-coding | protein-coding | |
| Description | leucine-rich repeat and calponin homology domain-containing protein 1leucine-rich repeats and calponin homology (CH) domain containing 1neuronal protein 81 | probable cation-transporting ATPase 13A3ATPase family homolog up-regulated in senescence cells 1ATPase type 13A3 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9Y2L9 | Q9H7F0 | |
| Ensembl transtripts involved in fusion gene | ENST00000311191, ENST00000389797, ENST00000389798, | ENST00000439040, ENST00000256031, | |
| Fusion gene scores | * DoF score | 12 X 7 X 5=420 | 7 X 9 X 5=315 |
| # samples | 13 | 11 | |
| ** MAII score | log2(13/420*10)=-1.69187770463767 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(11/315*10)=-1.51784830486262 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: LRCH1 [Title/Abstract] AND ATP13A3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | LRCH1(47127838)-ATP13A3(194126845), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | LRCH1-ATP13A3 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. LRCH1-ATP13A3 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. LRCH1-ATP13A3 seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | LRCH1 | GO:0034260 | negative regulation of GTPase activity | 28028151 |
 Fusion gene breakpoints across LRCH1 (5'-gene) Fusion gene breakpoints across LRCH1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
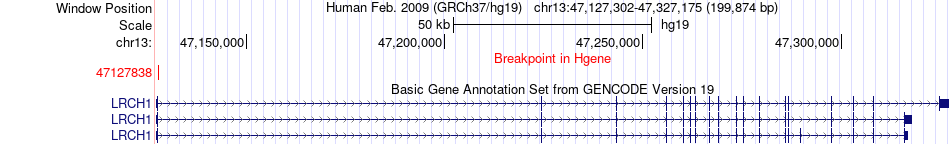 |
 Fusion gene breakpoints across ATP13A3 (3'-gene) Fusion gene breakpoints across ATP13A3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
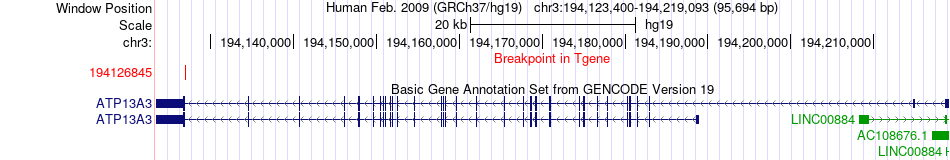 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | Non-Cancer | 173N | LRCH1 | chr13 | 47127838 | + | ATP13A3 | chr3 | 194126845 | - |
Top |
Fusion Gene ORF analysis for LRCH1-ATP13A3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| Frame-shift | ENST00000311191 | ENST00000439040 | LRCH1 | chr13 | 47127838 | + | ATP13A3 | chr3 | 194126845 | - |
| Frame-shift | ENST00000389797 | ENST00000439040 | LRCH1 | chr13 | 47127838 | + | ATP13A3 | chr3 | 194126845 | - |
| Frame-shift | ENST00000389798 | ENST00000439040 | LRCH1 | chr13 | 47127838 | + | ATP13A3 | chr3 | 194126845 | - |
| In-frame | ENST00000311191 | ENST00000256031 | LRCH1 | chr13 | 47127838 | + | ATP13A3 | chr3 | 194126845 | - |
| In-frame | ENST00000389797 | ENST00000256031 | LRCH1 | chr13 | 47127838 | + | ATP13A3 | chr3 | 194126845 | - |
| In-frame | ENST00000389798 | ENST00000256031 | LRCH1 | chr13 | 47127838 | + | ATP13A3 | chr3 | 194126845 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000311191 | LRCH1 | chr13 | 47127838 | + | ENST00000256031 | ATP13A3 | chr3 | 194126845 | - | 3973 | 536 | 615 | 34 | 193 |
| ENST00000389798 | LRCH1 | chr13 | 47127838 | + | ENST00000256031 | ATP13A3 | chr3 | 194126845 | - | 3941 | 504 | 583 | 2 | 193 |
| ENST00000389797 | LRCH1 | chr13 | 47127838 | + | ENST00000256031 | ATP13A3 | chr3 | 194126845 | - | 3881 | 444 | 523 | 2 | 174 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000311191 | ENST00000256031 | LRCH1 | chr13 | 47127838 | + | ATP13A3 | chr3 | 194126845 | - | 0.6142494 | 0.38575056 |
| ENST00000389798 | ENST00000256031 | LRCH1 | chr13 | 47127838 | + | ATP13A3 | chr3 | 194126845 | - | 0.61021346 | 0.38978657 |
| ENST00000389797 | ENST00000256031 | LRCH1 | chr13 | 47127838 | + | ATP13A3 | chr3 | 194126845 | - | 0.6088785 | 0.39112148 |
Top |
Fusion Genomic Features for LRCH1-ATP13A3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
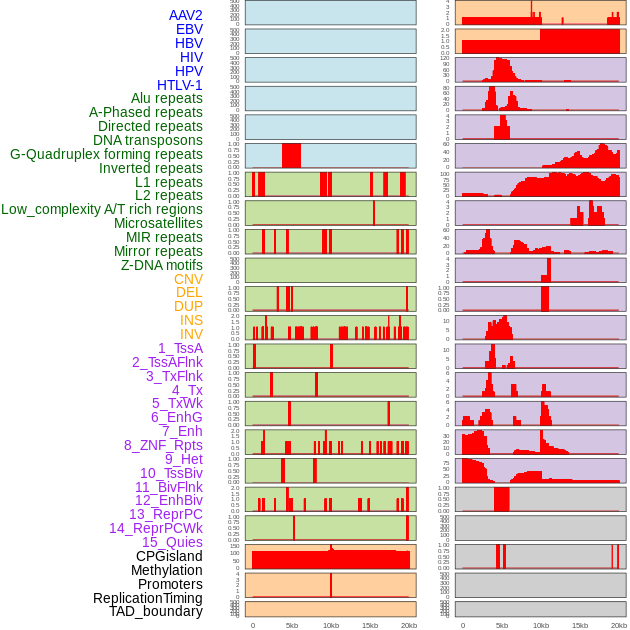 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for LRCH1-ATP13A3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr13:47127838/chr3:194126845) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| LRCH1 | ATP13A3 |
| FUNCTION: Acts as a negative regulator of GTPase CDC42 by sequestering CDC42-guanine exchange factor DOCK8. Probably by preventing CDC42 activation, negatively regulates CD4(+) T-cell migration. {ECO:0000269|PubMed:28028151}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000311191 | + | 1 | 19 | 21_36 | 102 | 697.0 | Compositional bias | Note=His-rich |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000311191 | + | 1 | 19 | 37_54 | 102 | 697.0 | Compositional bias | Note=Gly-rich |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000389797 | + | 1 | 20 | 21_36 | 102 | 764.0 | Compositional bias | Note=His-rich |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000389797 | + | 1 | 20 | 37_54 | 102 | 764.0 | Compositional bias | Note=Gly-rich |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000389798 | + | 1 | 19 | 21_36 | 102 | 729.0 | Compositional bias | Note=His-rich |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000389798 | + | 1 | 19 | 37_54 | 102 | 729.0 | Compositional bias | Note=Gly-rich |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000311191 | + | 1 | 19 | 576_692 | 102 | 697.0 | Domain | Calponin-homology (CH) |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000389797 | + | 1 | 20 | 576_692 | 102 | 764.0 | Domain | Calponin-homology (CH) |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000389798 | + | 1 | 19 | 576_692 | 102 | 729.0 | Domain | Calponin-homology (CH) |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000311191 | + | 1 | 19 | 121_143 | 102 | 697.0 | Repeat | Note=LRR 2 |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000311191 | + | 1 | 19 | 144_166 | 102 | 697.0 | Repeat | Note=LRR 3 |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000311191 | + | 1 | 19 | 167_187 | 102 | 697.0 | Repeat | Note=LRR 4 |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000311191 | + | 1 | 19 | 189_210 | 102 | 697.0 | Repeat | Note=LRR 5 |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000311191 | + | 1 | 19 | 212_234 | 102 | 697.0 | Repeat | Note=LRR 6 |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000311191 | + | 1 | 19 | 235_255 | 102 | 697.0 | Repeat | Note=LRR 7 |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000311191 | + | 1 | 19 | 257_278 | 102 | 697.0 | Repeat | Note=LRR 8 |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000311191 | + | 1 | 19 | 283_304 | 102 | 697.0 | Repeat | Note=LRR 9 |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000311191 | + | 1 | 19 | 98_119 | 102 | 697.0 | Repeat | Note=LRR 1 |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000389797 | + | 1 | 20 | 121_143 | 102 | 764.0 | Repeat | Note=LRR 2 |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000389797 | + | 1 | 20 | 144_166 | 102 | 764.0 | Repeat | Note=LRR 3 |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000389797 | + | 1 | 20 | 167_187 | 102 | 764.0 | Repeat | Note=LRR 4 |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000389797 | + | 1 | 20 | 189_210 | 102 | 764.0 | Repeat | Note=LRR 5 |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000389797 | + | 1 | 20 | 212_234 | 102 | 764.0 | Repeat | Note=LRR 6 |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000389797 | + | 1 | 20 | 235_255 | 102 | 764.0 | Repeat | Note=LRR 7 |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000389797 | + | 1 | 20 | 257_278 | 102 | 764.0 | Repeat | Note=LRR 8 |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000389797 | + | 1 | 20 | 283_304 | 102 | 764.0 | Repeat | Note=LRR 9 |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000389797 | + | 1 | 20 | 98_119 | 102 | 764.0 | Repeat | Note=LRR 1 |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000389798 | + | 1 | 19 | 121_143 | 102 | 729.0 | Repeat | Note=LRR 2 |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000389798 | + | 1 | 19 | 144_166 | 102 | 729.0 | Repeat | Note=LRR 3 |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000389798 | + | 1 | 19 | 167_187 | 102 | 729.0 | Repeat | Note=LRR 4 |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000389798 | + | 1 | 19 | 189_210 | 102 | 729.0 | Repeat | Note=LRR 5 |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000389798 | + | 1 | 19 | 212_234 | 102 | 729.0 | Repeat | Note=LRR 6 |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000389798 | + | 1 | 19 | 235_255 | 102 | 729.0 | Repeat | Note=LRR 7 |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000389798 | + | 1 | 19 | 257_278 | 102 | 729.0 | Repeat | Note=LRR 8 |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000389798 | + | 1 | 19 | 283_304 | 102 | 729.0 | Repeat | Note=LRR 9 |
| Hgene | LRCH1 | chr13:47127838 | chr3:194126845 | ENST00000389798 | + | 1 | 19 | 98_119 | 102 | 729.0 | Repeat | Note=LRR 1 |
| Tgene | ATP13A3 | chr13:47127838 | chr3:194126845 | ENST00000256031 | 30 | 32 | 231_234 | 1161 | 1227.0 | Compositional bias | Note=Poly-Tyr | |
| Tgene | ATP13A3 | chr13:47127838 | chr3:194126845 | ENST00000439040 | 31 | 33 | 231_234 | 1161 | 1227.0 | Compositional bias | Note=Poly-Tyr | |
| Tgene | ATP13A3 | chr13:47127838 | chr3:194126845 | ENST00000256031 | 30 | 32 | 1000_1020 | 1161 | 1227.0 | Transmembrane | Helical | |
| Tgene | ATP13A3 | chr13:47127838 | chr3:194126845 | ENST00000256031 | 30 | 32 | 1074_1094 | 1161 | 1227.0 | Transmembrane | Helical | |
| Tgene | ATP13A3 | chr13:47127838 | chr3:194126845 | ENST00000256031 | 30 | 32 | 1106_1126 | 1161 | 1227.0 | Transmembrane | Helical | |
| Tgene | ATP13A3 | chr13:47127838 | chr3:194126845 | ENST00000256031 | 30 | 32 | 1144_1164 | 1161 | 1227.0 | Transmembrane | Helical | |
| Tgene | ATP13A3 | chr13:47127838 | chr3:194126845 | ENST00000256031 | 30 | 32 | 206_226 | 1161 | 1227.0 | Transmembrane | Helical | |
| Tgene | ATP13A3 | chr13:47127838 | chr3:194126845 | ENST00000256031 | 30 | 32 | 233_253 | 1161 | 1227.0 | Transmembrane | Helical | |
| Tgene | ATP13A3 | chr13:47127838 | chr3:194126845 | ENST00000256031 | 30 | 32 | 29_49 | 1161 | 1227.0 | Transmembrane | Helical | |
| Tgene | ATP13A3 | chr13:47127838 | chr3:194126845 | ENST00000256031 | 30 | 32 | 410_430 | 1161 | 1227.0 | Transmembrane | Helical | |
| Tgene | ATP13A3 | chr13:47127838 | chr3:194126845 | ENST00000256031 | 30 | 32 | 449_469 | 1161 | 1227.0 | Transmembrane | Helical | |
| Tgene | ATP13A3 | chr13:47127838 | chr3:194126845 | ENST00000256031 | 30 | 32 | 941_961 | 1161 | 1227.0 | Transmembrane | Helical | |
| Tgene | ATP13A3 | chr13:47127838 | chr3:194126845 | ENST00000256031 | 30 | 32 | 963_983 | 1161 | 1227.0 | Transmembrane | Helical | |
| Tgene | ATP13A3 | chr13:47127838 | chr3:194126845 | ENST00000439040 | 31 | 33 | 1000_1020 | 1161 | 1227.0 | Transmembrane | Helical | |
| Tgene | ATP13A3 | chr13:47127838 | chr3:194126845 | ENST00000439040 | 31 | 33 | 1074_1094 | 1161 | 1227.0 | Transmembrane | Helical | |
| Tgene | ATP13A3 | chr13:47127838 | chr3:194126845 | ENST00000439040 | 31 | 33 | 1106_1126 | 1161 | 1227.0 | Transmembrane | Helical | |
| Tgene | ATP13A3 | chr13:47127838 | chr3:194126845 | ENST00000439040 | 31 | 33 | 1144_1164 | 1161 | 1227.0 | Transmembrane | Helical | |
| Tgene | ATP13A3 | chr13:47127838 | chr3:194126845 | ENST00000439040 | 31 | 33 | 206_226 | 1161 | 1227.0 | Transmembrane | Helical | |
| Tgene | ATP13A3 | chr13:47127838 | chr3:194126845 | ENST00000439040 | 31 | 33 | 233_253 | 1161 | 1227.0 | Transmembrane | Helical | |
| Tgene | ATP13A3 | chr13:47127838 | chr3:194126845 | ENST00000439040 | 31 | 33 | 29_49 | 1161 | 1227.0 | Transmembrane | Helical | |
| Tgene | ATP13A3 | chr13:47127838 | chr3:194126845 | ENST00000439040 | 31 | 33 | 410_430 | 1161 | 1227.0 | Transmembrane | Helical | |
| Tgene | ATP13A3 | chr13:47127838 | chr3:194126845 | ENST00000439040 | 31 | 33 | 449_469 | 1161 | 1227.0 | Transmembrane | Helical | |
| Tgene | ATP13A3 | chr13:47127838 | chr3:194126845 | ENST00000439040 | 31 | 33 | 941_961 | 1161 | 1227.0 | Transmembrane | Helical | |
| Tgene | ATP13A3 | chr13:47127838 | chr3:194126845 | ENST00000439040 | 31 | 33 | 963_983 | 1161 | 1227.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for LRCH1-ATP13A3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >49633_49633_1_LRCH1-ATP13A3_LRCH1_chr13_47127838_ENST00000311191_ATP13A3_chr3_194126845_ENST00000256031_length(transcript)=3973nt_BP=536nt CCGCAGTCCTTAGCTTCCCGGGGACAGGAAACCTTCAAGACCGAGCTGCCACGGCCGCCTCCCCGCCCGCCCCCCATTCTACGCGCCTGC CCACACCCTCCTCCCCTCCTTCCAGCGCCTTTCGGTGGAGCACTGCGGCACTCAGCCCGAGCTGCCGTTTTCCCCTCGCGGGGAACGCTG TGACCCCCCCGCAGGAGCGGCGGGGCGGGGTGGGGGGGCCCGGGAGAAGATGGCGACGCCGGGAAGCGAACCCCAACCTTTCGTCCCGGC CCTTTCGGTAGCTACTCTGCACCCACTTCATCATCCCCACCACCACCACCACCACCATCAGCACCACGGAGGAACCGGCGCCCCCGGCGG GGCGGGTGGTGGCGGCGGTGGCAGCGGGGGCTTCAACCTGCCCTTGAACCGGGGTCTGGAGCGCGCGCTTGAGGAGGCGGCCAACTCCGG GGGGCTGAACCTGAGCGCCAGGAAATTGAAGGAATTTCCCCGTACCGCAGCCCCCGGGCACGACCTCTCGGACACGGTGCAGGCAGGAGT CAGTGGATCGGTGGGGAAAATGCTGCTTACCCTGGGCCCTGGGCTGTAGAAAGAAGACACCAAAGGCAAAGTACATGTATCTGGCGCAGG AGCTCTTGGTTGATCCAGAATGGCCACCAAAACCTCAGACAACCACAGAAGCTAAAGCTTTAGTTAAGGAGAATGGATCATGTCAAATCA TCACCATAACATAGCAGTGAATCAGTCTCAGTGGTATTGCTGATAGCAGTATTCAGGAATATGTGATTTTAGGAGTTTCTGATCCTGTGT GTCAGAATGGCACTAGTTCAGTTTATGTCCCTTCTGATATAGTAGCTTATTTGACAGCTTTGCTCTTCCTTAAAATAAAAACAGAAAAAT ATATCGTCCTAACAGTTAAATTAACAATCAATCCATAAAGTCCTATATCTTCATTCAGCAACCCAAATATTACATACATTTCCAGAATTT TCTTGATTGTTACTTTCAGTGATATTCTTTATATTGGGTACAGGAGAAGTTTGGTGTTTGGTAGGTTTTTCAACATTAGTTTTTGAGACT AGTTTACCTCTTCACATTTATGCTCACAACCCTCTTGTTAGAAAAGTCTGTGTTTATATACAGGCTGTAAGTTTGTGATTGATAAAAAGA AGATGAGTGTTAATTAGCCTCCAGTGAAAATATACTGAAAGCCTGTTTTCATTTGATTCCAATGTTTCTTCCAAAGAATTCTGTATAAAC ATATGCCAATTCCCTATGATGGTCTAGAGTTAGGAATGAGTGTTTATGGTGTTGCTTATAGAACAACTCAGGTAATCTCCATTTCTGGTT TTATATTTTCTGTACAAACTGCCTGGGTTTTATTTTTCTAATCAGCAAGGTGCTTCACTGCCTTCTTGAGACGCCTCTCAAAGCTCTTAA ATGGCTCCTGTGCTATGTGTGGTGTTGGCAGTCTAATTTGCTTCTGTTAAATGTTGTAGAACCTTTTTCACTAGGAAATAAGATTCATTT CTTTCGGCAGTAGATGTAGATTCATCTTTTAACGTTTCTTCAAATTTGTTTCTGTCAGGCTTTGTGTTATTTTAAATGGTTTTTTAAAAT TTTCTTCTATGTTTTCAATTACCTAAAGACATAGGATAATAGTTTTTTTTAAGTTAGAATTTTACCTCATAAAATTTTTTGAGGTTTGAT GTATGTCTCTGTCTTATCAATAATGAGGCTTAAAAAATACTGGATTTGAATGGCTGCCGTTTTTTCAAAGCAATATGAATTTGATGAGTT TGTTTTATGCCATTAGGTGGCGCCAGAGGTCAGAACATGTCTATTTTGAATTGGATCGTTACAAATGAGCATATTTGATGCGGAAATTTC TGGGAGAAAAAAAATTGAGGAAATAAAGTTAAAAAATTGACATTCATTGAGCCAAAAGAGATGTGGAGAAACATTTTTCACCTTTCTGTT TGGCCTGATTAACATTTAAATTCTTGCCAAAATTAAGGTAACTTTTAAAAAACACCTTTTATAGGTGGATCCAGCAGTCTGGCAACGCCC ACAGTTACCACAACACAGAAAACTGATCGTGCTATAAAATGGACGCTAAACTATGAAAACAGTGTGACATTGTTCTCTGTTCTTCCAGAG CCAGTAACATGCTTGCTCGTGCTTTCTACTTCTAGCTGATCATTCTTTTCCCAACATATATTTACAAATTTACCAAATTTTACCTAGAAT TTTAGGACCAAATGGTTCTCACTCTTTATGCTGCAAAGACCTGGATGATGTTTGGTAACTATAGAAAAATAGAAATTACACTCAGGATCA CTGTTACTGCTATTGCCACTGATGATTCCTGCAAAAATATAATCGAAGTTTTCCATCAAATGTATAATATGCTATTAATACACATTAGAT GATAACAGTTGTTCCATGAATGATTCTATGAAGCTATGCATCTTAGACCTCTTGAGCTGTGAATTAGCACTATTTTCTATAGTTACTTAT TCTCTGGATCATTTTATAATTTCCATATTAATTTCAAATATGCTCGTGTTATTCTTCAGTGATTTCCACAATTGTGCATTTTATTCTTTG GTTTAAGTACTGAAGCATATAATGAAAGTAATTGCTAAGTAGCAGCTTAAAAATTCAATTATCCGATTGTATTTAACATCTTTAAGAGCA TGATCATAAAGAGCTATTTTTGACACCCCCCCCCCACTTTTTTAACATTTAGAGTTAGTAAGGGTTTTATATCTCTTCTGTCCATATTGT TTTCAAAGGAATGAGGTGTTTAGGTGGCTGGAAAAGCATTTGTAGGAAGTTAGATTTGAATATAGACAAGGTGGGTTATTCACGTTGAGA ATGTTATTTGAAGAATGCCTGTGAAGCCAGGTGTGGGTTCTACTCAGTGCCATAGATAGACTGAGTCTTCTCTCGTAGGTTACCATTACA TAGTAATTTTGATTCTGAATTTCACATTAAATTATTTGAGTTTATACAGACCTAAATTTTAAAATCTGTACATATATTATTTTGATGTAT TAAGATGAATATTGCTGATTTAAATTTTATTTATGCACATACTTAAAGGACAGAAATGTCTGGGAAAGTAATTGTTAAATAATGATATGT AACTTTTTAACTTTTTAAATAAATAACAAGATTTTTAATGTGTGTCTCCCTCAGGGTTGTTTAAAGTTTTTTTTCTCCCTCAAGTATAAA TAGTGGTAACTATATGTTTTGTATCTTCTAGCACCAACTGCTGTAAAGCAATGCTGCAAATAATGCTTGAATACAAGTGGCTAAGCCAAC AACAGAATAAATACTTTTATAGTAGTTTTATAATCCTGAAATTCGAAAGCTTTCCCAATTGCACTTGCATCTAAACAAAACTGTTGCAGT TTTTACTCTATTTATTTTGTTCCCCATGTTTATGAAAGTCCTGCACAGTTTCAAAGGCATGGTAAATAATATATCAATGTTTATGTAGTC TGTTACAGAAACAGCTATAGATAACATTATCCAGTGAAGAGCAAAATTCAAGCTTTAGAAAATATTCATGCATGCAATTTTGACATATCT AAAAATAGGTTTTTGTATATTTATGGTGGGAGGTGGTTGGGAACTTTTAACAAAATGGGGTGTTAATTTTTGTACAGTCTGTGGGCATTT ACACATTTTTAATGTATTAAAATTTGGTAATTATGTGTACATTAAATTAATAAAAGTTACTTCTAGTTATGATTTGTGAATTCCCTAAGA CCTTGGATTTTTTTAAGTAACTTTATATCAGAAATGATACTGCATCTTTATATTTTTAAAATTGTATTGCTGCTCAAGAATGGTACCCTC TTGTCAAAAAGGCATACATTCATAATTGTACATTCAGCATTGTAAATAATCTTATGAAACCTTTTTTGATTGAAGCTATTCAAAATAAAA >49633_49633_1_LRCH1-ATP13A3_LRCH1_chr13_47127838_ENST00000311191_ATP13A3_chr3_194126845_ENST00000256031_length(amino acids)=193AA_BP=0 MYFAFGVFFLQPRAQGKQHFPHRSTDSCLHRVREVVPGGCGTGKFLQFPGAQVQPPGVGRLLKRALQTPVQGQVEAPAATAATTRPAGGA GSSVVLMVVVVVVGMMKWVQSSYRKGRDERLGFASRRRHLLPGPPTPPRRSCGGVTAFPARGKRQLGLSAAVLHRKALEGGEEGVGRRVE -------------------------------------------------------------- >49633_49633_2_LRCH1-ATP13A3_LRCH1_chr13_47127838_ENST00000389797_ATP13A3_chr3_194126845_ENST00000256031_length(transcript)=3881nt_BP=444nt ACACCCTCCTCCCCTCCTTCCAGCGCCTTTCGGTGGAGCACTGCGGCACTCAGCCCGAGCTGCCGTTTTCCCCTCGCGGGGAACGCTGTG ACCCCCCCGCAGGAGCGGCGGGGCGGGGTGGGGGGGCCCGGGAGAAGATGGCGACGCCGGGAAGCGAACCCCAACCTTTCGTCCCGGCCC TTTCGGTAGCTACTCTGCACCCACTTCATCATCCCCACCACCACCACCACCACCATCAGCACCACGGAGGAACCGGCGCCCCCGGCGGGG CGGGTGGTGGCGGCGGTGGCAGCGGGGGCTTCAACCTGCCCTTGAACCGGGGTCTGGAGCGCGCGCTTGAGGAGGCGGCCAACTCCGGGG GGCTGAACCTGAGCGCCAGGAAATTGAAGGAATTTCCCCGTACCGCAGCCCCCGGGCACGACCTCTCGGACACGGTGCAGGCAGGAGTCA GTGGATCGGTGGGGAAAATGCTGCTTACCCTGGGCCCTGGGCTGTAGAAAGAAGACACCAAAGGCAAAGTACATGTATCTGGCGCAGGAG CTCTTGGTTGATCCAGAATGGCCACCAAAACCTCAGACAACCACAGAAGCTAAAGCTTTAGTTAAGGAGAATGGATCATGTCAAATCATC ACCATAACATAGCAGTGAATCAGTCTCAGTGGTATTGCTGATAGCAGTATTCAGGAATATGTGATTTTAGGAGTTTCTGATCCTGTGTGT CAGAATGGCACTAGTTCAGTTTATGTCCCTTCTGATATAGTAGCTTATTTGACAGCTTTGCTCTTCCTTAAAATAAAAACAGAAAAATAT ATCGTCCTAACAGTTAAATTAACAATCAATCCATAAAGTCCTATATCTTCATTCAGCAACCCAAATATTACATACATTTCCAGAATTTTC TTGATTGTTACTTTCAGTGATATTCTTTATATTGGGTACAGGAGAAGTTTGGTGTTTGGTAGGTTTTTCAACATTAGTTTTTGAGACTAG TTTACCTCTTCACATTTATGCTCACAACCCTCTTGTTAGAAAAGTCTGTGTTTATATACAGGCTGTAAGTTTGTGATTGATAAAAAGAAG ATGAGTGTTAATTAGCCTCCAGTGAAAATATACTGAAAGCCTGTTTTCATTTGATTCCAATGTTTCTTCCAAAGAATTCTGTATAAACAT ATGCCAATTCCCTATGATGGTCTAGAGTTAGGAATGAGTGTTTATGGTGTTGCTTATAGAACAACTCAGGTAATCTCCATTTCTGGTTTT ATATTTTCTGTACAAACTGCCTGGGTTTTATTTTTCTAATCAGCAAGGTGCTTCACTGCCTTCTTGAGACGCCTCTCAAAGCTCTTAAAT GGCTCCTGTGCTATGTGTGGTGTTGGCAGTCTAATTTGCTTCTGTTAAATGTTGTAGAACCTTTTTCACTAGGAAATAAGATTCATTTCT TTCGGCAGTAGATGTAGATTCATCTTTTAACGTTTCTTCAAATTTGTTTCTGTCAGGCTTTGTGTTATTTTAAATGGTTTTTTAAAATTT TCTTCTATGTTTTCAATTACCTAAAGACATAGGATAATAGTTTTTTTTAAGTTAGAATTTTACCTCATAAAATTTTTTGAGGTTTGATGT ATGTCTCTGTCTTATCAATAATGAGGCTTAAAAAATACTGGATTTGAATGGCTGCCGTTTTTTCAAAGCAATATGAATTTGATGAGTTTG TTTTATGCCATTAGGTGGCGCCAGAGGTCAGAACATGTCTATTTTGAATTGGATCGTTACAAATGAGCATATTTGATGCGGAAATTTCTG GGAGAAAAAAAATTGAGGAAATAAAGTTAAAAAATTGACATTCATTGAGCCAAAAGAGATGTGGAGAAACATTTTTCACCTTTCTGTTTG GCCTGATTAACATTTAAATTCTTGCCAAAATTAAGGTAACTTTTAAAAAACACCTTTTATAGGTGGATCCAGCAGTCTGGCAACGCCCAC AGTTACCACAACACAGAAAACTGATCGTGCTATAAAATGGACGCTAAACTATGAAAACAGTGTGACATTGTTCTCTGTTCTTCCAGAGCC AGTAACATGCTTGCTCGTGCTTTCTACTTCTAGCTGATCATTCTTTTCCCAACATATATTTACAAATTTACCAAATTTTACCTAGAATTT TAGGACCAAATGGTTCTCACTCTTTATGCTGCAAAGACCTGGATGATGTTTGGTAACTATAGAAAAATAGAAATTACACTCAGGATCACT GTTACTGCTATTGCCACTGATGATTCCTGCAAAAATATAATCGAAGTTTTCCATCAAATGTATAATATGCTATTAATACACATTAGATGA TAACAGTTGTTCCATGAATGATTCTATGAAGCTATGCATCTTAGACCTCTTGAGCTGTGAATTAGCACTATTTTCTATAGTTACTTATTC TCTGGATCATTTTATAATTTCCATATTAATTTCAAATATGCTCGTGTTATTCTTCAGTGATTTCCACAATTGTGCATTTTATTCTTTGGT TTAAGTACTGAAGCATATAATGAAAGTAATTGCTAAGTAGCAGCTTAAAAATTCAATTATCCGATTGTATTTAACATCTTTAAGAGCATG ATCATAAAGAGCTATTTTTGACACCCCCCCCCCACTTTTTTAACATTTAGAGTTAGTAAGGGTTTTATATCTCTTCTGTCCATATTGTTT TCAAAGGAATGAGGTGTTTAGGTGGCTGGAAAAGCATTTGTAGGAAGTTAGATTTGAATATAGACAAGGTGGGTTATTCACGTTGAGAAT GTTATTTGAAGAATGCCTGTGAAGCCAGGTGTGGGTTCTACTCAGTGCCATAGATAGACTGAGTCTTCTCTCGTAGGTTACCATTACATA GTAATTTTGATTCTGAATTTCACATTAAATTATTTGAGTTTATACAGACCTAAATTTTAAAATCTGTACATATATTATTTTGATGTATTA AGATGAATATTGCTGATTTAAATTTTATTTATGCACATACTTAAAGGACAGAAATGTCTGGGAAAGTAATTGTTAAATAATGATATGTAA CTTTTTAACTTTTTAAATAAATAACAAGATTTTTAATGTGTGTCTCCCTCAGGGTTGTTTAAAGTTTTTTTTCTCCCTCAAGTATAAATA GTGGTAACTATATGTTTTGTATCTTCTAGCACCAACTGCTGTAAAGCAATGCTGCAAATAATGCTTGAATACAAGTGGCTAAGCCAACAA CAGAATAAATACTTTTATAGTAGTTTTATAATCCTGAAATTCGAAAGCTTTCCCAATTGCACTTGCATCTAAACAAAACTGTTGCAGTTT TTACTCTATTTATTTTGTTCCCCATGTTTATGAAAGTCCTGCACAGTTTCAAAGGCATGGTAAATAATATATCAATGTTTATGTAGTCTG TTACAGAAACAGCTATAGATAACATTATCCAGTGAAGAGCAAAATTCAAGCTTTAGAAAATATTCATGCATGCAATTTTGACATATCTAA AAATAGGTTTTTGTATATTTATGGTGGGAGGTGGTTGGGAACTTTTAACAAAATGGGGTGTTAATTTTTGTACAGTCTGTGGGCATTTAC ACATTTTTAATGTATTAAAATTTGGTAATTATGTGTACATTAAATTAATAAAAGTTACTTCTAGTTATGATTTGTGAATTCCCTAAGACC TTGGATTTTTTTAAGTAACTTTATATCAGAAATGATACTGCATCTTTATATTTTTAAAATTGTATTGCTGCTCAAGAATGGTACCCTCTT GTCAAAAAGGCATACATTCATAATTGTACATTCAGCATTGTAAATAATCTTATGAAACCTTTTTTGATTGAAGCTATTCAAAATAAAAAT >49633_49633_2_LRCH1-ATP13A3_LRCH1_chr13_47127838_ENST00000389797_ATP13A3_chr3_194126845_ENST00000256031_length(amino acids)=174AA_BP=0 MYFAFGVFFLQPRAQGKQHFPHRSTDSCLHRVREVVPGGCGTGKFLQFPGAQVQPPGVGRLLKRALQTPVQGQVEAPAATAATTRPAGGA -------------------------------------------------------------- >49633_49633_3_LRCH1-ATP13A3_LRCH1_chr13_47127838_ENST00000389798_ATP13A3_chr3_194126845_ENST00000256031_length(transcript)=3941nt_BP=504nt CTTCAAGACCGAGCTGCCACGGCCGCCTCCCCGCCCGCCCCCCATTCTACGCGCCTGCCCACACCCTCCTCCCCTCCTTCCAGCGCCTTT CGGTGGAGCACTGCGGCACTCAGCCCGAGCTGCCGTTTTCCCCTCGCGGGGAACGCTGTGACCCCCCCGCAGGAGCGGCGGGGCGGGGTG GGGGGGCCCGGGAGAAGATGGCGACGCCGGGAAGCGAACCCCAACCTTTCGTCCCGGCCCTTTCGGTAGCTACTCTGCACCCACTTCATC ATCCCCACCACCACCACCACCACCATCAGCACCACGGAGGAACCGGCGCCCCCGGCGGGGCGGGTGGTGGCGGCGGTGGCAGCGGGGGCT TCAACCTGCCCTTGAACCGGGGTCTGGAGCGCGCGCTTGAGGAGGCGGCCAACTCCGGGGGGCTGAACCTGAGCGCCAGGAAATTGAAGG AATTTCCCCGTACCGCAGCCCCCGGGCACGACCTCTCGGACACGGTGCAGGCAGGAGTCAGTGGATCGGTGGGGAAAATGCTGCTTACCC TGGGCCCTGGGCTGTAGAAAGAAGACACCAAAGGCAAAGTACATGTATCTGGCGCAGGAGCTCTTGGTTGATCCAGAATGGCCACCAAAA CCTCAGACAACCACAGAAGCTAAAGCTTTAGTTAAGGAGAATGGATCATGTCAAATCATCACCATAACATAGCAGTGAATCAGTCTCAGT GGTATTGCTGATAGCAGTATTCAGGAATATGTGATTTTAGGAGTTTCTGATCCTGTGTGTCAGAATGGCACTAGTTCAGTTTATGTCCCT TCTGATATAGTAGCTTATTTGACAGCTTTGCTCTTCCTTAAAATAAAAACAGAAAAATATATCGTCCTAACAGTTAAATTAACAATCAAT CCATAAAGTCCTATATCTTCATTCAGCAACCCAAATATTACATACATTTCCAGAATTTTCTTGATTGTTACTTTCAGTGATATTCTTTAT ATTGGGTACAGGAGAAGTTTGGTGTTTGGTAGGTTTTTCAACATTAGTTTTTGAGACTAGTTTACCTCTTCACATTTATGCTCACAACCC TCTTGTTAGAAAAGTCTGTGTTTATATACAGGCTGTAAGTTTGTGATTGATAAAAAGAAGATGAGTGTTAATTAGCCTCCAGTGAAAATA TACTGAAAGCCTGTTTTCATTTGATTCCAATGTTTCTTCCAAAGAATTCTGTATAAACATATGCCAATTCCCTATGATGGTCTAGAGTTA GGAATGAGTGTTTATGGTGTTGCTTATAGAACAACTCAGGTAATCTCCATTTCTGGTTTTATATTTTCTGTACAAACTGCCTGGGTTTTA TTTTTCTAATCAGCAAGGTGCTTCACTGCCTTCTTGAGACGCCTCTCAAAGCTCTTAAATGGCTCCTGTGCTATGTGTGGTGTTGGCAGT CTAATTTGCTTCTGTTAAATGTTGTAGAACCTTTTTCACTAGGAAATAAGATTCATTTCTTTCGGCAGTAGATGTAGATTCATCTTTTAA CGTTTCTTCAAATTTGTTTCTGTCAGGCTTTGTGTTATTTTAAATGGTTTTTTAAAATTTTCTTCTATGTTTTCAATTACCTAAAGACAT AGGATAATAGTTTTTTTTAAGTTAGAATTTTACCTCATAAAATTTTTTGAGGTTTGATGTATGTCTCTGTCTTATCAATAATGAGGCTTA AAAAATACTGGATTTGAATGGCTGCCGTTTTTTCAAAGCAATATGAATTTGATGAGTTTGTTTTATGCCATTAGGTGGCGCCAGAGGTCA GAACATGTCTATTTTGAATTGGATCGTTACAAATGAGCATATTTGATGCGGAAATTTCTGGGAGAAAAAAAATTGAGGAAATAAAGTTAA AAAATTGACATTCATTGAGCCAAAAGAGATGTGGAGAAACATTTTTCACCTTTCTGTTTGGCCTGATTAACATTTAAATTCTTGCCAAAA TTAAGGTAACTTTTAAAAAACACCTTTTATAGGTGGATCCAGCAGTCTGGCAACGCCCACAGTTACCACAACACAGAAAACTGATCGTGC TATAAAATGGACGCTAAACTATGAAAACAGTGTGACATTGTTCTCTGTTCTTCCAGAGCCAGTAACATGCTTGCTCGTGCTTTCTACTTC TAGCTGATCATTCTTTTCCCAACATATATTTACAAATTTACCAAATTTTACCTAGAATTTTAGGACCAAATGGTTCTCACTCTTTATGCT GCAAAGACCTGGATGATGTTTGGTAACTATAGAAAAATAGAAATTACACTCAGGATCACTGTTACTGCTATTGCCACTGATGATTCCTGC AAAAATATAATCGAAGTTTTCCATCAAATGTATAATATGCTATTAATACACATTAGATGATAACAGTTGTTCCATGAATGATTCTATGAA GCTATGCATCTTAGACCTCTTGAGCTGTGAATTAGCACTATTTTCTATAGTTACTTATTCTCTGGATCATTTTATAATTTCCATATTAAT TTCAAATATGCTCGTGTTATTCTTCAGTGATTTCCACAATTGTGCATTTTATTCTTTGGTTTAAGTACTGAAGCATATAATGAAAGTAAT TGCTAAGTAGCAGCTTAAAAATTCAATTATCCGATTGTATTTAACATCTTTAAGAGCATGATCATAAAGAGCTATTTTTGACACCCCCCC CCCACTTTTTTAACATTTAGAGTTAGTAAGGGTTTTATATCTCTTCTGTCCATATTGTTTTCAAAGGAATGAGGTGTTTAGGTGGCTGGA AAAGCATTTGTAGGAAGTTAGATTTGAATATAGACAAGGTGGGTTATTCACGTTGAGAATGTTATTTGAAGAATGCCTGTGAAGCCAGGT GTGGGTTCTACTCAGTGCCATAGATAGACTGAGTCTTCTCTCGTAGGTTACCATTACATAGTAATTTTGATTCTGAATTTCACATTAAAT TATTTGAGTTTATACAGACCTAAATTTTAAAATCTGTACATATATTATTTTGATGTATTAAGATGAATATTGCTGATTTAAATTTTATTT ATGCACATACTTAAAGGACAGAAATGTCTGGGAAAGTAATTGTTAAATAATGATATGTAACTTTTTAACTTTTTAAATAAATAACAAGAT TTTTAATGTGTGTCTCCCTCAGGGTTGTTTAAAGTTTTTTTTCTCCCTCAAGTATAAATAGTGGTAACTATATGTTTTGTATCTTCTAGC ACCAACTGCTGTAAAGCAATGCTGCAAATAATGCTTGAATACAAGTGGCTAAGCCAACAACAGAATAAATACTTTTATAGTAGTTTTATA ATCCTGAAATTCGAAAGCTTTCCCAATTGCACTTGCATCTAAACAAAACTGTTGCAGTTTTTACTCTATTTATTTTGTTCCCCATGTTTA TGAAAGTCCTGCACAGTTTCAAAGGCATGGTAAATAATATATCAATGTTTATGTAGTCTGTTACAGAAACAGCTATAGATAACATTATCC AGTGAAGAGCAAAATTCAAGCTTTAGAAAATATTCATGCATGCAATTTTGACATATCTAAAAATAGGTTTTTGTATATTTATGGTGGGAG GTGGTTGGGAACTTTTAACAAAATGGGGTGTTAATTTTTGTACAGTCTGTGGGCATTTACACATTTTTAATGTATTAAAATTTGGTAATT ATGTGTACATTAAATTAATAAAAGTTACTTCTAGTTATGATTTGTGAATTCCCTAAGACCTTGGATTTTTTTAAGTAACTTTATATCAGA AATGATACTGCATCTTTATATTTTTAAAATTGTATTGCTGCTCAAGAATGGTACCCTCTTGTCAAAAAGGCATACATTCATAATTGTACA >49633_49633_3_LRCH1-ATP13A3_LRCH1_chr13_47127838_ENST00000389798_ATP13A3_chr3_194126845_ENST00000256031_length(amino acids)=193AA_BP=0 MYFAFGVFFLQPRAQGKQHFPHRSTDSCLHRVREVVPGGCGTGKFLQFPGAQVQPPGVGRLLKRALQTPVQGQVEAPAATAATTRPAGGA GSSVVLMVVVVVVGMMKWVQSSYRKGRDERLGFASRRRHLLPGPPTPPRRSCGGVTAFPARGKRQLGLSAAVLHRKALEGGEEGVGRRVE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for LRCH1-ATP13A3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for LRCH1-ATP13A3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for LRCH1-ATP13A3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
