
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:LRCH3-FMN2 (FusionGDB2 ID:49647) |
Fusion Gene Summary for LRCH3-FMN2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: LRCH3-FMN2 | Fusion gene ID: 49647 | Hgene | Tgene | Gene symbol | LRCH3 | FMN2 | Gene ID | 84859 | 56776 |
| Gene name | leucine rich repeats and calponin homology domain containing 3 | formin 2 | |
| Synonyms | - | - | |
| Cytomap | 3q29 | 1q43 | |
| Type of gene | protein-coding | protein-coding | |
| Description | DISP complex protein LRCH3leucine-rich repeat and calponin homology domain-containing protein 3leucine-rich repeats and calponin homology (CH) domain containing 3 | formin-2 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | Q9NZ56 | |
| Ensembl transtripts involved in fusion gene | ENST00000334859, ENST00000414675, ENST00000425562, ENST00000438796, ENST00000441090, ENST00000536618, ENST00000493726, | ENST00000496950, ENST00000319653, ENST00000545751, ENST00000543681, | |
| Fusion gene scores | * DoF score | 9 X 9 X 5=405 | 8 X 8 X 7=448 |
| # samples | 12 | 10 | |
| ** MAII score | log2(12/405*10)=-1.75488750216347 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(10/448*10)=-2.16349873228288 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: LRCH3 [Title/Abstract] AND FMN2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | LRCH3(197592342)-FMN2(240635672), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | FMN2 | GO:0006974 | cellular response to DNA damage stimulus | 26287480 |
| Tgene | FMN2 | GO:0070649 | formin-nucleated actin cable assembly | 26287480 |
| Tgene | FMN2 | GO:2000781 | positive regulation of double-strand break repair | 26287480 |
 Fusion gene breakpoints across LRCH3 (5'-gene) Fusion gene breakpoints across LRCH3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
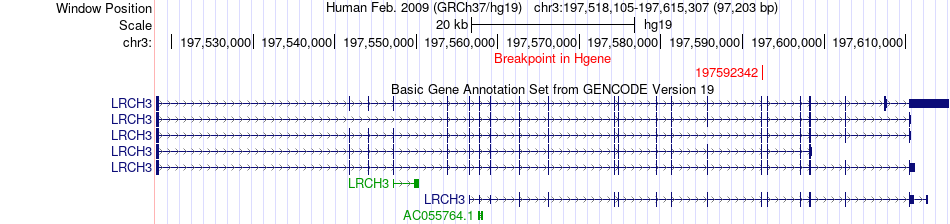 |
 Fusion gene breakpoints across FMN2 (3'-gene) Fusion gene breakpoints across FMN2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
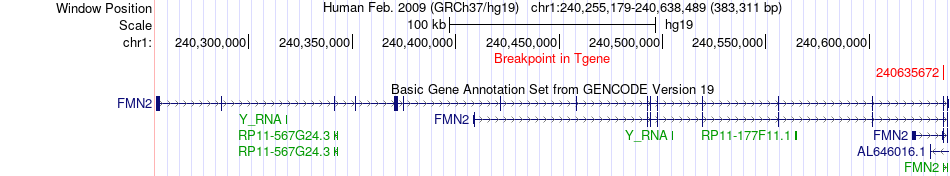 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | UCS | TCGA-N9-A4Q1-01A | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
Top |
Fusion Gene ORF analysis for LRCH3-FMN2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000334859 | ENST00000496950 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| 5CDS-3UTR | ENST00000414675 | ENST00000496950 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| 5CDS-3UTR | ENST00000425562 | ENST00000496950 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| 5CDS-3UTR | ENST00000438796 | ENST00000496950 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| 5CDS-3UTR | ENST00000441090 | ENST00000496950 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| 5CDS-3UTR | ENST00000536618 | ENST00000496950 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| Frame-shift | ENST00000334859 | ENST00000319653 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| Frame-shift | ENST00000334859 | ENST00000545751 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| Frame-shift | ENST00000414675 | ENST00000319653 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| Frame-shift | ENST00000414675 | ENST00000545751 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| Frame-shift | ENST00000425562 | ENST00000319653 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| Frame-shift | ENST00000425562 | ENST00000545751 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| Frame-shift | ENST00000438796 | ENST00000319653 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| Frame-shift | ENST00000438796 | ENST00000545751 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| Frame-shift | ENST00000441090 | ENST00000319653 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| Frame-shift | ENST00000441090 | ENST00000545751 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| Frame-shift | ENST00000536618 | ENST00000319653 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| Frame-shift | ENST00000536618 | ENST00000545751 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| In-frame | ENST00000334859 | ENST00000543681 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| In-frame | ENST00000414675 | ENST00000543681 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| In-frame | ENST00000425562 | ENST00000543681 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| In-frame | ENST00000438796 | ENST00000543681 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| In-frame | ENST00000441090 | ENST00000543681 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| In-frame | ENST00000536618 | ENST00000543681 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| intron-3CDS | ENST00000493726 | ENST00000319653 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| intron-3CDS | ENST00000493726 | ENST00000543681 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| intron-3CDS | ENST00000493726 | ENST00000545751 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
| intron-3UTR | ENST00000493726 | ENST00000496950 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000438796 | LRCH3 | chr3 | 197592342 | - | ENST00000543681 | FMN2 | chr1 | 240635672 | + | 2030 | 1809 | 14 | 1834 | 606 |
| ENST00000414675 | LRCH3 | chr3 | 197592342 | - | ENST00000543681 | FMN2 | chr1 | 240635672 | + | 1845 | 1624 | 15 | 1649 | 544 |
| ENST00000441090 | LRCH3 | chr3 | 197592342 | - | ENST00000543681 | FMN2 | chr1 | 240635672 | + | 1539 | 1318 | 15 | 1343 | 442 |
| ENST00000334859 | LRCH3 | chr3 | 197592342 | - | ENST00000543681 | FMN2 | chr1 | 240635672 | + | 1991 | 1770 | 5 | 1795 | 596 |
| ENST00000425562 | LRCH3 | chr3 | 197592342 | - | ENST00000543681 | FMN2 | chr1 | 240635672 | + | 1986 | 1765 | 0 | 1790 | 596 |
| ENST00000536618 | LRCH3 | chr3 | 197592342 | - | ENST00000543681 | FMN2 | chr1 | 240635672 | + | 1145 | 924 | 5 | 949 | 314 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000438796 | ENST00000543681 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + | 0.00127222 | 0.99872774 |
| ENST00000414675 | ENST00000543681 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + | 0.001392255 | 0.9986078 |
| ENST00000441090 | ENST00000543681 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + | 0.006043869 | 0.99395615 |
| ENST00000334859 | ENST00000543681 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + | 0.001236426 | 0.99876356 |
| ENST00000425562 | ENST00000543681 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + | 0.00122798 | 0.99877197 |
| ENST00000536618 | ENST00000543681 | LRCH3 | chr3 | 197592342 | - | FMN2 | chr1 | 240635672 | + | 0.020451898 | 0.9795481 |
Top |
Fusion Genomic Features for LRCH3-FMN2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
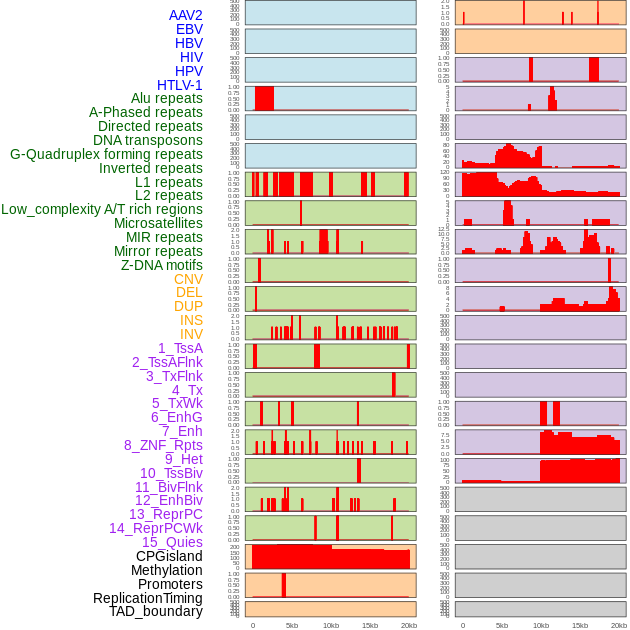 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for LRCH3-FMN2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:197592342/chr1:240635672) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | FMN2 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Actin-binding protein that is involved in actin cytoskeleton assembly and reorganization (PubMed:22330775, PubMed:21730168). Acts as an actin nucleation factor and promotes assembly of actin filaments together with SPIRE1 and SPIRE2 (PubMed:22330775, PubMed:21730168). Involved in intracellular vesicle transport along actin fibers, providing a novel link between actin cytoskeleton dynamics and intracellular transport (By similarity). Required for asymmetric spindle positioning, asymmetric oocyte division and polar body extrusion during female germ cell meiosis (By similarity). Plays a role in responses to DNA damage, cellular stress and hypoxia by protecting CDKN1A against degradation, and thereby plays a role in stress-induced cell cycle arrest (PubMed:23375502). Also acts in the nucleus: together with SPIRE1 and SPIRE2, promotes assembly of nuclear actin filaments in response to DNA damage in order to facilitate movement of chromatin and repair factors after DNA damage (PubMed:26287480). Protects cells against apoptosis by protecting CDKN1A against degradation (PubMed:23375502). {ECO:0000250|UniProtKB:Q9JL04, ECO:0000269|PubMed:21730168, ECO:0000269|PubMed:22330775, ECO:0000269|PubMed:23375502, ECO:0000269|PubMed:26287480}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000334859 | - | 16 | 19 | 384_389 | 588 | 713.0 | Compositional bias | Note=Poly-Glu |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000425562 | - | 16 | 21 | 384_389 | 588 | 778.0 | Compositional bias | Note=Poly-Glu |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000438796 | - | 16 | 22 | 384_389 | 588 | 2334.0 | Compositional bias | Note=Poly-Glu |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000334859 | - | 16 | 19 | 105_127 | 588 | 713.0 | Repeat | LRR 3 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000334859 | - | 16 | 19 | 128_150 | 588 | 713.0 | Repeat | LRR 4 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000334859 | - | 16 | 19 | 152_172 | 588 | 713.0 | Repeat | LRR 5 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000334859 | - | 16 | 19 | 173_195 | 588 | 713.0 | Repeat | LRR 6 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000334859 | - | 16 | 19 | 196_218 | 588 | 713.0 | Repeat | LRR 7 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000334859 | - | 16 | 19 | 220_239 | 588 | 713.0 | Repeat | LRR 8 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000334859 | - | 16 | 19 | 240_264 | 588 | 713.0 | Repeat | LRR 9 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000334859 | - | 16 | 19 | 266_290 | 588 | 713.0 | Repeat | LRR 10 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000334859 | - | 16 | 19 | 56_79 | 588 | 713.0 | Repeat | LRR 1 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000334859 | - | 16 | 19 | 81_104 | 588 | 713.0 | Repeat | LRR 2 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000425562 | - | 16 | 21 | 105_127 | 588 | 778.0 | Repeat | LRR 3 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000425562 | - | 16 | 21 | 128_150 | 588 | 778.0 | Repeat | LRR 4 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000425562 | - | 16 | 21 | 152_172 | 588 | 778.0 | Repeat | LRR 5 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000425562 | - | 16 | 21 | 173_195 | 588 | 778.0 | Repeat | LRR 6 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000425562 | - | 16 | 21 | 196_218 | 588 | 778.0 | Repeat | LRR 7 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000425562 | - | 16 | 21 | 220_239 | 588 | 778.0 | Repeat | LRR 8 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000425562 | - | 16 | 21 | 240_264 | 588 | 778.0 | Repeat | LRR 9 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000425562 | - | 16 | 21 | 266_290 | 588 | 778.0 | Repeat | LRR 10 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000425562 | - | 16 | 21 | 56_79 | 588 | 778.0 | Repeat | LRR 1 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000425562 | - | 16 | 21 | 81_104 | 588 | 778.0 | Repeat | LRR 2 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000438796 | - | 16 | 22 | 105_127 | 588 | 2334.0 | Repeat | LRR 3 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000438796 | - | 16 | 22 | 128_150 | 588 | 2334.0 | Repeat | LRR 4 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000438796 | - | 16 | 22 | 152_172 | 588 | 2334.0 | Repeat | LRR 5 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000438796 | - | 16 | 22 | 173_195 | 588 | 2334.0 | Repeat | LRR 6 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000438796 | - | 16 | 22 | 196_218 | 588 | 2334.0 | Repeat | LRR 7 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000438796 | - | 16 | 22 | 220_239 | 588 | 2334.0 | Repeat | LRR 8 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000438796 | - | 16 | 22 | 240_264 | 588 | 2334.0 | Repeat | LRR 9 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000438796 | - | 16 | 22 | 266_290 | 588 | 2334.0 | Repeat | LRR 10 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000438796 | - | 16 | 22 | 56_79 | 588 | 2334.0 | Repeat | LRR 1 |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000438796 | - | 16 | 22 | 81_104 | 588 | 2334.0 | Repeat | LRR 2 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000334859 | - | 16 | 19 | 652_765 | 588 | 713.0 | Domain | Calponin-homology (CH) |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000425562 | - | 16 | 21 | 652_765 | 588 | 778.0 | Domain | Calponin-homology (CH) |
| Hgene | LRCH3 | chr3:197592342 | chr1:240635672 | ENST00000438796 | - | 16 | 22 | 652_765 | 588 | 2334.0 | Domain | Calponin-homology (CH) |
| Tgene | FMN2 | chr3:197592342 | chr1:240635672 | ENST00000319653 | 15 | 18 | 1567_1597 | 1686 | 1723.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | FMN2 | chr3:197592342 | chr1:240635672 | ENST00000319653 | 15 | 18 | 1677_1699 | 1686 | 1723.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | FMN2 | chr3:197592342 | chr1:240635672 | ENST00000319653 | 15 | 18 | 193_231 | 1686 | 1723.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | FMN2 | chr3:197592342 | chr1:240635672 | ENST00000319653 | 15 | 18 | 670_706 | 1686 | 1723.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | FMN2 | chr3:197592342 | chr1:240635672 | ENST00000319653 | 15 | 18 | 200_227 | 1686 | 1723.0 | Compositional bias | Note=Gln-rich | |
| Tgene | FMN2 | chr3:197592342 | chr1:240635672 | ENST00000319653 | 15 | 18 | 758_1268 | 1686 | 1723.0 | Compositional bias | Note=Pro-rich | |
| Tgene | FMN2 | chr3:197592342 | chr1:240635672 | ENST00000319653 | 15 | 18 | 1283_1698 | 1686 | 1723.0 | Domain | FH2 |
Top |
Fusion Gene Sequence for LRCH3-FMN2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >49647_49647_1_LRCH3-FMN2_LRCH3_chr3_197592342_ENST00000334859_FMN2_chr1_240635672_ENST00000543681_length(transcript)=1991nt_BP=1770nt GGGAAATGGCGGCCGCGGGCTTGGTCGCTGTGGCAGCGGCTGCCGAGTACTCTGGCACGGTAGCGTCGGGAGGTAACCTCCCTGGTGTTC ACTGCGGCCCAAGCTCCGGGGCAGGCCCTGGTTTTGGCCCGGGCTCGTGGAGCCGCTCTCTCGATCGAGCCCTGGAGGAGGCGGCGGTCA CTGGGGTGCTGAGCCTGAGCGGCCGGAAACTGAGGGAGTTTCCCCGGGGAGCGGCCAACCACGACCTGACGGACACCACCCGGGCGGACC TGTCGCGAAATCGCCTTTCAGAAATTCCTATAGAAGCATGTCACTTTGTTTCTCTGGAAAATCTCAACTTGTACCAAAATTGTATTCGTT ATATTCCAGAGGCAATTTTAAACCTACAAGCTCTAACATTCTTAAATATTAGTCGGAACCAACTGTCAACATTGCCGGTACACTTGTGTA ATTTGCCATTGAAAGTCTTAATTGCTAGTAATAACAAATTGGTGTCACTTCCAGAAGAAATTGGACACCTTAGACATTTGATGGAACTTG ATGTGAGCTGCAATGAAATTCAAACTATACCTTCCCAAATTGGTAACCTGGAGGCCTTGAGAGACCTTAATGTAAGAAGAAATCACCTAG TACATTTGCCTGAAGAGCTGGCGGAGTTGCCTTTGATACGGTTAGACTTCTCATGCAATAAAATTACCACAATCCCTGTTTGTTATCGGA ACCTCAGGCACCTACAGACGATCACCCTAGATAACAATCCACTACAATCACCTCCTGCACAGATATGTATAAAAGGCAAAGTCCACATAT TTAAATACCTGAACATACAAGCTTGTAAGATTGCTCCAGATCTGCCGGATTATGATAGGAGACCGTTGGGTTTTGGCTCCTGCCATGAAG AACTGTACTCAAGTCGCCCTTATGGAGCCCTTGATTCAGGCTTCAATAGTGTGGACAGTGGTGATAAGAGATGGTCAGGGAATGAACCTA CAGATGAATTTTCAGATCTGCCTCTTCGAGTAGCAGAGATTACTAAAGAACAAAGACTACGAAGAGAAAGCCAGTACCAAGAGAACCGCG GCAGTTTGGTAGTAACAAACGGCGGAGTGGAACATGATCTGGATCAGATTGACTACATAGACAGCTGCACCGCAGAGGAAGAGGAGGCCG AGGTGAGACAGCCCAAGGGACCAGACCCAGACAGCCTTAGTTCACAGTTTATGGCGTATATTGAACAGCGGCGAATCTCTCATGAGGGTT CACCAGTAAAGCCAGTAGCCATTAGGGAGTTTCAAAAAACAGAAGATATGAGAAGATATTTACATCAAAACAGGGTTCCAGCTGAGCCAT CTTCCCTCCTGTCACTATCAGCAAGTCACAATCAGCTGTCACACACAGACCTGGAACTTCATCAGAGAAGGGAGCAGTTAGTAGAGCGCA CTCGGAGAGAGGCTCAGCTTGCTGCCCTGCAGTATGAGGAGGAGAAAATAAGGACCAAGCAGATCCAGAGAGATGCTGTCCTGGACTTTG TCAAACAAAAAGCATCACAAAGTCCACAAAAACAGCACCCGCTCCTAGATGGCGTAGATGGTGAGTGCCCCTTCCCATCCAGAAGGTCTC AGCACACTGATGATAGTGCCTTGTGCATGTCGCTGTCAGGGTTGAATCAAGTGGGCTGTGCTGCTACCCTGCCTCATTCTTCTGCCTTCA CGCCTCTTAAGAGTGATGACAGACCTAATGCTCTATTAAGTTCACCTGCAACAGAAACAGGTAAAAGAAGCCGAAGAGGTGTGTAGACAG AAGAAAGGAAAATCACTTTATAAAATAAAACCAAGACATGACTCTGGAATTAAAGCAAAGATAAGCATGAAAACTTGAACAATGAAAAGC AGAATGAAAATGAGTCATTGCAACGACTTTCACAAAATTCAGCTGACCTGAGAGTGGGAGGGAAACTACCGTCATTCTGCTCATGTTTCT >49647_49647_1_LRCH3-FMN2_LRCH3_chr3_197592342_ENST00000334859_FMN2_chr1_240635672_ENST00000543681_length(amino acids)=596AA_BP= MAAAGLVAVAAAAEYSGTVASGGNLPGVHCGPSSGAGPGFGPGSWSRSLDRALEEAAVTGVLSLSGRKLREFPRGAANHDLTDTTRADLS RNRLSEIPIEACHFVSLENLNLYQNCIRYIPEAILNLQALTFLNISRNQLSTLPVHLCNLPLKVLIASNNKLVSLPEEIGHLRHLMELDV SCNEIQTIPSQIGNLEALRDLNVRRNHLVHLPEELAELPLIRLDFSCNKITTIPVCYRNLRHLQTITLDNNPLQSPPAQICIKGKVHIFK YLNIQACKIAPDLPDYDRRPLGFGSCHEELYSSRPYGALDSGFNSVDSGDKRWSGNEPTDEFSDLPLRVAEITKEQRLRRESQYQENRGS LVVTNGGVEHDLDQIDYIDSCTAEEEEAEVRQPKGPDPDSLSSQFMAYIEQRRISHEGSPVKPVAIREFQKTEDMRRYLHQNRVPAEPSS LLSLSASHNQLSHTDLELHQRREQLVERTRREAQLAALQYEEEKIRTKQIQRDAVLDFVKQKASQSPQKQHPLLDGVDGECPFPSRRSQH -------------------------------------------------------------- >49647_49647_2_LRCH3-FMN2_LRCH3_chr3_197592342_ENST00000414675_FMN2_chr1_240635672_ENST00000543681_length(transcript)=1845nt_BP=1624nt GTTGTCGGCTGGGAAATGGCGGCCGCGGGCTTGGTCGCTGTGGCAGCGGCTGCCGAGTACTCTGGCACGGTAGCGTCGGGAGGTAACCTC CCTGGTGTTCACTGCGGCCCAAGCTCCGGGGCAGGCCCTGGTTTTGGCCCGGGCTCGTGGAGCCGCTCTCTCGATCGAGCCCTGGAGGAG GCGGCGGTCACTGGGGTGCTGAGCCTGAGCGGCCGGAAACTGAGGGAGTTTCCCCGGGGAGCGGCCAACCACGACCTGACGGACACCACC CGGGCGGACCTGTCGCGAAATCGCCTTTCAGAAATTCCTATAGAAGCATGTCACTTTGTTTCTCTGGAAAATCTCAACTTGTACCAAAAT TGTATTCGTTATATTCCAGAGGCAATTTTAAACCTACAAGCTCTAACATTCTTAAATATTAGTCGGAACCAACTGTCAACATTGCCGGTA CACTTGTGTAATTTGCCATTGAAAGTCTTAATTGCTAGTAATAACAAATTGGTGTCACTTCCAGAAGAAATTGGACACCTTAGACATTTG ATGGAACTTGATGTGAGCTGCAATGAAATTCAAACTATACCTTCCCAAATTGGTAACCTGGAGGCCTTGAGAGACCTTAATGTAAGAAGA AATCACCTAGTACATTTGCCTGAAGAGCTGGCGGAGTTGCCTTTGATACGGTTAGACTTCTCATGCAATAAAATTACCACAATCCCTGTT TGTTATCGGAACCTCAGGCACCTACAGACGATCACCCTAGATAACAATCCACTACAATCACCTCCTGCACAGATATGTATAAAAGGCAAA GTCCACATATTTAAATACCTGAACATACAAGCTTGTAAGATTGCTCCAGATCTGCCGGATTATGATAGGAGACCGTTGGGTTTTGGCTCC TGCCATGAAGAACTGTACTCAAGTCGCCCTTATGGAGCCCTTGATTCAGGCTTCAATAGTGTGGACAGTGGTGATAAGAGATGGTCAGGG AATGAACCTACAGATGAATTTTCAGATCTGCCTCTTCGAGTAGCAGAGATTACTAAAGAACAAAGACTACGAAGAGAAAGCCAGTACCAA GAGAACCGCGGCAGTTTGGTAGTAACAAACGGCGGAGTGGAACATGATCTGGATCAGATTGACTACATAGACAGCTGCACCGCAGAGGAA GAGGAGGCCGAGGGTTCACCAGTAAAGCCAGTAGCCATTAGGGAGTTTCAAAAAACAGAAGATATGAGAAGATATTTACATCAAAACAGG GTTCCAGCTGAGCCATCTTCCCTCCTGTCACTATCAGCAAGTCACAATCAGCTGTCACACACAGACCTGGAACTTCATCAGAGAAGGGAG CAGTTAGTAGAGCGCACTCGGAGAGAGGCTCAGCTTGCTGCCCTGCAGTATGAGGAGGAGAAAATAAGGACCAAGCAGATCCAGAGAGAT GCTGTCCTGGACTTTGTCAAACAAAAAGCATCACAAAGTCCACAAAAACAGCACCCGCTCCTAGATGGCGTAGATGGTGAGTGCCCCTTC CCATCCAGAAGGTCTCAGCACACTGATGATAGTGCCTTGTGCATGAGTGATGACAGACCTAATGCTCTATTAAGTTCACCTGCAACAGAA ACAGGTAAAAGAAGCCGAAGAGGTGTGTAGACAGAAGAAAGGAAAATCACTTTATAAAATAAAACCAAGACATGACTCTGGAATTAAAGC AAAGATAAGCATGAAAACTTGAACAATGAAAAGCAGAATGAAAATGAGTCATTGCAACGACTTTCACAAAATTCAGCTGACCTGAGAGTG >49647_49647_2_LRCH3-FMN2_LRCH3_chr3_197592342_ENST00000414675_FMN2_chr1_240635672_ENST00000543681_length(amino acids)=544AA_BP= MAAAGLVAVAAAAEYSGTVASGGNLPGVHCGPSSGAGPGFGPGSWSRSLDRALEEAAVTGVLSLSGRKLREFPRGAANHDLTDTTRADLS RNRLSEIPIEACHFVSLENLNLYQNCIRYIPEAILNLQALTFLNISRNQLSTLPVHLCNLPLKVLIASNNKLVSLPEEIGHLRHLMELDV SCNEIQTIPSQIGNLEALRDLNVRRNHLVHLPEELAELPLIRLDFSCNKITTIPVCYRNLRHLQTITLDNNPLQSPPAQICIKGKVHIFK YLNIQACKIAPDLPDYDRRPLGFGSCHEELYSSRPYGALDSGFNSVDSGDKRWSGNEPTDEFSDLPLRVAEITKEQRLRRESQYQENRGS LVVTNGGVEHDLDQIDYIDSCTAEEEEAEGSPVKPVAIREFQKTEDMRRYLHQNRVPAEPSSLLSLSASHNQLSHTDLELHQRREQLVER TRREAQLAALQYEEEKIRTKQIQRDAVLDFVKQKASQSPQKQHPLLDGVDGECPFPSRRSQHTDDSALCMSDDRPNALLSSPATETGKRS -------------------------------------------------------------- >49647_49647_3_LRCH3-FMN2_LRCH3_chr3_197592342_ENST00000425562_FMN2_chr1_240635672_ENST00000543681_length(transcript)=1986nt_BP=1765nt ATGGCGGCCGCGGGCTTGGTCGCTGTGGCAGCGGCTGCCGAGTACTCTGGCACGGTAGCGTCGGGAGGTAACCTCCCTGGTGTTCACTGC GGCCCAAGCTCCGGGGCAGGCCCTGGTTTTGGCCCGGGCTCGTGGAGCCGCTCTCTCGATCGAGCCCTGGAGGAGGCGGCGGTCACTGGG GTGCTGAGCCTGAGCGGCCGGAAACTGAGGGAGTTTCCCCGGGGAGCGGCCAACCACGACCTGACGGACACCACCCGGGCGGACCTGTCG CGAAATCGCCTTTCAGAAATTCCTATAGAAGCATGTCACTTTGTTTCTCTGGAAAATCTCAACTTGTACCAAAATTGTATTCGTTATATT CCAGAGGCAATTTTAAACCTACAAGCTCTAACATTCTTAAATATTAGTCGGAACCAACTGTCAACATTGCCGGTACACTTGTGTAATTTG CCATTGAAAGTCTTAATTGCTAGTAATAACAAATTGGTGTCACTTCCAGAAGAAATTGGACACCTTAGACATTTGATGGAACTTGATGTG AGCTGCAATGAAATTCAAACTATACCTTCCCAAATTGGTAACCTGGAGGCCTTGAGAGACCTTAATGTAAGAAGAAATCACCTAGTACAT TTGCCTGAAGAGCTGGCGGAGTTGCCTTTGATACGGTTAGACTTCTCATGCAATAAAATTACCACAATCCCTGTTTGTTATCGGAACCTC AGGCACCTACAGACGATCACCCTAGATAACAATCCACTACAATCACCTCCTGCACAGATATGTATAAAAGGCAAAGTCCACATATTTAAA TACCTGAACATACAAGCTTGTAAGATTGCTCCAGATCTGCCGGATTATGATAGGAGACCGTTGGGTTTTGGCTCCTGCCATGAAGAACTG TACTCAAGTCGCCCTTATGGAGCCCTTGATTCAGGCTTCAATAGTGTGGACAGTGGTGATAAGAGATGGTCAGGGAATGAACCTACAGAT GAATTTTCAGATCTGCCTCTTCGAGTAGCAGAGATTACTAAAGAACAAAGACTACGAAGAGAAAGCCAGTACCAAGAGAACCGCGGCAGT TTGGTAGTAACAAACGGCGGAGTGGAACATGATCTGGATCAGATTGACTACATAGACAGCTGCACCGCAGAGGAAGAGGAGGCCGAGGTG AGACAGCCCAAGGGACCAGACCCAGACAGCCTTAGTTCACAGTTTATGGCGTATATTGAACAGCGGCGAATCTCTCATGAGGGTTCACCA GTAAAGCCAGTAGCCATTAGGGAGTTTCAAAAAACAGAAGATATGAGAAGATATTTACATCAAAACAGGGTTCCAGCTGAGCCATCTTCC CTCCTGTCACTATCAGCAAGTCACAATCAGCTGTCACACACAGACCTGGAACTTCATCAGAGAAGGGAGCAGTTAGTAGAGCGCACTCGG AGAGAGGCTCAGCTTGCTGCCCTGCAGTATGAGGAGGAGAAAATAAGGACCAAGCAGATCCAGAGAGATGCTGTCCTGGACTTTGTCAAA CAAAAAGCATCACAAAGTCCACAAAAACAGCACCCGCTCCTAGATGGCGTAGATGGTGAGTGCCCCTTCCCATCCAGAAGGTCTCAGCAC ACTGATGATAGTGCCTTGTGCATGTCGCTGTCAGGGTTGAATCAAGTGGGCTGTGCTGCTACCCTGCCTCATTCTTCTGCCTTCACGCCT CTTAAGAGTGATGACAGACCTAATGCTCTATTAAGTTCACCTGCAACAGAAACAGGTAAAAGAAGCCGAAGAGGTGTGTAGACAGAAGAA AGGAAAATCACTTTATAAAATAAAACCAAGACATGACTCTGGAATTAAAGCAAAGATAAGCATGAAAACTTGAACAATGAAAAGCAGAAT GAAAATGAGTCATTGCAACGACTTTCACAAAATTCAGCTGACCTGAGAGTGGGAGGGAAACTACCGTCATTCTGCTCATGTTTCTTCTTG >49647_49647_3_LRCH3-FMN2_LRCH3_chr3_197592342_ENST00000425562_FMN2_chr1_240635672_ENST00000543681_length(amino acids)=596AA_BP= MAAAGLVAVAAAAEYSGTVASGGNLPGVHCGPSSGAGPGFGPGSWSRSLDRALEEAAVTGVLSLSGRKLREFPRGAANHDLTDTTRADLS RNRLSEIPIEACHFVSLENLNLYQNCIRYIPEAILNLQALTFLNISRNQLSTLPVHLCNLPLKVLIASNNKLVSLPEEIGHLRHLMELDV SCNEIQTIPSQIGNLEALRDLNVRRNHLVHLPEELAELPLIRLDFSCNKITTIPVCYRNLRHLQTITLDNNPLQSPPAQICIKGKVHIFK YLNIQACKIAPDLPDYDRRPLGFGSCHEELYSSRPYGALDSGFNSVDSGDKRWSGNEPTDEFSDLPLRVAEITKEQRLRRESQYQENRGS LVVTNGGVEHDLDQIDYIDSCTAEEEEAEVRQPKGPDPDSLSSQFMAYIEQRRISHEGSPVKPVAIREFQKTEDMRRYLHQNRVPAEPSS LLSLSASHNQLSHTDLELHQRREQLVERTRREAQLAALQYEEEKIRTKQIQRDAVLDFVKQKASQSPQKQHPLLDGVDGECPFPSRRSQH -------------------------------------------------------------- >49647_49647_4_LRCH3-FMN2_LRCH3_chr3_197592342_ENST00000438796_FMN2_chr1_240635672_ENST00000543681_length(transcript)=2030nt_BP=1809nt GCGCATGCGCTGAGCTGGCGGGCCCGAGTGTTGTCGGCTGGGAAATGGCGGCCGCGGGCTTGGTCGCTGTGGCAGCGGCTGCCGAGTACT CTGGCACGGTAGCGTCGGGAGGTAACCTCCCTGGTGTTCACTGCGGCCCAAGCTCCGGGGCAGGCCCTGGTTTTGGCCCGGGCTCGTGGA GCCGCTCTCTCGATCGAGCCCTGGAGGAGGCGGCGGTCACTGGGGTGCTGAGCCTGAGCGGCCGGAAACTGAGGGAGTTTCCCCGGGGAG CGGCCAACCACGACCTGACGGACACCACCCGGGCGGACCTGTCGCGAAATCGCCTTTCAGAAATTCCTATAGAAGCATGTCACTTTGTTT CTCTGGAAAATCTCAACTTGTACCAAAATTGTATTCGTTATATTCCAGAGGCAATTTTAAACCTACAAGCTCTAACATTCTTAAATATTA GTCGGAACCAACTGTCAACATTGCCGGTACACTTGTGTAATTTGCCATTGAAAGTCTTAATTGCTAGTAATAACAAATTGGTGTCACTTC CAGAAGAAATTGGACACCTTAGACATTTGATGGAACTTGATGTGAGCTGCAATGAAATTCAAACTATACCTTCCCAAATTGGTAACCTGG AGGCCTTGAGAGACCTTAATGTAAGAAGAAATCACCTAGTACATTTGCCTGAAGAGCTGGCGGAGTTGCCTTTGATACGGTTAGACTTCT CATGCAATAAAATTACCACAATCCCTGTTTGTTATCGGAACCTCAGGCACCTACAGACGATCACCCTAGATAACAATCCACTACAATCAC CTCCTGCACAGATATGTATAAAAGGCAAAGTCCACATATTTAAATACCTGAACATACAAGCTTGTAAGATTGCTCCAGATCTGCCGGATT ATGATAGGAGACCGTTGGGTTTTGGCTCCTGCCATGAAGAACTGTACTCAAGTCGCCCTTATGGAGCCCTTGATTCAGGCTTCAATAGTG TGGACAGTGGTGATAAGAGATGGTCAGGGAATGAACCTACAGATGAATTTTCAGATCTGCCTCTTCGAGTAGCAGAGATTACTAAAGAAC AAAGACTACGAAGAGAAAGCCAGTACCAAGAGAACCGCGGCAGTTTGGTAGTAACAAACGGCGGAGTGGAACATGATCTGGATCAGATTG ACTACATAGACAGCTGCACCGCAGAGGAAGAGGAGGCCGAGGTGAGACAGCCCAAGGGACCAGACCCAGACAGCCTTAGTTCACAGTTTA TGGCGTATATTGAACAGCGGCGAATCTCTCATGAGGGTTCACCAGTAAAGCCAGTAGCCATTAGGGAGTTTCAAAAAACAGAAGATATGA GAAGATATTTACATCAAAACAGGGTTCCAGCTGAGCCATCTTCCCTCCTGTCACTATCAGCAAGTCACAATCAGCTGTCACACACAGACC TGGAACTTCATCAGAGAAGGGAGCAGTTAGTAGAGCGCACTCGGAGAGAGGCTCAGCTTGCTGCCCTGCAGTATGAGGAGGAGAAAATAA GGACCAAGCAGATCCAGAGAGATGCTGTCCTGGACTTTGTCAAACAAAAAGCATCACAAAGTCCACAAAAACAGCACCCGCTCCTAGATG GCGTAGATGGTGAGTGCCCCTTCCCATCCAGAAGGTCTCAGCACACTGATGATAGTGCCTTGTGCATGTCGCTGTCAGGGTTGAATCAAG TGGGCTGTGCTGCTACCCTGCCTCATTCTTCTGCCTTCACGCCTCTTAAGAGTGATGACAGACCTAATGCTCTATTAAGTTCACCTGCAA CAGAAACAGGTAAAAGAAGCCGAAGAGGTGTGTAGACAGAAGAAAGGAAAATCACTTTATAAAATAAAACCAAGACATGACTCTGGAATT AAAGCAAAGATAAGCATGAAAACTTGAACAATGAAAAGCAGAATGAAAATGAGTCATTGCAACGACTTTCACAAAATTCAGCTGACCTGA >49647_49647_4_LRCH3-FMN2_LRCH3_chr3_197592342_ENST00000438796_FMN2_chr1_240635672_ENST00000543681_length(amino acids)=606AA_BP= MAGPSVVGWEMAAAGLVAVAAAAEYSGTVASGGNLPGVHCGPSSGAGPGFGPGSWSRSLDRALEEAAVTGVLSLSGRKLREFPRGAANHD LTDTTRADLSRNRLSEIPIEACHFVSLENLNLYQNCIRYIPEAILNLQALTFLNISRNQLSTLPVHLCNLPLKVLIASNNKLVSLPEEIG HLRHLMELDVSCNEIQTIPSQIGNLEALRDLNVRRNHLVHLPEELAELPLIRLDFSCNKITTIPVCYRNLRHLQTITLDNNPLQSPPAQI CIKGKVHIFKYLNIQACKIAPDLPDYDRRPLGFGSCHEELYSSRPYGALDSGFNSVDSGDKRWSGNEPTDEFSDLPLRVAEITKEQRLRR ESQYQENRGSLVVTNGGVEHDLDQIDYIDSCTAEEEEAEVRQPKGPDPDSLSSQFMAYIEQRRISHEGSPVKPVAIREFQKTEDMRRYLH QNRVPAEPSSLLSLSASHNQLSHTDLELHQRREQLVERTRREAQLAALQYEEEKIRTKQIQRDAVLDFVKQKASQSPQKQHPLLDGVDGE -------------------------------------------------------------- >49647_49647_5_LRCH3-FMN2_LRCH3_chr3_197592342_ENST00000441090_FMN2_chr1_240635672_ENST00000543681_length(transcript)=1539nt_BP=1318nt GTTGTCGGCTGGGAAATGGCGGCCGCGGGCTTGGTCGCTGTGGCAGCGGCTGCCGAGTACTCTGGCACGGTAGCGTCGGGAGGTAACCTC CCTGGTGTTCACTGCGGCCCAAGCTCCGGGGCAGGCCCTGGTTTTGGCCCGGGCTCGTGGAGCCGCTCTCTCGATCGAGCCCTGGAGGAG GCGGCGGTCACTGGGGTGCTGAGCCTGAGCGGCCGGAAACTGAGGGAGTTTCCCCGGGGAGCGGCCAACCACGACCTGACGGACACCACC CGGGCGGAGCTGGCGGAGTTGCCTTTGATACGGTTAGACTTCTCATGCAATAAAATTACCACAATCCCTGTTTGTTATCGGAACCTCAGG CACCTACAGACGATCACCCTAGATAACAATCCACTACAATCACCTCCTGCACAGATATGTATAAAAGGCAAAGTCCACATATTTAAATAC CTGAACATACAAGCTTGTAAGATTGCTCCAGATCTGCCGGATTATGATAGGAGACCGTTGGGTTTTGGCTCCTGCCATGAAGAACTGTAC TCAAGTCGCCCTTATGGAGCCCTTGATTCAGGCTTCAATAGTGTGGACAGTGGTGATAAGAGATGGTCAGGGAATGAACCTACAGATGAA TTTTCAGATCTGCCTCTTCGAGTAGCAGAGATTACTAAAGAACAAAGACTACGAAGAGAAAGCCAGTACCAAGAGAACCGCGGCAGTTTG GTAGTAACAAACGGCGGAGTGGAACATGATCTGGATCAGATTGACTACATAGACAGCTGCACCGCAGAGGAAGAGGAGGCCGAGGGTTCA CCAGTAAAGCCAGTAGCCATTAGGGAGTTTCAAAAAACAGAAGATATGAGAAGATATTTACATCAAAACAGGGTTCCAGCTGAGCCATCT TCCCTCCTGTCACTATCAGCAAGTCACAATCAGCTGTCACACACAGACCTGGAACTTCATCAGAGAAGGGAGCAGTTAGTAGAGCGCACT CGGAGAGAGGCTCAGCTTGCTGCCCTGCAGTATGAGGAGGAGAAAATAAGGACCAAGCAGATCCAGAGAGATGCTGTCCTGGACTTTGTC AAACAAAAAGCATCACAAAGTCCACAAAAACAGCACCCGCTCCTAGATGGCGTAGATGGTGAGTGCCCCTTCCCATCCAGAAGGTCTCAG CACACTGATGATAGTGCCTTGTGCATGTCGCTGTCAGGGTTGAATCAAGTGGGCTGTGCTGCTACCCTGCCTCATTCTTCTGCCTTCACG CCTCTTAAGAGTGATGACAGACCTAATGCTCTATTAAGTTCACCTGCAACAGAAACAGGTAAAAGAAGCCGAAGAGGTGTGTAGACAGAA GAAAGGAAAATCACTTTATAAAATAAAACCAAGACATGACTCTGGAATTAAAGCAAAGATAAGCATGAAAACTTGAACAATGAAAAGCAG AATGAAAATGAGTCATTGCAACGACTTTCACAAAATTCAGCTGACCTGAGAGTGGGAGGGAAACTACCGTCATTCTGCTCATGTTTCTTC >49647_49647_5_LRCH3-FMN2_LRCH3_chr3_197592342_ENST00000441090_FMN2_chr1_240635672_ENST00000543681_length(amino acids)=442AA_BP= MAAAGLVAVAAAAEYSGTVASGGNLPGVHCGPSSGAGPGFGPGSWSRSLDRALEEAAVTGVLSLSGRKLREFPRGAANHDLTDTTRAELA ELPLIRLDFSCNKITTIPVCYRNLRHLQTITLDNNPLQSPPAQICIKGKVHIFKYLNIQACKIAPDLPDYDRRPLGFGSCHEELYSSRPY GALDSGFNSVDSGDKRWSGNEPTDEFSDLPLRVAEITKEQRLRRESQYQENRGSLVVTNGGVEHDLDQIDYIDSCTAEEEEAEGSPVKPV AIREFQKTEDMRRYLHQNRVPAEPSSLLSLSASHNQLSHTDLELHQRREQLVERTRREAQLAALQYEEEKIRTKQIQRDAVLDFVKQKAS -------------------------------------------------------------- >49647_49647_6_LRCH3-FMN2_LRCH3_chr3_197592342_ENST00000536618_FMN2_chr1_240635672_ENST00000543681_length(transcript)=1145nt_BP=924nt CAGATCTGCCGGATTATGATAGGAGACCGTTGGGTTTTGGCTCCTGCCATGAAGAACTGTACTCAAGTCGCCCTTATGGAGCCCTTGATT CAGGCTTCAATAGTGTGGACAGTGGTGATAAGAGATGGTCAGGGAATGAACCTACAGATGAATTTTCAGATCTGCCTCTTCGAGTAGCAG AGATTACTAAAGAACAAAGACTACGAAGAGAAAGCCAGTACCAAGAGAACCGCGGCAGTTTGGTAGTAACAAACGGCGGAGTGGAACATG ATCTGGATCAGATTGACTACATAGACAGCTGCACCGCAGAGGAAGAGGAGGCCGAGGTGAGACAGCCCAAGGGACCAGACCCAGACAGCC TTAGTTCACAGTTTATGGCGTATATTGAACAGCGGCGAATCTCTCATGAGGGTTCACCAGTAAAGCCAGTAGCCATTAGGGAGTTTCAAA AAACAGAAGATATGAGAAGATATTTACATCAAAACAGGGTTCCAGCTGAGCCATCTTCCCTCCTGTCACTATCAGCAAGTCACAATCAGC TGTCACACACAGACCTGGAACTTCATCAGAGAAGGGAGCAGTTAGTAGAGCGCACTCGGAGAGAGGCTCAGCTTGCTGCCCTGCAGTATG AGGAGGAGAAAATAAGGACCAAGCAGATCCAGAGAGATGCTGTCCTGGACTTTGTCAAACAAAAAGCATCACAAAGTCCACAAAAACAGC ACCCGCTCCTAGATGGCGTAGATGGTGAGTGCCCCTTCCCATCCAGAAGGTCTCAGCACACTGATGATAGTGCCTTGTGCATGTCGCTGT CAGGGTTGAATCAAGTGGGCTGTGCTGCTACCCTGCCTCATTCTTCTGCCTTCACGCCTCTTAAGAGTGATGACAGACCTAATGCTCTAT TAAGTTCACCTGCAACAGAAACAGGTAAAAGAAGCCGAAGAGGTGTGTAGACAGAAGAAAGGAAAATCACTTTATAAAATAAAACCAAGA CATGACTCTGGAATTAAAGCAAAGATAAGCATGAAAACTTGAACAATGAAAAGCAGAATGAAAATGAGTCATTGCAACGACTTTCACAAA >49647_49647_6_LRCH3-FMN2_LRCH3_chr3_197592342_ENST00000536618_FMN2_chr1_240635672_ENST00000543681_length(amino acids)=314AA_BP= MPDYDRRPLGFGSCHEELYSSRPYGALDSGFNSVDSGDKRWSGNEPTDEFSDLPLRVAEITKEQRLRRESQYQENRGSLVVTNGGVEHDL DQIDYIDSCTAEEEEAEVRQPKGPDPDSLSSQFMAYIEQRRISHEGSPVKPVAIREFQKTEDMRRYLHQNRVPAEPSSLLSLSASHNQLS HTDLELHQRREQLVERTRREAQLAALQYEEEKIRTKQIQRDAVLDFVKQKASQSPQKQHPLLDGVDGECPFPSRRSQHTDDSALCMSLSG -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for LRCH3-FMN2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for LRCH3-FMN2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for LRCH3-FMN2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
