
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:LRP1B-HECW2 (FusionGDB2 ID:49730) |
Fusion Gene Summary for LRP1B-HECW2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: LRP1B-HECW2 | Fusion gene ID: 49730 | Hgene | Tgene | Gene symbol | LRP1B | HECW2 | Gene ID | 53353 | 57520 |
| Gene name | LDL receptor related protein 1B | HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 | |
| Synonyms | LRP-1B|LRP-DIT|LRPDIT | NDHSAL|NEDL2 | |
| Cytomap | 2q22.1-q22.2 | 2q32.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | low-density lipoprotein receptor-related protein 1BLRP-deleted in tumorslow density lipoprotein receptor related protein-deleted in tumorlow density lipoprotein receptor-related protein 1Blow density lipoprotein-related protein 1B (deleted in tumors) | E3 ubiquitin-protein ligase HECW2HECT-type E3 ubiquitin transferase HECW2NEDD4-like E3 ubiquitin-protein ligase 2NEDD4-related E3 ubiquitin ligase NEDL2 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9NZR2 | Q9P2P5 | |
| Ensembl transtripts involved in fusion gene | ENST00000389484, ENST00000486364, | ENST00000409111, ENST00000260983, | |
| Fusion gene scores | * DoF score | 10 X 10 X 4=400 | 9 X 8 X 4=288 |
| # samples | 10 | 9 | |
| ** MAII score | log2(10/400*10)=-2 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/288*10)=-1.67807190511264 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: LRP1B [Title/Abstract] AND HECW2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | LRP1B(142567848)-HECW2(197208488), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across LRP1B (5'-gene) Fusion gene breakpoints across LRP1B (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
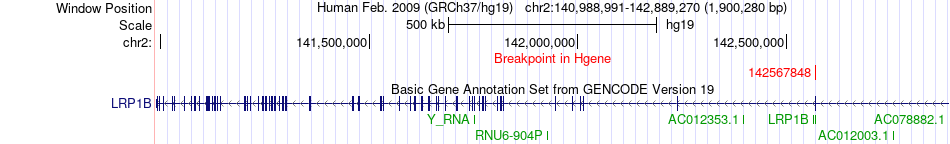 |
 Fusion gene breakpoints across HECW2 (3'-gene) Fusion gene breakpoints across HECW2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
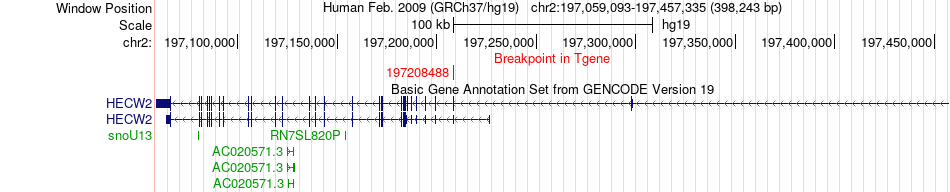 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUSC | TCGA-22-5482-01A | LRP1B | chr2 | 142567848 | - | HECW2 | chr2 | 197208488 | - |
| ChimerDB4 | LUSC | TCGA-22-5482 | LRP1B | chr2 | 142567848 | - | HECW2 | chr2 | 197208488 | - |
Top |
Fusion Gene ORF analysis for LRP1B-HECW2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000389484 | ENST00000409111 | LRP1B | chr2 | 142567848 | - | HECW2 | chr2 | 197208488 | - |
| 5UTR-3CDS | ENST00000486364 | ENST00000260983 | LRP1B | chr2 | 142567848 | - | HECW2 | chr2 | 197208488 | - |
| 5UTR-5UTR | ENST00000486364 | ENST00000409111 | LRP1B | chr2 | 142567848 | - | HECW2 | chr2 | 197208488 | - |
| In-frame | ENST00000389484 | ENST00000260983 | LRP1B | chr2 | 142567848 | - | HECW2 | chr2 | 197208488 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000389484 | LRP1B | chr2 | 142567848 | - | ENST00000260983 | HECW2 | chr2 | 197208488 | - | 12511 | 1177 | 861 | 5603 | 1580 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000389484 | ENST00000260983 | LRP1B | chr2 | 142567848 | - | HECW2 | chr2 | 197208488 | - | 0.000431296 | 0.99956864 |
Top |
Fusion Genomic Features for LRP1B-HECW2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
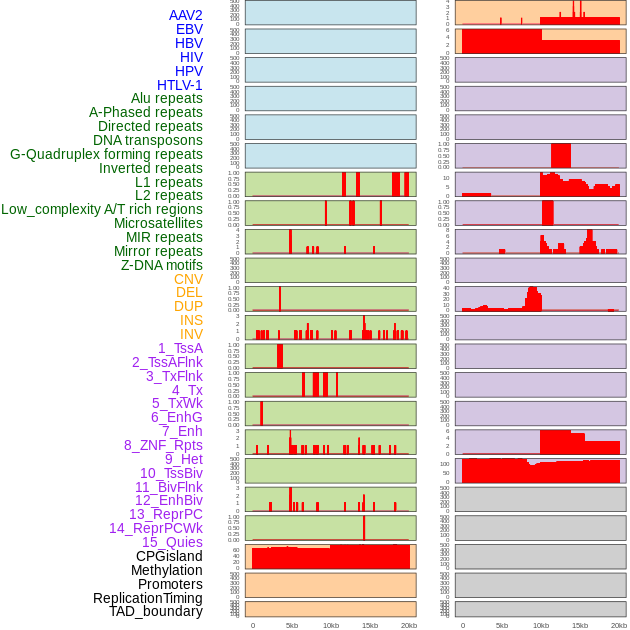 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for LRP1B-HECW2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:142567848/chr2:197208488) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| LRP1B | HECW2 |
| FUNCTION: Potential cell surface proteins that bind and internalize ligands in the process of receptor-mediated endocytosis. | FUNCTION: E3 ubiquitin-protein ligase that mediates ubiquitination of TP73. Acts to stabilize TP73 and enhance activation of transcription by TP73 (PubMed:12890487). Involved in the regulation of mitotic metaphase/anaphase transition (PubMed:24163370). {ECO:0000269|PubMed:12890487, ECO:0000269|PubMed:24163370}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 31_70 | 68 | 4600.0 | Domain | LDL-receptor class A 1 |
| Tgene | HECW2 | chr2:142567848 | chr2:197208488 | ENST00000260983 | 1 | 29 | 847_874 | 97 | 1573.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | HECW2 | chr2:142567848 | chr2:197208488 | ENST00000409111 | 0 | 27 | 847_874 | 0 | 1217.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | HECW2 | chr2:142567848 | chr2:197208488 | ENST00000260983 | 1 | 29 | 1237_1572 | 97 | 1573.0 | Domain | HECT | |
| Tgene | HECW2 | chr2:142567848 | chr2:197208488 | ENST00000260983 | 1 | 29 | 167_301 | 97 | 1573.0 | Domain | C2 | |
| Tgene | HECW2 | chr2:142567848 | chr2:197208488 | ENST00000260983 | 1 | 29 | 807_840 | 97 | 1573.0 | Domain | WW 1 | |
| Tgene | HECW2 | chr2:142567848 | chr2:197208488 | ENST00000260983 | 1 | 29 | 985_1018 | 97 | 1573.0 | Domain | WW 2 | |
| Tgene | HECW2 | chr2:142567848 | chr2:197208488 | ENST00000409111 | 0 | 27 | 1237_1572 | 0 | 1217.0 | Domain | HECT | |
| Tgene | HECW2 | chr2:142567848 | chr2:197208488 | ENST00000409111 | 0 | 27 | 167_301 | 0 | 1217.0 | Domain | C2 | |
| Tgene | HECW2 | chr2:142567848 | chr2:197208488 | ENST00000409111 | 0 | 27 | 807_840 | 0 | 1217.0 | Domain | WW 1 | |
| Tgene | HECW2 | chr2:142567848 | chr2:197208488 | ENST00000409111 | 0 | 27 | 985_1018 | 0 | 1217.0 | Domain | WW 2 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 1005_1043 | 68 | 4600.0 | Domain | LDL-receptor class A 7 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 1052_1089 | 68 | 4600.0 | Domain | LDL-receptor class A 8 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 1094_1132 | 68 | 4600.0 | Domain | LDL-receptor class A 9 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 1135_1174 | 68 | 4600.0 | Domain | LDL-receptor class A 10 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 116_154 | 68 | 4600.0 | Domain | EGF-like 1 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 1174_1213 | 68 | 4600.0 | Domain | EGF-like 5 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 1214_1253 | 68 | 4600.0 | Domain | EGF-like 6 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 1527_1570 | 68 | 4600.0 | Domain | EGF-like 7 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 155_194 | 68 | 4600.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 1834_1875 | 68 | 4600.0 | Domain | EGF-like 8 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 2143_2183 | 68 | 4600.0 | Domain | EGF-like 9 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 2464_2504 | 68 | 4600.0 | Domain | EGF-like 10 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 2509_2548 | 68 | 4600.0 | Domain | LDL-receptor class A 11 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 2551_2587 | 68 | 4600.0 | Domain | LDL-receptor class A 12 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 2590_2626 | 68 | 4600.0 | Domain | LDL-receptor class A 13 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 2629_2675 | 68 | 4600.0 | Domain | LDL-receptor class A 14 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 2681_2717 | 68 | 4600.0 | Domain | LDL-receptor class A 15 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 2719_2757 | 68 | 4600.0 | Domain | LDL-receptor class A 16 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 2760_2800 | 68 | 4600.0 | Domain | LDL-receptor class A 17 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 2804_2841 | 68 | 4600.0 | Domain | LDL-receptor class A 18 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 2844_2885 | 68 | 4600.0 | Domain | LDL-receptor class A 19 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 2890_2926 | 68 | 4600.0 | Domain | LDL-receptor class A 20 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 2927_2967 | 68 | 4600.0 | Domain | EGF-like 11 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 2968_3008 | 68 | 4600.0 | Domain | EGF-like 12%3B calcium-binding |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 3273_3314 | 68 | 4600.0 | Domain | EGF-like 13 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 3316_3353 | 68 | 4600.0 | Domain | LDL-receptor class A 21 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 3356_3392 | 68 | 4600.0 | Domain | LDL-receptor class A 22 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 3395_3432 | 68 | 4600.0 | Domain | LDL-receptor class A 23 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 3435_3472 | 68 | 4600.0 | Domain | LDL-receptor class A 24 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 3475_3511 | 68 | 4600.0 | Domain | LDL-receptor class A 25 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 3514_3550 | 68 | 4600.0 | Domain | LDL-receptor class A 26 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 3552_3588 | 68 | 4600.0 | Domain | LDL-receptor class A 27 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 3593_3629 | 68 | 4600.0 | Domain | LDL-receptor class A 28 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 3631_3668 | 68 | 4600.0 | Domain | LDL-receptor class A 29 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 3673_3711 | 68 | 4600.0 | Domain | LDL-receptor class A 30 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 3714_3752 | 68 | 4600.0 | Domain | LDL-receptor class A 31 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 3761_3797 | 68 | 4600.0 | Domain | LDL-receptor class A 32 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 3801_3843 | 68 | 4600.0 | Domain | EGF-like 14 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 3844_3881 | 68 | 4600.0 | Domain | EGF-like 15 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 4171_4208 | 68 | 4600.0 | Domain | EGF-like 16 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 4213_4249 | 68 | 4600.0 | Domain | EGF-like 17 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 4249_4285 | 68 | 4600.0 | Domain | EGF-like 18 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 4285_4321 | 68 | 4600.0 | Domain | EGF-like 19 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 4321_4357 | 68 | 4600.0 | Domain | EGF-like 20 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 4357_4392 | 68 | 4600.0 | Domain | EGF-like 21 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 4390_4427 | 68 | 4600.0 | Domain | EGF-like 22 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 471_517 | 68 | 4600.0 | Domain | EGF-like 3 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 76_114 | 68 | 4600.0 | Domain | LDL-receptor class A 2 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 794_834 | 68 | 4600.0 | Domain | EGF-like 4 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 844_882 | 68 | 4600.0 | Domain | LDL-receptor class A 3 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 885_923 | 68 | 4600.0 | Domain | LDL-receptor class A 4 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 926_963 | 68 | 4600.0 | Domain | LDL-receptor class A 5 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 966_1003 | 68 | 4600.0 | Domain | LDL-receptor class A 6 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 4492_4495 | 68 | 4600.0 | Motif | Endocytosis signal |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 4559_4562 | 68 | 4600.0 | Motif | Endocytosis signal |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 1300_1346 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 8 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 1347_1389 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 9 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 1390_1436 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 10 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 1437_1480 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 11 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 1481_1522 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 12 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 1618_1660 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 13 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 1661_1704 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 14 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 1705_1744 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 15 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 1745_1787 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 16 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 1922_1964 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 17 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 1965_2007 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 28 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 2008_2051 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 19 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 2052_2095 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 20 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 2239_2280 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 21 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 2281_2329 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 22 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 2330_2374 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 23 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 2375_2416 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 24 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 2417_2459 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 25 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 295_337 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 1 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 3055_3098 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 26 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 3099_3141 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 27 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 3142_3185 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 28 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 3186_3224 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 29 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 3225_3268 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 30 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 338_381 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 2 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 382_425 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 3 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 3933_3980 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 31 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 3981_4038 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 32 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 4039_4082 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 33 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 4083_4127 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 34 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 568_610 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 4 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 611_656 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 5 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 657_706 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 6 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 707_750 | 68 | 4600.0 | Repeat | Note=LDL-receptor class B 7 |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 25_4444 | 68 | 4600.0 | Topological domain | Extracellular |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 4468_4599 | 68 | 4600.0 | Topological domain | Cytoplasmic |
| Hgene | LRP1B | chr2:142567848 | chr2:197208488 | ENST00000389484 | - | 2 | 91 | 4445_4467 | 68 | 4600.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for LRP1B-HECW2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >49730_49730_1_LRP1B-HECW2_LRP1B_chr2_142567848_ENST00000389484_HECW2_chr2_197208488_ENST00000260983_length(transcript)=12511nt_BP=1177nt AAAGACAGAACCCCAGAGAAAAACGCTGCCAATTCGTTGCTTTATTGTTCCCTGCCTGGGGACCTCAATAGCCTTTTCCATTAACCTTCC CTTCTTACGCAACGGTTAATGACTTTGGGGGTTGTTTTGCTTTCTGTTTCTGCTGAGTCACTAAATTTTGCCTCTTTGTCCCCAGGTGCT GCTCAGCATAAAAGTTAAAAGTGCAATTCAGGAAGTACTGGGATTCTGTGTAGAGCCGAGGAAACCATTTCCCTAAGAGAAGCTCTGTTC CTTGGCTTGTCCTTCCTTCCCGGGAAGGAAGCTTCCGAGGAACGAAGGGAGAAGCTTTGTTTTGCCTGCAGAAGCAGCCCTGTGCTCGGC TGAGGGTTCTCAGCTGGCTGTGAACTGCGGAGCATTGTAGGCGCCTGGCTGGCTCAGGCCAATGCAGAAGTCTCTCCCTTCTCCAAAGAC CCAAATCCCCACAGAACCAGCTTCGAGTTACTTTCCCTTCAAGGGGATTAAAATAATTGTGATTTGTGGCGCTCTCCGTTCGCGGTGGTA TTTTCCTGTTGTGTTAAATGCCTCTTATTAAGTAATAGATGTGATTTATGTGAACGACGAAGGGGTGTGTGGTGGATTCGGTGATTAATC AGTGAATTCCCATCCGCTGGCATCTCTCACTGCCCCTCTTGCGTGATGTAAGATCAGACGTACCCTGCATTGAAAAGTCAAGACACACGG GCGTCTCGCTCGCGCTCACACACGCTCTGCCTCCTCTCTCCCGCACGCGCGCATCCCTCCACCTTCCACATCCTGCTCCAGGCAGGAGAA GGCTGACTGGCTGGACTCATTGAGCTGAAGAATTTCCAGTGACATTTGTAAATGACGCCGCTCGATTCCAGGCTCCAAGCGGCCCCTGCC GCCGCCGCCGCCGCCGGGCCGAAGGTGCCGCCGAGCAGTCTCCAGCGCAGGCTTCCTTACCGGGCGACCACAATGTCCGAGTTTCTCCTC GCCTTACTCACTCTCTCGGGATTATTGCCGATTGCCAGGGTGCTGACCGTGGGAGCCGACCGAGATCAGCAGTTGTGTGATCCTGGTGAA TTTCTTTGCCACGATCACGTGACTTGTGTCTCCCAGAGCTGGCTGTGTGATGGGGACCCTGACTGCCCTGATGATTCAGACGAGTCTTTA GATACCTATGAGAATTCTCCAGCCAACTTCTGGGATTCTAAGAACAGGGGTGTGACTGGAACACAAAAAGGGCAAATTGTATGGAGAATT GAGCCTGGGCCCTATTTCATGGAACCGGAGATAAAAATCTGTTTTAAATATTACCACGGCATTAGTGGAGCCCTGCGAGCCACGACCCCC TGCATCACCGTGAAGAACCCAGCTGTGATGATGGGGGCAGAAGGCATGGAGGGAGGTGCTTCAGGAAACCTGCATTCTCGAAAACTTGTT AGCTTTACATTGTCAGATCTTAGGGCAGTTGGGCTAAAGAAAGGGATGTTCTTCAATCCTGACCCTTATCTTAAGATGTCAATTCAGCCA GGAAAGAAGAGCAGTTTCCCCACCTGTGCCCACCACGGGCAGGAGAGACGGTCTACTATCATCAGTAACACCACCAATCCAATTTGGCAC CGAGAGAAATATTCCTTTTTTGCACTTCTTACTGATGTCTTAGAAATTGAAATTAAAGACAAATTTGCCAAGAGCCGTCCCATCATCAAG CGTTTTCTGGGGAAACTAACCATTCCAGTCCAGAGGCTGCTGGAGCGACAAGCCATCGGTGATCAAATGCTCAGCTACAACCTTGGCAGA AGGCTCCCAGCTGACCACGTGAGTGGGTACCTCCAGTTTAAAGTGGAGGTTACGTCTTCTGTTCATGAAGATGCCTCTCCAGAAGCTGTT GGCACAATACTTGGAGTCAATTCTGTGAATGGAGACTTAGGTAGCCCTTCCGATGACGAGGACATGCCAGGGAGCCATCACGACAGCCAG GTGTGCTCTAATGGGCCAGTTTCTGAGGACAGTGCTGCCGATGGAACCCCCAAGCATTCATTCAGGACTAGCTCCACTCTGGAAATAGAC ACAGAGGAATTAACCTCTACGTCTTCAAGGACCTCACCTCCCAGAGGACGTCAGGATTCACTCAATGATTACTTAGATGCTATCGAACAC AATGGCCACTCCAGGCCGGGGACAGCGACCTGCTCTGAGAGGTCCATGGGTGCCTCTCCGAAACTGAGGAGTAGTTTTCCCACTGACACA AGACTCAATGCCATGCTTCATATTGATTCAGATGAAGAAGATCACGAGTTCCAGCAAGACCTGGGTTACCCATCTTCCTTGGAGGAGGAG GGAGGCCTGATCATGTTTAGCAGGGCATCCAGAGCTGATGATGGAAGCCTGACATCTCAGACAAAGCTGGAGGACAACCCTGTTGAGAAT GAGGAAGCCTCCACACACGAAGCTGCTTCCTTTGAGGATAAGCCAGAAAATCTTCCAGAGCTTGCAGAGAGCTCCTTACCTGCAGGCCCA GCCCCAGAGGAAGGTGAAGGCGGCCCAGAGCCTCAACCCAGTGCTGACCAGGGCAGTGCTGAACTGTGTGGCTCTCAAGAGGTAGATCAG CCCACAAGTGGCGCAGACACAGGGACTTCCGATGCATCAGGAGGAAGCCGAAGAGCCGTCAGTGAGACCGAGTCTCTCGATCAGGGCTCT GAGCCTTCCCAGGTGTCCTCTGAAACAGAACCCAGTGATCCTGCCAGGACAGAGAGTGTGAGCGAAGCCAGCACCAGGCCTGAGGGAGAG AGTGACCTGGAATGCGCTGACAGCTCCTGCAATGAGAGTGTGACCACGCAGCTGTCCTCTGTGGACACACGGTGCTCCTCTCTTGAGAGC GCACGGTTTCCTGAAACCCCAGCCTTTTCTTCTCAGGAGGAGGAAGACGGAGCCTGTGCAGCAGAGCCCACCAGCAGTGGCCCTGCCGAA GGGTCGCAGGAATCCGTGTGCACTGCTGGTTCTTTACCTGTGGTACAGGTGCCCAGTGGGGAGGATGAAGGGCCAGGGGCAGAATCGGCC ACTGTACCTGACCAGGAGGAGCTGGGGGAGGTCTGGCAGCGGAGGGGGAGCCTGGAGGGAGCTGCAGCTGCTGCCGAGAGCCCACCGCAA GAAGAAGGCAGTGCTGGGGAGGCCCAAGGCACCTGTGAAGGGGCAACTGCCCAGGAGGAGGGCGCTACTGGAGGTTCTCAAGCCAACGGC CACCAGCCACTGCGATCACTACCTTCAGTGCGCCAGGATGTTAGCCGGTACCAGAGGGTGGACGAGGCTCTCCCACCAAACTGGGAGGCA CGCATTGACAGCCACGGCAGGATCTTCTACGTGGATCACGTAAACAGAACCACGACGTGGCAGCGACCGACAGCTCCCCCAGCCCCGCAG GTGCTGCAGAGATCTAACTCCATACAGCAAATGGAGCAGCTGAACCGGCGATATCAGAGTATCCGCAGAACCATGACCAATGAGAGGCCT GAGGAAAATACCAATGCTATTGATGGGGCAGGGGAGGAAGCTGACTTTCACCAAGCCAGTGCAGATTTCCGACGGGAGAACATCCTGCCT CACTCTACCTCCAGATCCAGGATCACGTTGCTGTTACAGTCTCCCCCCGTGAAGTTCCTCATCAGCCCAGAGTTCTTCACCGTGCTGCAT TCTAACCCTAGTGCCTACCGCATGTTTACAAACAACACGTGTTTGAAGCACATGATCACCAAAGTCCGGAGGGACACCCACCACTTTGAA CGCTACCAGCATAACCGCGACCTTGTGGGATTCCTCAACATGTTCGCGAACAAACAGCTAGAGCTGCCGCGGGGATGGGAAATGAAACAT GATCACCAGGGCAAGGCATTCTTTGTGGACCACAACTCCCGCACCACCACTTTCATTGATCCCCGGCTCCCACTTCAGAGCAGTAGACCC ACAAGTGCGCTGGTTCATCGGCAACACCTGACAAGGCAACGCAGCCACAGTGCGGGTGAGGTAGGAGAAGATTCTCGACATGCAGGACCA CCAGTTCTTCCCAGGCCATCCAGTACATTCAATACAGTCAGTAGGCCACAGTACCAGGACATGGTTCCAGTGGCTTACAATGACAAGATT GTTGCATTTCTACGTCAACCCAATATCTTTGAAATTCTACAGGAGCGTCAACCTGATCTTACCAGGAATCACTCACTCAGGGAGAAGATC CAATTTATCCGAACTGAAGGGACTCCAGGATTGGTGCGCCTTTCAAGCGATGCAGACCTTGTTATGTTGCTGAGCTTATTTGAAGAAGAG ATAATGTCGTATGTGCCTCCACATGCCTTACTCCACCCCAGCTACTGTCAGTCCCCACGTGGCTCTCCCGTGTCATCTCCTCAGAACTCG CCAGGTACCCAGCGTGCCAATGCCCGGGCTCCAGCCCCTTACAAGCGGGATTTCGAAGCCAAACTGAGGAACTTTTACAGGAAGTTAGAG ACTAAAGGATATGGACAAGGCCCAGGGAAGTTAAAGTTAATTATCCGAAGAGATCACTTACTAGAAGATGCTTTTAATCAGATTATGGGC TACTCCAGAAAAGACCTGCAGAGAAATAAGCTATATGTCACCTTCGTTGGGGAGGAAGGGCTGGATTACAGTGGGCCTTCTAGAGAGTTT TTCTTCCTGGTATCCAGAGAACTCTTTAACCCATATTATGGCTTATTTGAATATTCAGCCAATGACACATACACAGTACAAATAAGTCCT ATGTCTGCTTTTGTAGACAATCACCATGAATGGTTCCGATTCAGTGGTAGGATCCTTGGTCTTGCACTAATACACCAGTATTTGTTGGAT GCCTTCTTCACACGGCCCTTTTATAAGGCTCTTCTCAGAATTCTATGTGACCTGAGTGACCTAGAATACCTTGATGAAGAGTTCCATCAG AGCCTGCAGTGGATGAAAGACAATGATATCCATGACATCCTAGACCTCACGTTCACTGTGAACGAAGAAGTATTTGGGCAGATAACTGAA CGAGAATTAAAGCCAGGGGGTGCCAATATCCCAGTTACAGAGAAGAACAAGAAGGAGTACATCGAGAGGATGGTGAAGTGGAGGATTGAG AGGGGTGTTGTACAGCAAACAGAGAGCTTAGTGCGTGGCTTCTATGAGGTGGTGGATGCCAGGCTGGTATCTGTTTTTGATGCAAGAGAA CTGGAATTGGTCATCGCAGGCACAGCTGAAATAGACCTAAGTGATTGGAGAAACAACACAGAATATAGAGGAGGATACCATGACAATCAT ATTGTAATTCGGTGGTTCTGGGCTGCAGTGGAAAGATTCAACAATGAACAACGACTAAGGTTGTTACAGTTTGTTACAGGCACATCCAGC ATTCCCTATGAAGGATTTGCTTCACTCCGAGGGAGTAACGGCCCAAGAAGATTCTGTGTGGAGAAATGGGGGAAAATCACTGCTCTTCCC AGAGCGCATACATGTTTTAACCGTCTGGATCTGCCTCCCTACCCATCCTTTTCCATGCTTTATGAAAAACTGTTGACAGCAGTTGAAGAA ACCAGTACTTTTGGACTTGAGTGACCTGGAAGCTGAATGCCCATCTCTGTGGACAGGCAGGTTCAGAAGCTGCCTTCTAGAAGAATGATT GAACATTGGAAGTTTCAAGAGGATGCTTCCTTTAGGATAAAGCTATGTGCTGTTGTTTTCCAGGAACAAGTGCTCTGTCACATTTGGGGA CTGGAGATGAGTCCTCTTGGAAGGATTTGGGTGAGCTTGATGCCCAGGGAACAACCCAACCGTCTTTCAATCAACAGTTCTTGACTGCCA AACTTTTTCCATTTGTTATGTTCCAAGACAAAGATGAACCCATACATGATCAGCTCCACGGTAATTTTTAGGGACTCAGGAGAATCTTGA AACTTACCCTTGAACGTGGTTCAAGCCAAACTGGCAGCATTTGGCCCAATCTCCAAATTAGAGCAAGTTAAATAATATAATAAAAGTAAA TATATTTCCTGAAAGTACATTCATTTAAGCCCTAAGTTATAACAGAATATTCATTTCTTGCTTATGAGTGCCTGCATGGTGTGCACCATA GGTTTCCGCTTTCATGGGACATGAGTGAAAATGAAACCAAGTCAATATGAGGTACCTTTACAGATTTGCAATAAGATGGTCTGTGACAAT GTATATGCAAGTGGTATGTGTGTAATTATGGCTAAAGACAAACCATTATTCAGTGAATTACTAATGACAGATTTTATGCTTTATAATGCA TGAAAACAATTTTAAAATAACTAGCAATTAATCACAGCATATCAGGAAAAAGTACACAGTGAGTTCTGTTTATTTTTTGTAGGTTCATTA TGTTTATGTTCTTTAAGATGTATATAAGAACCTACCTATCATGCTGTATGTATCACTCATTCCATTTTCATGTTCCATGCATACTCGGGC ATCATGCTAATATGTATCCTTTTAAGCACTCTCAAGGAAACAAAAGGGCCTTTTATTTTTATAAAGGTAAAAAAAATTCCCCAAATATTT TGCACTGAATGTACCAAAGGTGAAGGGACATTACAATATGACTAACAGCAACTCCATCACTTGAGAAGTATAATAGAAAATAGCTTCTAA ATCAAACTTCCTTCACAGTGCCGTGTCTACCACTACAAGGACTGTGCATCTAAGTAATAATTTTTTAAGATTCACTATATGTGATAGTAT GATATGCATTTATTTAAAATGCATTAGACTCTCTTCCATCCATCAAATACTTTACAGGATGGCATTTAATACAGATATTTCGTATTTCCC CCACTGCTTTTTATTTGTACAGCATCATTAAACACTAAGCTCAGTTAAGGAGCCATCAGCAACACTGAAGAGATCAGTAGTAAGAATTCC ATTTTCCCTCATCAGTGAAGACACCACAAATTGAAACTCAGAACTATATTTCTAAGCCTGCATTTTCACTGATGCATAATTTTCTTATTA ATATTAAGAGACAGTTTTTCTATGGCATCTCCAAAACTGCATGACATCACTAGTCTTACTTCTGCTTAATTTTATGAGAAGGTATTCTTC ATTTTAATTGCTTTTGGGATTACTCCACATCTTTGTTTATTTCTTGACTAATCAGATTTTCAATAGAGTGAAGTTAAATTGGGGGTCATA AAAGCATTGGATTGACATATGGTTTGCCAGCCTATGGGTTTACAGGCATTGCCCAAACATTTCTTTGAGATCTATATTTATAAGCAGCCA TGGAATTCCTATTATGGGATGTTGGCAATCTTACATTTTATAGAGGTCATATGCATAGTTTTCATAGGTGTTTTGTAAGAACTGATTGCT CTCCTGTGAGTTAAGCTATGTTTACTACTGGGACCCTCAAGAGGAATACCACTTATGTTACACTCCTGCACTAAAGGCACGTACTGCAGT GTGAAGAAATGTTCTGAAAAAGGGTTATAGAAATCTGGAAATAAGAAAGGAAGAGCTCTCTGTATTCTATAATTGGAAGAGAAAAAAAGA AAAACTTTTAACTGGAAATGTTAGTTTGTACTTATTGATCATGAATACAAGTATATATTTAATTTTGCAAACTTTTCACTTGTTTGTATT CTTTTCATTATAAGGAAAAGTAGTATAACACACTTTTTTTTAAGTCACATATTTGATGGTATACAAGATGTTTCAGGGCTGGCATTCTCC AAGCAGCAGATAAAGGTTGCATTTCTTTCAAAAATACATTCAAAGACCATGTTATCCTTCATTTTGGTTTGAGACATTTAGTTATGGTAT TTCATATTATTAGGTAAAAGATATTTGAAATAAAATGTATCTCTTTAATTATTTCTAACAAATATATTTCTTTATTAAGGCATCCAACTT TTTCCCAAATAACAGTACACTTTAATGTATGTAGACATTCTTAATTAGTATAATCATCTAGTTGATACTGTTCTCAGATTGGGAATTATC AAAGATGTACTTATCATATCAGAACACCATGCTAACTTATCTATTATTCTTTCTTAGTAAAACAAAACAGCCAGTAATAACTATATAAAA TATTAAATAACATTTCCACAGAATAAATTAGACTTTCATGTACTCTACAATTAGAAATATCCATAAATATTTTCTTCTGGACTGTGAACC CAGATAACTAGGCTTTCTGGTCTTATTTTAAGTGATGCTCTTTATACTATAGAAGACACTCTTAATACATCATATATGTATGCAGAACAT CTCTTAGATGAGAGAACAAGCACATCATTGGCATTCCCTCTGTGCCAGTTTGATGTTTGTTTGACGATGGAGCTACAAGAGTCTAAATCT GCCTTTGAAAAGCTCAGCTCACAGTGTAGTAGAGTAGACTCCAAGGTTCAAAAAAGTGTTTTTAATAAAATTTGGTAAGTTCTACAATCA AGGAATGAAGAGAGAAGAGAACAGCTTTTTCCATCTGGGAGATTCAATAATGGCTTCATATATAGATGATAGTGTGGCTAAAAAAAATTA CATCAATGCTGTTTCTATTGGGAAGCAATGGCAAGTAATAGTATGTCAATACTGGTCAAGTGTTGAAGCCAGGATGCACTTACACAACGA GAGTGTAAAGTGTGTAGGGAGCTGATCTGTGAAGGAGCAACTCAAAGATTTTGAAATTATCAGTGAAATGCCAAAACAGGGAAAGTTTCC CTGTTTCCCTTCATCTATTCTACAATGCACCATTGGTTTTCAGTGTGAAAAATCCCAATCCCTGAGCCCAAACCACTTCTGGGGTCAAAA CATTTTTCTTGTGCACAAAATGAATGGATCTTGGTCAAAGAAGTATGGTCACAATTTTCTAGCATATATTGCGTTTATAATTTAGCAGAA ATTCTGCAATAAATAAAGGAAAATCTGTTTTACCTGTGCAACAATCAGAAATGGTAAAAATGCATTCAAGTGTTTGAAAGAAATGGTAGA ATATGCTTGCATAGTGCTTTATAGTTTCAAAGTGCTGTCAGGTACATCGTCCGTTTAGCTCCTCACGCACTTTGTAGGGTACAAGCATCA TGTTCCATCTGGGTTGTGGCTGATTTTCATGGATGGCTTGTGGCATCAGTTTCATTACTTCATAGTCACAATTCTGTGTTTCATCTTGAG AAATGTGGAGACTAATGTTTGTACCAGTATATTCCACATTCAGTGAGGTTTCCAAGATGCTTCCTCCAAACTCTCACTTTCTTAAACAGA TACATTTAAAATATGACACCTTCTGAAAAGTCCATAAAGAAATTAGTATATTCATGGAAAAAATTTCCCCGTAGACTTATTTGCATTTCA TGACTTGACATGAGAAATTTAAAAAACAGAAGGCAAAATGTGATTGCTTTTGATGGATTTATACAGAATAGTCATTTAATTTTCTCAAGT ATTATTTGGAAACAAGATAATGGAAAACATTAATCAGAAGCCTTATTTTATTTTTAAATTTTTAATAGGGATGGATCTAGTAAGCCCTAA TAGCTGAGTTCCAGTGATAAAATTCACAAATTTTATTTATGATTGGACCACTTCTAACAAGGCAAATATCTTATAATGAAGATAGGAGTC TGTTCAATTTAACCTTATTCAGGAAACTCCGTATTTATGTGAAATGATGCAATGCCATTATTAAGAGGTATCTGTAACCCTGCATAATAA AAACATGCATAAGAAGAATGTGGTCTAATCTCCAAACTGAGTCTCAGTTTCAGGGCCTTTTGCTTTCTTTGTTTGTTTGTTTGAGACAGT CTGGCTCTGTCATCCAGGCTGGAGTGCAGTGGCACATGAGCTCACTGCAACCTCCACCTCCCAGGCTCAAGCGATCCTCCCACCTCAGCC TCCTGAGTAACTGGGACTACAGGCGCATACCACCACACCTGGCTAATTTTTGTATTTTTGTAGAGACAAGGCCCAGGCACGTCTCAAACT CTTGAGCTCAAGCGATCCGCCCGCCTTGGCCTCCCAAAGTGCTAGGAGTAATTATAGGTGTGAGCCACCATGCTGGCAGTTGTTTTTGTT TATTTGTGATAAGGTCTGGCTGTCACCCATGCTGGAGTGCAGTGGCATGATCTTGGCTCACTGCAACCTCTGCCTTCCAGGCTCAAGCCA GCCTCCCACCTCAGCCTCCGAGTAGCTAGGACTACAGGCATGCACCACCATGCCCAGCTAATTTTTTTATATTTTTTGTAGACACAGGGT TTTGCCACGTTGCCCAGGCTGGTCTTGAACTCCTGAGCTCAAGCAGTCCTCCCACCTTGGCCTCCCGAAGTGCTGGGATTACAGACGTGA GCCTCTGGGCCTTGCTCTGTTTTGTCTGCTTTCTGATTTAGTAGGTGAGGGACAGCACTCTGGACCTTAGCCATTTTACTTCCATCTTAC TGCCGTGAGTTTGATGAGAATGTCACTTTTAGACTCTAATTTTCAAAAACTTAGTTTCTTTCCCATTCATTTGTGTTTTCCAAATTAATT TGTTTGTCTAGTATGGATGCAAAGGAATGCTGAAGACATTAAAGCTTTCTGAATTGTGGCTTAAGGCCAGCTACTGCCCACTGAAAGGAT GATAAAAATAAAAAAGGAGGAAAGAGGAGCCCAGCAGTTGTGCCACCCCGCAAGAATATAATGCTAATTATTGACCAACTGTCAAGATTC AGAGTGGTAACTGCATTCCGAGGAGCTGTCAGGAAGGATGACCGTGTGTGTGCTGTCCATATCATAGCCATCTGGGTCAGAAGGAGCGAA AGGATGGTTAGGACAGGAGTGTAGTGGAGGGAAAGTTAAAACAACGGTAGATGGATCATCTTAGGGCTTGCTGGTGGGGGTAGGAACATA CTTCAATTTGGTTTCAGACTGAGAAGAAGAAAGCTATGCTCCCTGATGAAGGCACAGTTTGATCTACTAGATGGGAAATACCCTCATTTT ATAACTTCAGTAATTTAACATTTTCTGTATTGCAAAGTGGAATTTAGTCCCAAAGTATTAAAGCTAACCATAGCTTGATCAGGAGATTTG ACAATTTAGCATAAAATTATGTTGTGGATTTTTTTATTTAAGAATTATTGTTACTATTATTGAACACCATGTATGAAATGAAAGAAAGGG TGCTTTTATTTCATTATACTTATTTGGTTTTTTTAGCGGACATCCCTTGACATTAGAATAGCATTTGAGACATTTTCATGATTTCTAAAG ATAACTTAAATGCTCTCTGTTAATCTGGAAAGTGAAATCCTAAGAGTGAGGTTGCAAATGGAGACTAAATTAGTGTTTCCAAGTCAGGAA GTTAATCAAGACCGGAAATAGATTTGCTTAGATTTTATCTGCAGAGTCGTTTATTTGGCTTTCGTAATTGAACAGCTCAGCAGAATAAGA TGGCCCTAGTTGTATGGTTTCTGGGGAACACAGGAGAATTAAATTTATATATGTGATTTTTATGTGTAGTGCCCTTGGGCAGGTGATATC TTTCTATATGACTTTTAAGATATTACAACCTGAGGAGTGTGGATAGCCGTGTTGTGTGATTCAAACTGAATCAGCTATTACTGATGCAGG TAAGGAACCATCAGTCAACTTCTGGTTACTATCACCTTCATAAAAAAGAATGGATGATGTTCAGGAATTACCTTGGAAGGATTAACCAGG AATTAACACAGAATGGGATGTTCTTTTAGGACCCTAATGCTCTAAAAGATAAAATTACAACCTATTTTTCTTTATATGATAGCATAAGTA AAATTTGGGGCTGTTAATATGTTGGAAAGTGCTTAAAATTACATGTTTTTATAATAGGGATGCAGGCATTCCAAAATAAATGAATAAAAA TACATGCATCTCTCTCGCCTCTTACTGAAAGTATTAATCTAATTTTATAGTAATGATTACTTTTTAATATTAAGGCTCTGTAAAAAAATT ATTTCTAAGATTTCAGTAGAACACAGGTGGTGTATTTTTTTAAGCTTTTAATTTCTCTTCACCAACCTCAGTCATTACGTTCACCACACA GTGTGTCACAGTTAATAGTGGTATTTCAGTTTTTGCGTTGATTTAAGCTTCATATTTATTTAATGTGACTTTTGATGGAGAATAATAATG GGAGATGCTTGCTGTTAGCATTAAGGTTATACAGCTCTCACTTCACCTTATATGTAATTTTTTTTTCAAAGTAAATAATGCAGATTGTTG ACATCCTAGAAATGGTTTCTGGGTGGTATTATATGGGGCCTAGGACTCTGAACTGGGACTAGTTGTTTATTTTCTGGGAAGTCTGTATTC TTATTCTCAACTGAACCCCTGACTCTTTTGTTGTATTTTTTATGATTCCGAAATGCTTTGAGGGAAAGATATTTTAATGTGTACAATTTG TTTTATTTTCATACTGATCTCTATTTTTGTTTAAAACCATGTTGCATCATGAAATCACTTAATCCAAAATAAATCTTCTTTAAAAAAAAT >49730_49730_1_LRP1B-HECW2_LRP1B_chr2_142567848_ENST00000389484_HECW2_chr2_197208488_ENST00000260983_length(amino acids)=1580AA_BP=106 MTPLDSRLQAAPAAAAAAGPKVPPSSLQRRLPYRATTMSEFLLALLTLSGLLPIARVLTVGADRDQQLCDPGEFLCHDHVTCVSQSWLCD GDPDCPDDSDESLDTYENSPANFWDSKNRGVTGTQKGQIVWRIEPGPYFMEPEIKICFKYYHGISGALRATTPCITVKNPAVMMGAEGME GGASGNLHSRKLVSFTLSDLRAVGLKKGMFFNPDPYLKMSIQPGKKSSFPTCAHHGQERRSTIISNTTNPIWHREKYSFFALLTDVLEIE IKDKFAKSRPIIKRFLGKLTIPVQRLLERQAIGDQMLSYNLGRRLPADHVSGYLQFKVEVTSSVHEDASPEAVGTILGVNSVNGDLGSPS DDEDMPGSHHDSQVCSNGPVSEDSAADGTPKHSFRTSSTLEIDTEELTSTSSRTSPPRGRQDSLNDYLDAIEHNGHSRPGTATCSERSMG ASPKLRSSFPTDTRLNAMLHIDSDEEDHEFQQDLGYPSSLEEEGGLIMFSRASRADDGSLTSQTKLEDNPVENEEASTHEAASFEDKPEN LPELAESSLPAGPAPEEGEGGPEPQPSADQGSAELCGSQEVDQPTSGADTGTSDASGGSRRAVSETESLDQGSEPSQVSSETEPSDPART ESVSEASTRPEGESDLECADSSCNESVTTQLSSVDTRCSSLESARFPETPAFSSQEEEDGACAAEPTSSGPAEGSQESVCTAGSLPVVQV PSGEDEGPGAESATVPDQEELGEVWQRRGSLEGAAAAAESPPQEEGSAGEAQGTCEGATAQEEGATGGSQANGHQPLRSLPSVRQDVSRY QRVDEALPPNWEARIDSHGRIFYVDHVNRTTTWQRPTAPPAPQVLQRSNSIQQMEQLNRRYQSIRRTMTNERPEENTNAIDGAGEEADFH QASADFRRENILPHSTSRSRITLLLQSPPVKFLISPEFFTVLHSNPSAYRMFTNNTCLKHMITKVRRDTHHFERYQHNRDLVGFLNMFAN KQLELPRGWEMKHDHQGKAFFVDHNSRTTTFIDPRLPLQSSRPTSALVHRQHLTRQRSHSAGEVGEDSRHAGPPVLPRPSSTFNTVSRPQ YQDMVPVAYNDKIVAFLRQPNIFEILQERQPDLTRNHSLREKIQFIRTEGTPGLVRLSSDADLVMLLSLFEEEIMSYVPPHALLHPSYCQ SPRGSPVSSPQNSPGTQRANARAPAPYKRDFEAKLRNFYRKLETKGYGQGPGKLKLIIRRDHLLEDAFNQIMGYSRKDLQRNKLYVTFVG EEGLDYSGPSREFFFLVSRELFNPYYGLFEYSANDTYTVQISPMSAFVDNHHEWFRFSGRILGLALIHQYLLDAFFTRPFYKALLRILCD LSDLEYLDEEFHQSLQWMKDNDIHDILDLTFTVNEEVFGQITERELKPGGANIPVTEKNKKEYIERMVKWRIERGVVQQTESLVRGFYEV VDARLVSVFDARELELVIAGTAEIDLSDWRNNTEYRGGYHDNHIVIRWFWAAVERFNNEQRLRLLQFVTGTSSIPYEGFASLRGSNGPRR -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for LRP1B-HECW2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | HECW2 | chr2:142567848 | chr2:197208488 | ENST00000260983 | 1 | 29 | 737_1068 | 97.33333333333333 | 1573.0 | TP73 | |
| Tgene | HECW2 | chr2:142567848 | chr2:197208488 | ENST00000409111 | 0 | 27 | 737_1068 | 0 | 1217.0 | TP73 |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for LRP1B-HECW2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for LRP1B-HECW2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
