
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:LRP1-FAM19A2 (FusionGDB2 ID:49743) |
Fusion Gene Summary for LRP1-FAM19A2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: LRP1-FAM19A2 | Fusion gene ID: 49743 | Hgene | Tgene | Gene symbol | LRP1 | FAM19A2 | Gene ID | 10438 | 338811 |
| Gene name | C1D nuclear receptor corepressor | TAFA chemokine like family member 2 | |
| Synonyms | LRP1|Rrp47|SUN-CoR|SUNCOR|hC1D | FAM19A2|TAFA-2 | |
| Cytomap | 2p14 | 12q14.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | nuclear nucleic acid-binding protein C1DC1D DNA-binding proteinC1D nuclear receptor co-repressornuclear DNA-binding proteinsmall unique nuclear receptor co-repressorsmall unique nuclear receptor corepressor | chemokine-like protein TAFA-2family with sequence similarity 19 (chemokine (C-C motif)-like), member A2family with sequence similarity 19 member A2, C-C motif chemokine likeprotein FAM19A2 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q07954 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000243077, ENST00000338962, ENST00000553277, ENST00000553446, ENST00000554174, | ENST00000549456, ENST00000416284, ENST00000551449, ENST00000551619, ENST00000550003, | |
| Fusion gene scores | * DoF score | 23 X 27 X 12=7452 | 14 X 10 X 6=840 |
| # samples | 26 | 14 | |
| ** MAII score | log2(26/7452*10)=-4.84104414591318 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(14/840*10)=-2.58496250072116 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: LRP1 [Title/Abstract] AND FAM19A2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | LRP1(57569488)-FAM19A2(62104198), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across LRP1 (5'-gene) Fusion gene breakpoints across LRP1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
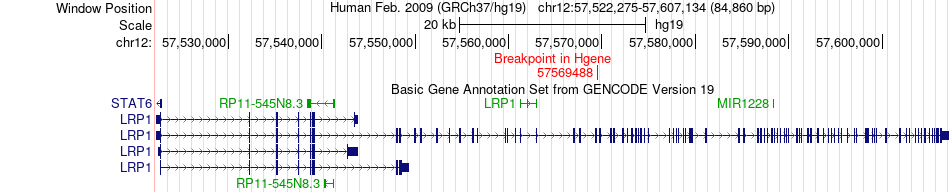 |
 Fusion gene breakpoints across FAM19A2 (3'-gene) Fusion gene breakpoints across FAM19A2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
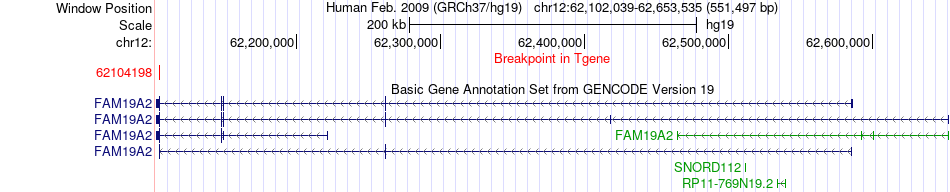 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-3B-A9HS-01A | LRP1 | chr12 | 57569488 | - | FAM19A2 | chr12 | 62104198 | - |
| ChimerDB4 | SARC | TCGA-3B-A9HS-01A | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - |
Top |
Fusion Gene ORF analysis for LRP1-FAM19A2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000243077 | ENST00000549456 | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - |
| Frame-shift | ENST00000243077 | ENST00000416284 | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - |
| Frame-shift | ENST00000243077 | ENST00000551449 | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - |
| Frame-shift | ENST00000243077 | ENST00000551619 | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - |
| In-frame | ENST00000243077 | ENST00000550003 | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - |
| intron-3CDS | ENST00000338962 | ENST00000416284 | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - |
| intron-3CDS | ENST00000338962 | ENST00000550003 | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - |
| intron-3CDS | ENST00000338962 | ENST00000551449 | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - |
| intron-3CDS | ENST00000338962 | ENST00000551619 | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - |
| intron-3CDS | ENST00000553277 | ENST00000416284 | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - |
| intron-3CDS | ENST00000553277 | ENST00000550003 | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - |
| intron-3CDS | ENST00000553277 | ENST00000551449 | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - |
| intron-3CDS | ENST00000553277 | ENST00000551619 | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - |
| intron-3CDS | ENST00000553446 | ENST00000416284 | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - |
| intron-3CDS | ENST00000553446 | ENST00000550003 | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - |
| intron-3CDS | ENST00000553446 | ENST00000551449 | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - |
| intron-3CDS | ENST00000553446 | ENST00000551619 | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - |
| intron-3CDS | ENST00000554174 | ENST00000416284 | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - |
| intron-3CDS | ENST00000554174 | ENST00000550003 | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - |
| intron-3CDS | ENST00000554174 | ENST00000551449 | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - |
| intron-3CDS | ENST00000554174 | ENST00000551619 | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - |
| intron-intron | ENST00000338962 | ENST00000549456 | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - |
| intron-intron | ENST00000553277 | ENST00000549456 | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - |
| intron-intron | ENST00000553446 | ENST00000549456 | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - |
| intron-intron | ENST00000554174 | ENST00000549456 | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000243077 | LRP1 | chr12 | 57569488 | + | ENST00000550003 | FAM19A2 | chr12 | 62104198 | - | 6400 | 4259 | 442 | 4302 | 1286 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000243077 | ENST00000550003 | LRP1 | chr12 | 57569488 | + | FAM19A2 | chr12 | 62104198 | - | 0.001492763 | 0.9985072 |
Top |
Fusion Genomic Features for LRP1-FAM19A2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
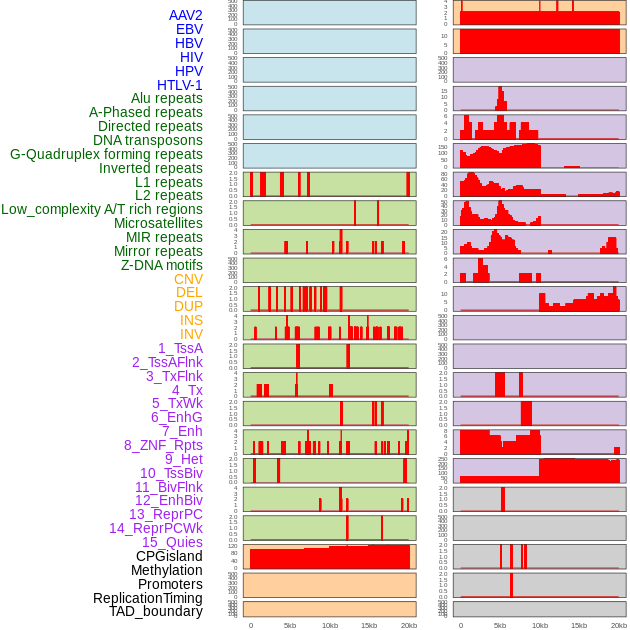 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for LRP1-FAM19A2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:57569488/chr12:62104198) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| LRP1 | . |
| FUNCTION: Endocytic receptor involved in endocytosis and in phagocytosis of apoptotic cells (PubMed:11907044, PubMed:12713657). Required for early embryonic development (By similarity). Involved in cellular lipid homeostasis. Involved in the plasma clearance of chylomicron remnants and activated LRPAP1 (alpha 2-macroglobulin), as well as the local metabolism of complexes between plasminogen activators and their endogenous inhibitors. Acts as an LRPAP1 alpha-2-macroglobulin receptor (PubMed:26142438, PubMed:1702392). Acts as TAU/MAPT receptor and controls the endocytosis of TAU/MAPT as well as its subsequent spread (PubMed:32296178). May modulate cellular events, such as APP metabolism, kinase-dependent intracellular signaling, neuronal calcium signaling as well as neurotransmission (PubMed:12888553). {ECO:0000250|UniProtKB:Q91ZX7, ECO:0000269|PubMed:11907044, ECO:0000269|PubMed:12713657, ECO:0000269|PubMed:12888553, ECO:0000269|PubMed:1702392, ECO:0000269|PubMed:26142438, ECO:0000269|PubMed:32296178}.; FUNCTION: (Microbial infection) Functions as a receptor for Pseudomonas aeruginosa exotoxin A. {ECO:0000269|PubMed:1618748}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 1013_1053 | 1264 | 4545.0 | Domain | LDL-receptor class A 7 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 1060_1099 | 1264 | 4545.0 | Domain | LDL-receptor class A 8 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 1102_1142 | 1264 | 4545.0 | Domain | LDL-receptor class A 9 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 111_149 | 1264 | 4545.0 | Domain | EGF-like 1 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 1143_1182 | 1264 | 4545.0 | Domain | LDL-receptor class A 10 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 1183_1222 | 1264 | 4545.0 | Domain | EGF-like 5 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 1223_1262 | 1264 | 4545.0 | Domain | EGF-like 6 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 150_189 | 1264 | 4545.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 25_66 | 1264 | 4545.0 | Domain | LDL-receptor class A 1 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 474_520 | 1264 | 4545.0 | Domain | EGF-like 3 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 70_110 | 1264 | 4545.0 | Domain | LDL-receptor class A 2 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 803_843 | 1264 | 4545.0 | Domain | EGF-like 4 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 852_892 | 1264 | 4545.0 | Domain | LDL-receptor class A 3 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 893_933 | 1264 | 4545.0 | Domain | LDL-receptor class A 4 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 934_973 | 1264 | 4545.0 | Domain | LDL-receptor class A 5 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 974_1013 | 1264 | 4545.0 | Domain | LDL-receptor class A 6 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 292_334 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 1 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 335_378 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 2 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 379_422 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 3 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 571_613 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 4 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 614_659 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 5 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 660_710 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 6 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 711_754 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 7 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 1536_1579 | 1264 | 4545.0 | Domain | EGF-like 7 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 1846_1887 | 1264 | 4545.0 | Domain | EGF-like 8 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 2155_2195 | 1264 | 4545.0 | Domain | EGF-like 9 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 2478_2518 | 1264 | 4545.0 | Domain | EGF-like 10 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 2522_2563 | 1264 | 4545.0 | Domain | LDL-receptor class A 11 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 2564_2602 | 1264 | 4545.0 | Domain | LDL-receptor class A 12 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 2603_2641 | 1264 | 4545.0 | Domain | LDL-receptor class A 13 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 2642_2690 | 1264 | 4545.0 | Domain | LDL-receptor class A 14 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 2694_2732 | 1264 | 4545.0 | Domain | LDL-receptor class A 15 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 2732_2771 | 1264 | 4545.0 | Domain | LDL-receptor class A 16 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 2772_2814 | 1264 | 4545.0 | Domain | LDL-receptor class A 17 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 2816_2855 | 1264 | 4545.0 | Domain | LDL-receptor class A 18 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 2856_2899 | 1264 | 4545.0 | Domain | LDL-receptor class A 19 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 2902_2940 | 1264 | 4545.0 | Domain | LDL-receptor class A 20 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 2941_2981 | 1264 | 4545.0 | Domain | EGF-like 11 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 2982_3022 | 1264 | 4545.0 | Domain | EGF-like 12%3B calcium-binding |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 3290_3331 | 1264 | 4545.0 | Domain | EGF-like 13 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 3332_3371 | 1264 | 4545.0 | Domain | LDL-receptor class A 21 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 3372_3410 | 1264 | 4545.0 | Domain | LDL-receptor class A 22 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 3411_3450 | 1264 | 4545.0 | Domain | LDL-receptor class A 23 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 3451_3491 | 1264 | 4545.0 | Domain | LDL-receptor class A 24 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 3492_3533 | 1264 | 4545.0 | Domain | LDL-receptor class A 25 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 3534_3572 | 1264 | 4545.0 | Domain | LDL-receptor class A 26 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 3573_3611 | 1264 | 4545.0 | Domain | LDL-receptor class A 27 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 3611_3649 | 1264 | 4545.0 | Domain | LDL-receptor class A 28 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 3652_3692 | 1264 | 4545.0 | Domain | LDL-receptor class A 29 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 3693_3733 | 1264 | 4545.0 | Domain | LDL-receptor class A 30 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 3739_3778 | 1264 | 4545.0 | Domain | LDL-receptor class A 31 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 3781_3823 | 1264 | 4545.0 | Domain | EGF-like 14 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 3824_3861 | 1264 | 4545.0 | Domain | EGF-like 15 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 4147_4183 | 1264 | 4545.0 | Domain | EGF-like 16 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 4196_4232 | 1264 | 4545.0 | Domain | EGF-like 17 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 4232_4268 | 1264 | 4545.0 | Domain | EGF-like 18 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 4268_4304 | 1264 | 4545.0 | Domain | EGF-like 19 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 4304_4340 | 1264 | 4545.0 | Domain | EGF-like 20 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 4340_4375 | 1264 | 4545.0 | Domain | EGF-like 21 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 4373_4409 | 1264 | 4545.0 | Domain | EGF-like 22 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 3940_3943 | 1264 | 4545.0 | Motif | Recognition site for proteolytical processing |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 4502_4507 | 1264 | 4545.0 | Motif | Note=NPXY motif |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 1309_1355 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 8 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 1356_1398 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 9 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 1399_1445 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 10 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 1446_1490 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 11 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 1491_1531 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 12 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 1627_1669 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 13 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 1670_1713 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 14 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 1714_1753 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 15 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 1754_1798 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 16 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 1934_1976 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 17 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 1977_2019 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 18 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 2020_2063 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 19 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 2064_2107 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 20 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 2253_2294 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 21 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 2295_2343 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 22 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 2344_2388 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 23 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 2389_2431 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 24 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 2432_2473 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 25 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 3069_3113 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 26 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 3114_3156 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 27 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 3157_3200 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 28 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 3201_3243 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 29 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 3244_3284 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 30 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 3912_3954 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 31 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 3970_4012 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 32 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 4013_4056 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 33 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 4057_4101 | 1264 | 4545.0 | Repeat | Note=LDL-receptor class B 34 |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 20_4419 | 1264 | 4545.0 | Topological domain | Extracellular |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 4420_4444 | 1264 | 4545.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for LRP1-FAM19A2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >49743_49743_1_LRP1-FAM19A2_LRP1_chr12_57569488_ENST00000243077_FAM19A2_chr12_62104198_ENST00000550003_length(transcript)=6400nt_BP=4259nt CAGCGGTGCGAGCTCCAGGCCCATGCACTGAGGAGGCGGAAACAAGGGGAGCCCCCAGAGCTCCATCAAGCCCCCTCCAAAGGCTCCCCT ACCCGGTCCACGCCCCCCACCCCCCCTCCCCGCCTCCTCCCAATTGTGCATTTTTGCAGCCGGAGGCGGCTCCGAGATGGGGCTGTGAGC TTCGCCCGGGGAGGGGGAAAGAGCAGCGAGGAGTGAAGCGGGGGGGTGGGGTGAAGGGTTTGGATTTCGGGGCAGGGGGCGCACCCCCGT CAGCAGGCCCTCCCCAAGGGGCTCGGAACTCTACCTCTTCACCCACGCCCCTGGTGCGCTTTGCCGAAGGAAAGAATAAGAACAGAGAAG GAGGAGGGGGAAAGGAGGAAAAGGGGGACCCCCCAACTGGGGGGGGTGAAGGAGAGAAGTAGCAGGACCAGAGGGGAAGGGGCTGCTGCT TGCATCAGCCCACACCATGCTGACCCCGCCGTTGCTCCTGCTGCTGCCCCTGCTCTCAGCTCTGGTCGCGGCGGCTATCGACGCCCCTAA GACTTGCAGCCCCAAGCAGTTTGCCTGCAGAGATCAAATAACCTGTATCTCAAAGGGCTGGCGGTGCGACGGTGAGAGGGACTGCCCAGA CGGATCTGACGAGGCCCCTGAGATTTGTCCACAGAGTAAGGCCCAGCGATGCCAGCCAAACGAGCATAACTGCCTGGGTACTGAGCTGTG TGTTCCCATGTCCCGCCTCTGCAATGGGGTCCAGGACTGCATGGACGGCTCAGATGAGGGGCCCCACTGCCGAGAGCTCCAAGGCAACTG CTCTCGCCTGGGCTGCCAGCACCATTGTGTCCCCACACTCGATGGGCCCACCTGCTACTGCAACAGCAGCTTTCAGCTTCAGGCAGATGG CAAGACCTGCAAAGATTTTGATGAGTGCTCAGTGTACGGCACCTGCAGCCAGCTATGCACCAACACAGACGGCTCCTTCATATGTGGCTG TGTTGAAGGATACCTCCTGCAGCCGGATAACCGCTCCTGCAAGGCCAAGAACGAGCCAGTAGACCGGCCCCCTGTGCTGTTGATAGCCAA CTCCCAGAACATCTTGGCCACGTACCTGAGTGGGGCCCAGGTGTCTACCATCACACCTACGAGCACGCGGCAGACCACAGCCATGGACTT CAGCTATGCCAACGAGACCGTATGCTGGGTGCATGTTGGGGACAGTGCTGCTCAGACGCAGCTCAAGTGTGCCCGCATGCCTGGCCTAAA GGGCTTCGTGGATGAGCACACCATCAACATCTCCCTCAGTCTGCACCACGTGGAACAGATGGCCATCGACTGGCTGACAGGCAACTTCTA CTTTGTGGATGACATCGATGATAGGATCTTTGTCTGCAACAGAAATGGGGACACATGTGTCACATTGCTAGACCTGGAACTCTACAACCC CAAGGGCATTGCCCTGGACCCTGCCATGGGGAAGGTGTTTTTCACTGACTATGGGCAGATCCCAAAGGTGGAACGCTGTGACATGGATGG GCAGAACCGCACCAAGCTCGTCGACAGCAAGATTGTGTTTCCTCATGGCATCACGCTGGACCTGGTCAGCCGCCTTGTCTACTGGGCAGA TGCCTATCTGGACTATATTGAAGTGGTGGACTATGAGGGCAAGGGCCGCCAGACCATCATCCAGGGCATCCTGATTGAGCACCTGTACGG CCTGACTGTGTTTGAGAATTATCTCTATGCCACCAACTCGGACAATGCCAATGCCCAGCAGAAGACGAGTGTGATCCGTGTGAACCGCTT TAACAGCACCGAGTACCAGGTTGTCACCCGGGTGGACAAGGGTGGTGCCCTCCACATCTACCACCAGAGGCGTCAGCCCCGAGTGAGGAG CCATGCCTGTGAAAACGACCAGTATGGGAAGCCGGGTGGCTGCTCTGACATCTGCCTGCTGGCCAACAGCCACAAGGCGCGGACCTGCCG CTGCCGTTCCGGCTTCAGCCTGGGCAGTGACGGGAAGTCATGCAAGAAGCCGGAGCATGAGCTGTTCCTCGTGTATGGCAAGGGCCGGCC AGGCATCATCCGGGGCATGGATATGGGGGCCAAGGTCCCGGATGAGCACATGATCCCCATTGAAAACCTCATGAACCCCCGAGCCCTGGA CTTCCACGCTGAGACCGGCTTCATCTACTTTGCCGACACCACCAGCTACCTCATTGGCCGCCAGAAGATTGATGGCACTGAGCGGGAGAC CATCCTGAAGGACGGCATCCACAATGTGGAGGGTGTGGCCGTGGACTGGATGGGAGACAATCTGTACTGGACGGACGATGGGCCCAAAAA GACAATCAGCGTGGCCAGGCTGGAGAAAGCTGCTCAGACCCGCAAGACTTTAATCGAGGGCAAAATGACACACCCCAGGGCTATTGTGGT GGATCCACTCAATGGGTGGATGTACTGGACAGACTGGGAGGAGGACCCCAAGGACAGTCGGCGTGGGCGGCTGGAGAGGGCGTGGATGGA TGGCTCACACCGAGACATCTTTGTCACCTCCAAGACAGTGCTTTGGCCCAATGGGCTAAGCCTGGACATCCCGGCTGGGCGCCTCTACTG GGTGGATGCCTTCTACGACCGCATCGAGACGATACTGCTCAATGGCACAGACCGGAAGATTGTGTATGAAGGTCCTGAGCTGAACCACGC CTTTGGCCTGTGTCACCATGGCAACTACCTCTTCTGGACTGAGTATCGGAGTGGCAGTGTCTACCGCTTGGAACGGGGTGTAGGAGGCGC ACCCCCCACTGTGACCCTTCTGCGCAGTGAGCGGCCCCCCATCTTTGAGATCCGAATGTATGATGCCCAGCAGCAGCAAGTTGGCACCAA CAAATGCCGGGTGAACAATGGCGGCTGCAGCAGCCTGTGCTTGGCCACCCCTGGGAGCCGCCAGTGCGCCTGTGCTGAGGACCAGGTGTT GGACGCAGACGGCGTCACTTGCTTGGCGAACCCATCCTACGTGCCTCCACCCCAGTGCCAGCCAGGCGAGTTTGCCTGTGCCAACAGCCG CTGCATCCAGGAGCGCTGGAAGTGTGACGGAGACAACGATTGCCTGGACAACAGTGATGAGGCCCCAGCCCTCTGCCATCAGCACACCTG CCCCTCGGACCGATTCAAGTGCGAGAACAACCGGTGCATCCCCAACCGCTGGCTCTGCGACGGGGACAATGACTGTGGGAACAGTGAAGA TGAGTCCAATGCCACTTGTTCAGCCCGCACCTGCCCCCCCAACCAGTTCTCCTGTGCCAGTGGCCGCTGCATCCCCATCTCCTGGACGTG TGATCTGGATGACGACTGTGGGGACCGCTCTGATGAGTCTGCTTCGTGTGCCTATCCCACCTGCTTCCCCCTGACTCAGTTTACCTGCAA CAATGGCAGATGTATCAACATCAACTGGAGATGCGACAATGACAATGACTGTGGGGACAACAGTGACGAAGCCGGCTGCAGCCACTCCTG TTCTAGCACCCAGTTCAAGTGCAACAGCGGGCGTTGCATCCCCGAGCACTGGACCTGCGATGGGGACAATGACTGCGGAGACTACAGTGA TGAGACACACGCCAACTGCACCAACCAGGCCACGAGGCCCCCTGGTGGCTGCCACACTGATGAGTTCCAGTGCCGGCTGGATGGACTATG CATCCCCCTGCGGTGGCGCTGCGATGGGGACACTGACTGCATGGACTCCAGCGATGAGAAGAGCTGTGAGGGAGTGACCCACGTCTGCGA TCCCAGTGTCAAGTTTGGCTGCAAGGACTCAGCTCGGTGCATCAGCAAAGCGTGGGTGTGTGATGGCGACAATGACTGTGAGGATAACTC GGACGAGGAGAACTGCGAGTCCCTGGCCTGCAGGCCACCCTCGCACCCTTGTGCCAACAACACCTCAGTCTGCCTGCCCCCTGACAAGCT GTGTGATGGCAACGACGACTGTGGCGACGGCTCAGATGAGGGCGAGCTCTGCGACCAGTGCTCTCTGAATAACGGTGGCTGCAGCCACAA CTGCTCAGTGGCACCTGGCGAAGGCATTGTGTGTTCCTGCCCTCTGGGCATGGAGCTGGGGCCCGACAACCACACCTGCCAGATCCAGAG CTACTGTGCCAAGCATCTCAAATGCAGCCAAAAGTGCGACCAGAACAAGTTCAGCGTGAAGTGCTCCTGCTACGAGGGCTGGGTCCTGGA ACCTGACGGCGAGAGCTGCCGCAGCCTGGGTAACCCATTAACCCAGGAGAAATCAAGTGATCCTCAAGGCTGATGACATTGAACATGCGC ATAGAAACTTAACTCAACTCCTGAGGTGATCTTGAAGATTTTTATACCACTTGAAAGAGGCGCTCAATAGTCTATTTCCAAGGGATTTCA TGGCCTCTTCTTGAAATCAAGACTTTTTAAAAGTCAGACATGAACTTGCATGTCATGAAGATTTCAGCAGATTTGAACTGTGTTCAACTT GTAAATTGTTAAAAGAATTTGAAGTCACTGTCTGAGGAGCTGGTGAAGAGTTGTTTTTTTCAGGGTGATGTTAGAGACAGTCACCTTTTG AGTTATTGGCTCCAGATGTGACTACTTTTCTTGTTTCTGCAAGCTGTATCCCAAGTGCACTGTCCTTCTGTCCTGGATGTGTTCCTGGGT CCTATGTTCATTTGCTAGTGGGACTACACATGGCTTTAATGACATTTCCTTTGAGAACTTTTCCTCTGGCATGGTGTAGACTGAGACAAT TTTATTTATATCCTAATCTTGGAGCTCAGAAAGCCTACATGTTTTAACATCTTAAAGTTGCTTTTGTTAAAGGAATGGAAATATATATCC ATTGGTAATAATGTTGGCAAGTAATAGTTATCTGAATAAATCAATCATATAAGAATGTATAGACAAGCTGACATATTTCCCTAAGGCTAA CAACACCCTGCTGAAGCTCTTTGTCAAATAGGTAGTAGTTAGAACTGGATTGCCATTTTCATTATATAATACTTTGTACCTCTAGAGCAC TCTCCCTTTCTGTTTTTTTTTAAGTGAGCTTTTCTTTAATTTTTTATGTTTACTTATTCCCTTCACAGAAATCAGCAGTGAGCAGTCAAG TTAATGGGTAGCCTTCAGTTTCAAAAAAATTGACAGGGATGCATGTGAGTTTCTGATTTCTTAGCTTGAACATTATTCACTTAGATTTCT TCCAGTATTTTTTAAAAAACTGTCCTATCTCATTTTAAAAGACTTTCTTTTGCTTGATCCCAATGACTGTTTGAATGCTTATATATTTGT TCAATCTGTTGATAGAAAAAATTGTTCATTTTCCTCAGTCTCAAATTTATAAATATTTGCTTACAGTTTTCCTATTCAAACAATTTGTTA GGCCAATATTTTGTGACATTTTTGTAGCGATTTTAACGTTTATGGTTTTGGTTCTACAGGAAAGTCATAAATATTTAAAGGCCTTAAACA TGTATGTACTTTTTTTTTCTAAGTTATAGAATGTATAATTTTGTACTACATTTATTTTGTTTCATTTGTGATATGAAGGGAGAGAAGAAA GAAAAGTGCATAGCCATTCTGTAACAATATTGTGTAAACCTATAGTTTGAAGGAATGCAAGGAGAAGGATTTCTGTGTTTTACTCATTTT AGGCTGTTCAGAAGATGCTTCAAAAATTGTCCTGTTAGAATTTCCATCATGGAAGGTGGTATGGAAGAAGGTATGGAAATACTTTGTATT CTAAAAACTCACTGACGTGGTCAGTTAGACATACGTTGGTTTCCAGGATAGAGGCCCATATATCCTGGGGAGCTTTGGTCTATTAGTTTG TGACAATATTCAAAGGCCAAAACACTACTCAGACACTTTCCTGGGAAGAGCAACTAAAAATGTAAAATTGGTTAAAAATAAAATCTGAAA AGTATGTATCTCACATTGAACTAAAATCCACTGTCTCATAAGTTCATGGAATGAAATGGCTTTCTGCCTCCATTTTAATCATGCATAAAA TGAATTAGATGGCTTTGAGTGGATTTTCACAATGGCTCAAGACTATATGAAATTATAAAAAAAAAGTTGCCCTGGGGTTTCTGCATCAAT TAGAATATCATTAATTTCTTTGTAACCAAGTGAAAAACTATACTTTTTGGAAATTATGAATTTGTCCTAGGTTTGTTTGAGATTTGAAAT TATACATCATGCTTCTCATTTTTTAAACTATGTTCTTTAAATCAACACTGGAAACTCTGTATTATATACAAGTGTAATACATGCATATAA TAGAAAAAAAACATGGAATTTCAAATATACTAACTAGATTATCCCCAGTAGATTAATGTTGTGACTATTCAGAAAAGGTGAATAAAATTG >49743_49743_1_LRP1-FAM19A2_LRP1_chr12_57569488_ENST00000243077_FAM19A2_chr12_62104198_ENST00000550003_length(amino acids)=1286AA_BP= MLLASAHTMLTPPLLLLLPLLSALVAAAIDAPKTCSPKQFACRDQITCISKGWRCDGERDCPDGSDEAPEICPQSKAQRCQPNEHNCLGT ELCVPMSRLCNGVQDCMDGSDEGPHCRELQGNCSRLGCQHHCVPTLDGPTCYCNSSFQLQADGKTCKDFDECSVYGTCSQLCTNTDGSFI CGCVEGYLLQPDNRSCKAKNEPVDRPPVLLIANSQNILATYLSGAQVSTITPTSTRQTTAMDFSYANETVCWVHVGDSAAQTQLKCARMP GLKGFVDEHTINISLSLHHVEQMAIDWLTGNFYFVDDIDDRIFVCNRNGDTCVTLLDLELYNPKGIALDPAMGKVFFTDYGQIPKVERCD MDGQNRTKLVDSKIVFPHGITLDLVSRLVYWADAYLDYIEVVDYEGKGRQTIIQGILIEHLYGLTVFENYLYATNSDNANAQQKTSVIRV NRFNSTEYQVVTRVDKGGALHIYHQRRQPRVRSHACENDQYGKPGGCSDICLLANSHKARTCRCRSGFSLGSDGKSCKKPEHELFLVYGK GRPGIIRGMDMGAKVPDEHMIPIENLMNPRALDFHAETGFIYFADTTSYLIGRQKIDGTERETILKDGIHNVEGVAVDWMGDNLYWTDDG PKKTISVARLEKAAQTRKTLIEGKMTHPRAIVVDPLNGWMYWTDWEEDPKDSRRGRLERAWMDGSHRDIFVTSKTVLWPNGLSLDIPAGR LYWVDAFYDRIETILLNGTDRKIVYEGPELNHAFGLCHHGNYLFWTEYRSGSVYRLERGVGGAPPTVTLLRSERPPIFEIRMYDAQQQQV GTNKCRVNNGGCSSLCLATPGSRQCACAEDQVLDADGVTCLANPSYVPPPQCQPGEFACANSRCIQERWKCDGDNDCLDNSDEAPALCHQ HTCPSDRFKCENNRCIPNRWLCDGDNDCGNSEDESNATCSARTCPPNQFSCASGRCIPISWTCDLDDDCGDRSDESASCAYPTCFPLTQF TCNNGRCININWRCDNDNDCGDNSDEAGCSHSCSSTQFKCNSGRCIPEHWTCDGDNDCGDYSDETHANCTNQATRPPGGCHTDEFQCRLD GLCIPLRWRCDGDTDCMDSSDEKSCEGVTHVCDPSVKFGCKDSARCISKAWVCDGDNDCEDNSDEENCESLACRPPSHPCANNTSVCLPP DKLCDGNDDCGDGSDEGELCDQCSLNNGGCSHNCSVAPGEGIVCSCPLGMELGPDNHTCQIQSYCAKHLKCSQKCDQNKFSVKCSCYEGW -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for LRP1-FAM19A2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | LRP1 | chr12:57569488 | chr12:62104198 | ENST00000243077 | + | 23 | 89 | 4445_4544 | 1264.3333333333333 | 4545.0 | MAFB |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for LRP1-FAM19A2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for LRP1-FAM19A2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
