
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:LRP5-HSCB (FusionGDB2 ID:49797) |
Fusion Gene Summary for LRP5-HSCB |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: LRP5-HSCB | Fusion gene ID: 49797 | Hgene | Tgene | Gene symbol | LRP5 | HSCB | Gene ID | 4041 | 150274 |
| Gene name | LDL receptor related protein 5 | HscB mitochondrial iron-sulfur cluster cochaperone | |
| Synonyms | BMND1|EVR1|EVR4|HBM|LR3|LRP-5|LRP-7|LRP7|OPPG|OPS|OPTA1|PCLD4|VBCH2 | DNAJC20|HSC20|JAC1 | |
| Cytomap | 11q13.2 | 22q12.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | low-density lipoprotein receptor-related protein 5low density lipoprotein receptor-related protein 5low density lipoprotein receptor-related protein 7 | iron-sulfur cluster co-chaperone protein HscBDnaJ (Hsp40) homolog, subfamily C, member 20HscB iron-sulfur cluster co-chaperone homologHscB mitochondrial iron-sulfur cluster co-chaperoneJ-type co-chaperone HSC20epididymis secretory sperm binding prote | |
| Modification date | 20200329 | 20200320 | |
| UniProtAcc | O75197 | Q8IWL3 | |
| Ensembl transtripts involved in fusion gene | ENST00000294304, ENST00000529481, | ENST00000495977, ENST00000216027, ENST00000398941, | |
| Fusion gene scores | * DoF score | 36 X 25 X 12=10800 | 12 X 9 X 8=864 |
| # samples | 41 | 18 | |
| ** MAII score | log2(41/10800*10)=-4.71926359243275 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(18/864*10)=-2.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: LRP5 [Title/Abstract] AND HSCB [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | LRP5(68214001)-HSCB(29139870), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | LRP5 | GO:0008284 | positive regulation of cell proliferation | 9790987 |
| Hgene | LRP5 | GO:0045840 | positive regulation of mitotic nuclear division | 9790987 |
| Hgene | LRP5 | GO:0045893 | positive regulation of transcription, DNA-templated | 15035989|17955262 |
| Hgene | LRP5 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 12857724 |
| Hgene | LRP5 | GO:0060070 | canonical Wnt signaling pathway | 11029007|12121999|12857724|15908424|24706814|25920554 |
 Fusion gene breakpoints across LRP5 (5'-gene) Fusion gene breakpoints across LRP5 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
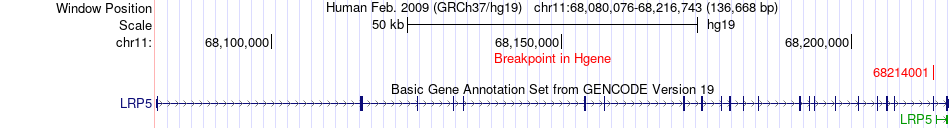 |
 Fusion gene breakpoints across HSCB (3'-gene) Fusion gene breakpoints across HSCB (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
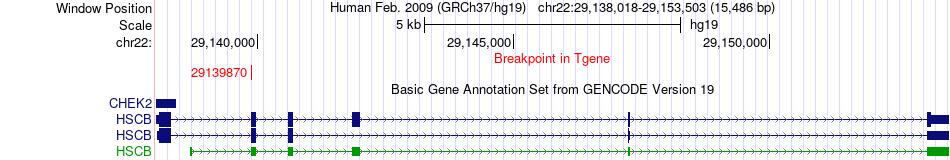 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | HNSC | TCGA-CV-6939-01A | LRP5 | chr11 | 68214001 | + | HSCB | chr22 | 29139870 | + |
Top |
Fusion Gene ORF analysis for LRP5-HSCB |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000294304 | ENST00000495977 | LRP5 | chr11 | 68214001 | + | HSCB | chr22 | 29139870 | + |
| In-frame | ENST00000294304 | ENST00000216027 | LRP5 | chr11 | 68214001 | + | HSCB | chr22 | 29139870 | + |
| In-frame | ENST00000294304 | ENST00000398941 | LRP5 | chr11 | 68214001 | + | HSCB | chr22 | 29139870 | + |
| intron-3CDS | ENST00000529481 | ENST00000216027 | LRP5 | chr11 | 68214001 | + | HSCB | chr22 | 29139870 | + |
| intron-3CDS | ENST00000529481 | ENST00000398941 | LRP5 | chr11 | 68214001 | + | HSCB | chr22 | 29139870 | + |
| intron-3UTR | ENST00000529481 | ENST00000495977 | LRP5 | chr11 | 68214001 | + | HSCB | chr22 | 29139870 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000294304 | LRP5 | chr11 | 68214001 | + | ENST00000216027 | HSCB | chr22 | 29139870 | + | 5505 | 4692 | 106 | 5163 | 1685 |
| ENST00000294304 | LRP5 | chr11 | 68214001 | + | ENST00000398941 | HSCB | chr22 | 29139870 | + | 5370 | 4692 | 106 | 4887 | 1593 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000294304 | ENST00000216027 | LRP5 | chr11 | 68214001 | + | HSCB | chr22 | 29139870 | + | 0.000990036 | 0.99900997 |
| ENST00000294304 | ENST00000398941 | LRP5 | chr11 | 68214001 | + | HSCB | chr22 | 29139870 | + | 0.001996776 | 0.9980032 |
Top |
Fusion Genomic Features for LRP5-HSCB |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| LRP5 | chr11 | 68214001 | + | HSCB | chr22 | 29139869 | + | 0.062854625 | 0.9371454 |
| LRP5 | chr11 | 68214001 | + | HSCB | chr22 | 29139869 | + | 0.062854625 | 0.9371454 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
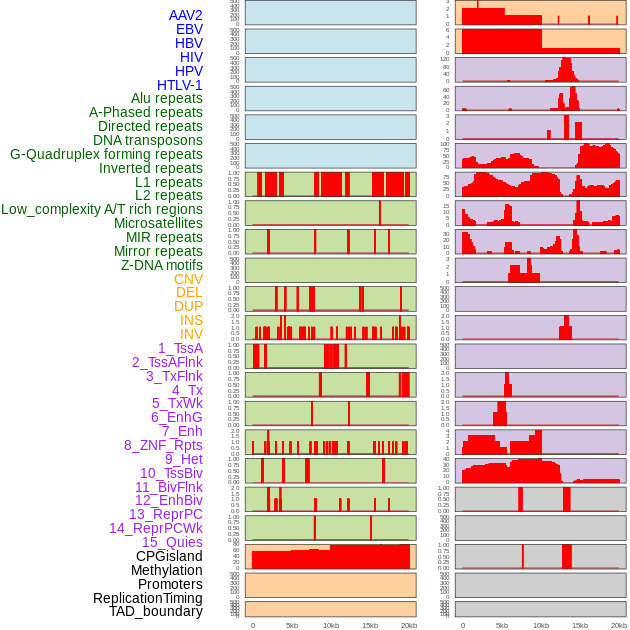 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
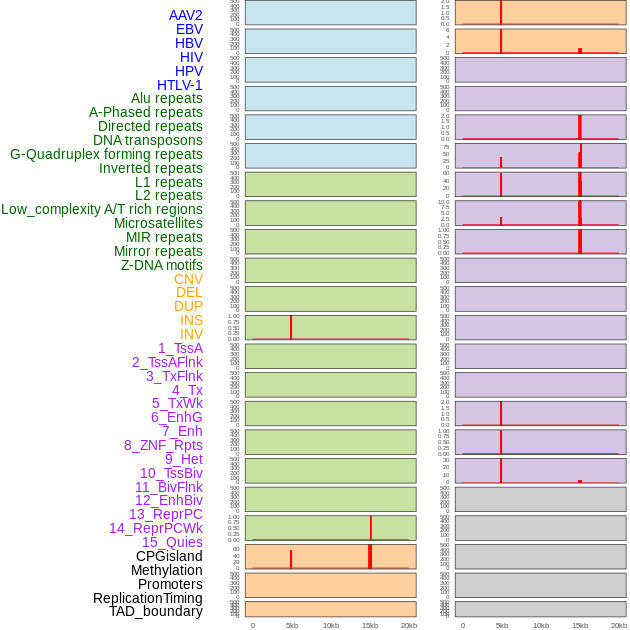 |
Top |
Fusion Protein Features for LRP5-HSCB |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr11:68214001/chr22:29139870) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| LRP5 | HSCB |
| FUNCTION: Acts as a coreceptor with members of the frizzled family of seven-transmembrane spanning receptors to transduce signal by Wnt proteins (PubMed:11336703, PubMed:11448771, PubMed:15778503, PubMed:11719191, PubMed:15908424, PubMed:16252235). Activates the canonical Wnt signaling pathway that controls cell fate determination and self-renewal during embryonic development and adult tissue regeneration (PubMed:11336703, PubMed:11719191). In particular, may play an important role in the development of the posterior patterning of the epiblast during gastrulation (By similarity). During bone development, regulates osteoblast proliferation and differentiation thus determining bone mass (PubMed:11719191). Mechanistically, the formation of the signaling complex between Wnt ligand, frizzled receptor and LRP5 coreceptor promotes the recruitment of AXIN1 to LRP5, stabilizing beta-catenin/CTNNB1 and activating TCF/LEF-mediated transcriptional programs (PubMed:11336703, PubMed:25920554, PubMed:24706814, PubMed:14731402). Acts as a coreceptor for non-Wnt proteins, such as norrin/NDP. Binding of norrin/NDP to frizzled 4/FZD4-LRP5 receptor complex triggers beta-catenin/CTNNB1-dependent signaling known to be required for retinal vascular development (PubMed:27228167, PubMed:16252235). Plays a role in controlling postnatal vascular regression in retina via macrophage-induced endothelial cell apoptosis (By similarity). {ECO:0000250|UniProtKB:Q91VN0, ECO:0000269|PubMed:11336703, ECO:0000269|PubMed:11448771, ECO:0000269|PubMed:11719191, ECO:0000269|PubMed:14731402, ECO:0000269|PubMed:15778503, ECO:0000269|PubMed:15908424, ECO:0000269|PubMed:16252235, ECO:0000269|PubMed:24706814, ECO:0000269|PubMed:25920554, ECO:0000269|PubMed:27228167}. | FUNCTION: Acts as a co-chaperone in iron-sulfur cluster assembly in both mitochondria and the cytoplasm (PubMed:20668094, PubMed:29309586). Required for incorporation of iron-sulfur clusters into SDHB, the iron-sulfur protein subunit of succinate dehydrogenase that is involved in complex II of the mitochondrial electron transport chain (PubMed:26749241). Recruited to SDHB by interaction with SDHAF1 which first binds SDHB and then recruits the iron-sulfur transfer complex formed by HSC20, HSPA9 and ISCU through direct binding to HSC20 (PubMed:26749241). Also mediates complex formation between components of the cytosolic iron-sulfur biogenesis pathway and the CIA targeting complex composed of CIAO1, DIPK1B/FAM69B and MMS19 by binding directly to the scaffold protein ISCU and to CIAO1 (PubMed:29309586). This facilitates iron-sulfur cluster insertion into a number of cytoplasmic and nuclear proteins including POLD1, ELP3, DPYD and PPAT (PubMed:29309586). {ECO:0000269|PubMed:20668094, ECO:0000269|PubMed:26749241, ECO:0000269|PubMed:29309586}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 1213_1254 | 1528 | 1616.0 | Domain | Note=EGF-like 4 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 1258_1296 | 1528 | 1616.0 | Domain | LDL-receptor class A 1 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 1297_1333 | 1528 | 1616.0 | Domain | LDL-receptor class A 2 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 1335_1371 | 1528 | 1616.0 | Domain | LDL-receptor class A 3 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 295_337 | 1528 | 1616.0 | Domain | Note=EGF-like 1 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 601_641 | 1528 | 1616.0 | Domain | Note=EGF-like 2 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 902_942 | 1528 | 1616.0 | Domain | Note=EGF-like 3 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 1500_1506 | 1528 | 1616.0 | Motif | Note=PPPSP motif A |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 32_288 | 1528 | 1616.0 | Region | Note=Beta-propeller 1 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 341_602 | 1528 | 1616.0 | Region | Note=Beta-propeller 2 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 644_903 | 1528 | 1616.0 | Region | Note=Beta-propeller 3 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 945_1212 | 1528 | 1616.0 | Region | Note=Beta-propeller 4 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 1036_1078 | 1528 | 1616.0 | Repeat | Note=LDL-receptor class B 17 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 1079_1123 | 1528 | 1616.0 | Repeat | Note=LDL-receptor class B 18 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 1124_1164 | 1528 | 1616.0 | Repeat | Note=LDL-receptor class B 19 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 1165_1207 | 1528 | 1616.0 | Repeat | Note=LDL-receptor class B 20 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 120_162 | 1528 | 1616.0 | Repeat | Note=LDL-receptor class B 2 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 123_126 | 1528 | 1616.0 | Repeat | Note=YWTD 2 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 163_206 | 1528 | 1616.0 | Repeat | Note=LDL-receptor class B 3 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 166_169 | 1528 | 1616.0 | Repeat | Note=YWTD 3 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 207_247 | 1528 | 1616.0 | Repeat | Note=LDL-receptor class B 4 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 248_290 | 1528 | 1616.0 | Repeat | Note=LDL-receptor class B 5 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 251_254 | 1528 | 1616.0 | Repeat | Note=YWTD 4 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 385_427 | 1528 | 1616.0 | Repeat | Note=LDL-receptor class B 6 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 388_391 | 1528 | 1616.0 | Repeat | Note=YWTD 5 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 428_470 | 1528 | 1616.0 | Repeat | Note=LDL-receptor class B 7 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 431_434 | 1528 | 1616.0 | Repeat | Note=YWTD 6 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 471_514 | 1528 | 1616.0 | Repeat | Note=LDL-receptor class B 8 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 474_477 | 1528 | 1616.0 | Repeat | Note=YWTD 7 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 515_557 | 1528 | 1616.0 | Repeat | Note=LDL-receptor class B 9 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 558_600 | 1528 | 1616.0 | Repeat | Note=LDL-receptor class B 10 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 559_562 | 1528 | 1616.0 | Repeat | Note=YWTD 8 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 687_729 | 1528 | 1616.0 | Repeat | Note=LDL-receptor class B 11 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 690_693 | 1528 | 1616.0 | Repeat | Note=YWTD 9 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 730_772 | 1528 | 1616.0 | Repeat | Note=LDL-receptor class B 12 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 75_119 | 1528 | 1616.0 | Repeat | Note=LDL-receptor class B 1 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 773_815 | 1528 | 1616.0 | Repeat | Note=LDL-receptor class B 13 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 78_81 | 1528 | 1616.0 | Repeat | Note=YWTD 1 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 816_855 | 1528 | 1616.0 | Repeat | Note=LDL-receptor class B 14 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 819_822 | 1528 | 1616.0 | Repeat | Note=YWTD 10 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 856_898 | 1528 | 1616.0 | Repeat | Note=LDL-receptor class B 15 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 859_862 | 1528 | 1616.0 | Repeat | Note=YWTD 11 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 989_1035 | 1528 | 1616.0 | Repeat | Note=LDL-receptor class B 16 |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 32_1384 | 1528 | 1616.0 | Topological domain | Extracellular |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 1385_1407 | 1528 | 1616.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 1495_1610 | 1528 | 1616.0 | Compositional bias | Note=Pro-rich |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 1538_1545 | 1528 | 1616.0 | Motif | Note=PPPSP motif B |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 1574_1581 | 1528 | 1616.0 | Motif | Note=PPPSP motif C |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 1591_1596 | 1528 | 1616.0 | Motif | Note=PPPSP motif D |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 1605_1612 | 1528 | 1616.0 | Motif | Note=PPPSP motif E |
| Hgene | LRP5 | chr11:68214001 | chr22:29139870 | ENST00000294304 | + | 22 | 23 | 1408_1615 | 1528 | 1616.0 | Topological domain | Cytoplasmic |
| Tgene | HSCB | chr11:68214001 | chr22:29139870 | ENST00000216027 | 0 | 6 | 72_144 | 78 | 236.0 | Domain | Note=J |
Top |
Fusion Gene Sequence for LRP5-HSCB |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >49797_49797_1_LRP5-HSCB_LRP5_chr11_68214001_ENST00000294304_HSCB_chr22_29139870_ENST00000216027_length(transcript)=5505nt_BP=4692nt GCCCGAGGGGGGAGGCGGAGGCGCCGGGAGCCGCGCGAGGAGCCGCCGCCGCCGCGCCATGGAGCCCGAGTGAGCGCGGCGCGGGCCCGT CCGGCCGCCGGACAACATGGAGGCAGCGCCGCCCGGGCCGCCGTGGCCGCTGCTGCTGCTGCTGCTGCTGCTGCTGGCGCTGTGCGGCTG CCCGGCCCCCGCCGCGGCCTCGCCGCTCCTGCTATTTGCCAACCGCCGGGACGTACGGCTGGTGGACGCCGGCGGAGTCAAGCTGGAGTC CACCATCGTGGTCAGCGGCCTGGAGGATGCGGCCGCAGTGGACTTCCAGTTTTCCAAGGGAGCCGTGTACTGGACAGACGTGAGCGAGGA GGCCATCAAGCAGACCTACCTGAACCAGACGGGGGCCGCCGTGCAGAACGTGGTCATCTCCGGCCTGGTCTCTCCCGACGGCCTCGCCTG CGACTGGGTGGGCAAGAAGCTGTACTGGACGGACTCAGAGACCAACCGCATCGAGGTGGCCAACCTCAATGGCACATCCCGGAAGGTGCT CTTCTGGCAGGACCTTGACCAGCCGAGGGCCATCGCCTTGGACCCCGCTCACGGGTACATGTACTGGACAGACTGGGGTGAGACGCCCCG GATTGAGCGGGCAGGGATGGATGGCAGCACCCGGAAGATCATTGTGGACTCGGACATTTACTGGCCCAATGGACTGACCATCGACCTGGA GGAGCAGAAGCTCTACTGGGCTGACGCCAAGCTCAGCTTCATCCACCGTGCCAACCTGGACGGCTCGTTCCGGCAGAAGGTGGTGGAGGG CAGCCTGACGCACCCCTTCGCCCTGACGCTCTCCGGGGACACTCTGTACTGGACAGACTGGCAGACCCGCTCCATCCATGCCTGCAACAA GCGCACTGGGGGGAAGAGGAAGGAGATCCTGAGTGCCCTCTACTCACCCATGGACATCCAGGTGCTGAGCCAGGAGCGGCAGCCTTTCTT CCACACTCGCTGTGAGGAGGACAATGGCGGCTGCTCCCACCTGTGCCTGCTGTCCCCAAGCGAGCCTTTCTACACATGCGCCTGCCCCAC GGGTGTGCAGCTGCAGGACAACGGCAGGACGTGTAAGGCAGGAGCCGAGGAGGTGCTGCTGCTGGCCCGGCGGACGGACCTACGGAGGAT CTCGCTGGACACGCCGGACTTCACCGACATCGTGCTGCAGGTGGACGACATCCGGCACGCCATTGCCATCGACTACGACCCGCTAGAGGG CTATGTCTACTGGACAGATGACGAGGTGCGGGCCATCCGCAGGGCGTACCTGGACGGGTCTGGGGCGCAGACGCTGGTCAACACCGAGAT CAACGACCCCGATGGCATCGCGGTCGACTGGGTGGCCCGAAACCTCTACTGGACCGACACGGGCACGGACCGCATCGAGGTGACGCGCCT CAACGGCACCTCCCGCAAGATCCTGGTGTCGGAGGACCTGGACGAGCCCCGAGCCATCGCACTGCACCCCGTGATGGGCCTCATGTACTG GACAGACTGGGGAGAGAACCCTAAAATCGAGTGTGCCAACTTGGATGGGCAGGAGCGGCGTGTGCTGGTCAATGCCTCCCTCGGGTGGCC CAACGGCCTGGCCCTGGACCTGCAGGAGGGGAAGCTCTACTGGGGAGACGCCAAGACAGACAAGATCGAGGTGATCAATGTTGATGGGAC GAAGAGGCGGACCCTCCTGGAGGACAAGCTCCCGCACATTTTTGGGTTCACGCTGCTGGGGGACTTCATCTACTGGACTGACTGGCAGCG CCGCAGCATCGAGCGGGTGCACAAGGTCAAGGCCAGCCGGGACGTCATCATTGACCAGCTGCCCGACCTGATGGGGCTCAAAGCTGTGAA TGTGGCCAAGGTCGTCGGAACCAACCCGTGTGCGGACAGGAACGGGGGGTGCAGCCACCTGTGCTTCTTCACACCCCACGCAACCCGGTG TGGCTGCCCCATCGGCCTGGAGCTGCTGAGTGACATGAAGACCTGCATCGTGCCTGAGGCCTTCTTGGTCTTCACCAGCAGAGCCGCCAT CCACAGGATCTCCCTCGAGACCAATAACAACGACGTGGCCATCCCGCTCACGGGCGTCAAGGAGGCCTCAGCCCTGGACTTTGATGTGTC CAACAACCACATCTACTGGACAGACGTCAGCCTGAAGACCATCAGCCGCGCCTTCATGAACGGGAGCTCGGTGGAGCACGTGGTGGAGTT TGGCCTTGACTACCCCGAGGGCATGGCCGTTGACTGGATGGGCAAGAACCTCTACTGGGCCGACACTGGGACCAACAGAATCGAAGTGGC GCGGCTGGACGGGCAGTTCCGGCAAGTCCTCGTGTGGAGGGACTTGGACAACCCGAGGTCGCTGGCCCTGGATCCCACCAAGGGCTACAT CTACTGGACCGAGTGGGGCGGCAAGCCGAGGATCGTGCGGGCCTTCATGGACGGGACCAACTGCATGACGCTGGTGGACAAGGTGGGCCG GGCCAACGACCTCACCATTGACTACGCTGACCAGCGCCTCTACTGGACCGACCTGGACACCAACATGATCGAGTCGTCCAACATGCTGGG TCAGGAGCGGGTCGTGATTGCCGACGATCTCCCGCACCCGTTCGGTCTGACGCAGTACAGCGATTATATCTACTGGACAGACTGGAATCT GCACAGCATTGAGCGGGCCGACAAGACTAGCGGCCGGAACCGCACCCTCATCCAGGGCCACCTGGACTTCGTGATGGACATCCTGGTGTT CCACTCCTCCCGCCAGGATGGCCTCAATGACTGTATGCACAACAACGGGCAGTGTGGGCAGCTGTGCCTTGCCATCCCCGGCGGCCACCG CTGCGGCTGCGCCTCACACTACACCCTGGACCCCAGCAGCCGCAACTGCAGCCCGCCCACCACCTTCTTGCTGTTCAGCCAGAAATCTGC CATCAGTCGGATGATCCCGGACGACCAGCACAGCCCGGATCTCATCCTGCCCCTGCATGGACTGAGGAACGTCAAAGCCATCGACTATGA CCCACTGGACAAGTTCATCTACTGGGTGGATGGGCGCCAGAACATCAAGCGAGCCAAGGACGACGGGACCCAGCCCTTTGTTTTGACCTC TCTGAGCCAAGGCCAAAACCCAGACAGGCAGCCCCACGACCTCAGCATCGACATCTACAGCCGGACACTGTTCTGGACGTGCGAGGCCAC CAATACCATCAACGTCCACAGGCTGAGCGGGGAAGCCATGGGGGTGGTGCTGCGTGGGGACCGCGACAAGCCCAGGGCCATCGTCGTCAA CGCGGAGCGAGGGTACCTGTACTTCACCAACATGCAGGACCGGGCAGCCAAGATCGAACGCGCAGCCCTGGACGGCACCGAGCGCGAGGT CCTCTTCACCACCGGCCTCATCCGCCCTGTGGCCCTGGTGGTGGACAACACACTGGGCAAGCTGTTCTGGGTGGACGCGGACCTGAAGCG CATTGAGAGCTGTGACCTGTCAGGGGCCAACCGCCTGACCCTGGAGGACGCCAACATCGTGCAGCCTCTGGGCCTGACCATCCTTGGCAA GCATCTCTACTGGATCGACCGCCAGCAGCAGATGATCGAGCGTGTGGAGAAGACCACCGGGGACAAGCGGACTCGCATCCAGGGCCGTGT CGCCCACCTCACTGGCATCCATGCAGTGGAGGAAGTCAGCCTGGAGGAGTTCTCAGCCCACCCATGTGCCCGTGACAATGGTGGCTGCTC CCACATCTGTATTGCCAAGGGTGATGGGACACCACGGTGCTCATGCCCAGTCCACCTCGTGCTCCTGCAGAACCTGCTGACCTGTGGAGA GCCGCCCACCTGCTCCCCGGACCAGTTTGCATGTGCCACAGGGGAGATCGACTGTATCCCCGGGGCCTGGCGCTGTGACGGCTTTCCCGA GTGCGATGACCAGAGCGACGAGGAGGGCTGCCCCGTGTGCTCCGCCGCCCAGTTCCCCTGCGCGCGGGGTCAGTGTGTGGACCTGCGCCT GCGCTGCGACGGCGAGGCAGACTGTCAGGACCGCTCAGACGAGGCGGACTGTGACGCCATCTGCCTGCCCAACCAGTTCCGGTGTGCGAG CGGCCAGTGTGTCCTCATCAAACAGCAGTGCGACTCCTTCCCCGACTGTATCGACGGCTCCGACGAGCTCATGTGTGAAATCACCAAGCC GCCCTCAGACGACAGCCCGGCCCACAGCAGTGCCATCGGGCCCGTCATTGGCATCATCCTCTCTCTCTTCGTCATGGGTGGTGTCTATTT TGTGTGCCAGCGCGTGGTGTGCCAGCGCTATGCGGGGGCCAACGGGCCCTTCCCGCACGAGTATGTCAGCGGGACCCCGCACGTGCCCCT CAATTTCATAGCCCCGGGCGGTTCCCAGCATGGCCCCTTCACAGGCATCGCATGCGGAAAGTCCATGATGAGCTCCGTGAGCCTGATGGG GGGCCGGGGCGGGGTGCCCCTCTACGACCGGAACCACGTCACAGGGGCCTCGTCCAGCAGCTCGTCCAGCACGAAGGCCACGCTGTACCC GCCGATCCTGAACCCGCCGCCCTCCCCGGCCACGGACCCCTCCCTGTACAACATGGACATGTTCTACTCTTCAAACATTCCGGCCACTGC GAGACCGTACAGCAACCGTTCCTTCAGAGTTGATACAGCGAAGCTCCAGCACAGGTACCAGCAACTGCAGCGTCTTGTCCACCCAGATTT CTTCAGCCAGAGGTCTCAGACTGAAAAGGACTTCTCAGAGAAGCATTCGACCCTGGTGAATGATGCCTATAAGACCCTCCTGGCCCCCCT GAGCAGAGGACTGTACCTTCTAAAGCTCCATGGAATAGAGATTCCTGAAAGGACAGATTATGAAATGGACAGGCAATTCCTCATAGAAAT AATGGAAATCAATGAAAAACTCGCAGAAGCTGAAAGTGAAGCTGCCATGAAAGAGATTGAATCCATTGTCAAAGCTAAACAGAAAGAATT TACTGACAATGTGAGCAGTGCTTTTGAACAAGATGACTTTGAAGAAGCCAAGGAAATTTTGACAAAGATGAGATACTTTTCAAATATAGA AGAAAAGATCAAGTTAAAGAAGATTCCCCTTTAATTGTGGATAGTTTAAAGTTTAAAAAATAAAGTTCTTGCTGGGCACAGTGGCTCACA CCTGTAATCCCAGCACTTTGGGAGGCTGAGGTGGGTGGATGACAAGGTCAGGAGTTCAAGACCAGCTTGGCCAACATAGTGAAACCCCGT CTCTGCTGAAAATACAAAAATTAGCCGGGCATGGTGGCGCGTGCCTGTAATCCCAGCTACTTGGTAGGCCGAGGCAGGAGAATCGCTTAA ACCCGTGAGGTGGAGGTTGCAGTGAGCAGAGATCACGCAACTGCACTCCAGCTTGGGCAACAGAGTGAGACTTAATCTTGAAAAATAAAT >49797_49797_1_LRP5-HSCB_LRP5_chr11_68214001_ENST00000294304_HSCB_chr22_29139870_ENST00000216027_length(amino acids)=1685AA_BP=1526 MEAAPPGPPWPLLLLLLLLLALCGCPAPAAASPLLLFANRRDVRLVDAGGVKLESTIVVSGLEDAAAVDFQFSKGAVYWTDVSEEAIKQT YLNQTGAAVQNVVISGLVSPDGLACDWVGKKLYWTDSETNRIEVANLNGTSRKVLFWQDLDQPRAIALDPAHGYMYWTDWGETPRIERAG MDGSTRKIIVDSDIYWPNGLTIDLEEQKLYWADAKLSFIHRANLDGSFRQKVVEGSLTHPFALTLSGDTLYWTDWQTRSIHACNKRTGGK RKEILSALYSPMDIQVLSQERQPFFHTRCEEDNGGCSHLCLLSPSEPFYTCACPTGVQLQDNGRTCKAGAEEVLLLARRTDLRRISLDTP DFTDIVLQVDDIRHAIAIDYDPLEGYVYWTDDEVRAIRRAYLDGSGAQTLVNTEINDPDGIAVDWVARNLYWTDTGTDRIEVTRLNGTSR KILVSEDLDEPRAIALHPVMGLMYWTDWGENPKIECANLDGQERRVLVNASLGWPNGLALDLQEGKLYWGDAKTDKIEVINVDGTKRRTL LEDKLPHIFGFTLLGDFIYWTDWQRRSIERVHKVKASRDVIIDQLPDLMGLKAVNVAKVVGTNPCADRNGGCSHLCFFTPHATRCGCPIG LELLSDMKTCIVPEAFLVFTSRAAIHRISLETNNNDVAIPLTGVKEASALDFDVSNNHIYWTDVSLKTISRAFMNGSSVEHVVEFGLDYP EGMAVDWMGKNLYWADTGTNRIEVARLDGQFRQVLVWRDLDNPRSLALDPTKGYIYWTEWGGKPRIVRAFMDGTNCMTLVDKVGRANDLT IDYADQRLYWTDLDTNMIESSNMLGQERVVIADDLPHPFGLTQYSDYIYWTDWNLHSIERADKTSGRNRTLIQGHLDFVMDILVFHSSRQ DGLNDCMHNNGQCGQLCLAIPGGHRCGCASHYTLDPSSRNCSPPTTFLLFSQKSAISRMIPDDQHSPDLILPLHGLRNVKAIDYDPLDKF IYWVDGRQNIKRAKDDGTQPFVLTSLSQGQNPDRQPHDLSIDIYSRTLFWTCEATNTINVHRLSGEAMGVVLRGDRDKPRAIVVNAERGY LYFTNMQDRAAKIERAALDGTEREVLFTTGLIRPVALVVDNTLGKLFWVDADLKRIESCDLSGANRLTLEDANIVQPLGLTILGKHLYWI DRQQQMIERVEKTTGDKRTRIQGRVAHLTGIHAVEEVSLEEFSAHPCARDNGGCSHICIAKGDGTPRCSCPVHLVLLQNLLTCGEPPTCS PDQFACATGEIDCIPGAWRCDGFPECDDQSDEEGCPVCSAAQFPCARGQCVDLRLRCDGEADCQDRSDEADCDAICLPNQFRCASGQCVL IKQQCDSFPDCIDGSDELMCEITKPPSDDSPAHSSAIGPVIGIILSLFVMGGVYFVCQRVVCQRYAGANGPFPHEYVSGTPHVPLNFIAP GGSQHGPFTGIACGKSMMSSVSLMGGRGGVPLYDRNHVTGASSSSSSSTKATLYPPILNPPPSPATDPSLYNMDMFYSSNIPATARPYSN RSFRVDTAKLQHRYQQLQRLVHPDFFSQRSQTEKDFSEKHSTLVNDAYKTLLAPLSRGLYLLKLHGIEIPERTDYEMDRQFLIEIMEINE -------------------------------------------------------------- >49797_49797_2_LRP5-HSCB_LRP5_chr11_68214001_ENST00000294304_HSCB_chr22_29139870_ENST00000398941_length(transcript)=5370nt_BP=4692nt GCCCGAGGGGGGAGGCGGAGGCGCCGGGAGCCGCGCGAGGAGCCGCCGCCGCCGCGCCATGGAGCCCGAGTGAGCGCGGCGCGGGCCCGT CCGGCCGCCGGACAACATGGAGGCAGCGCCGCCCGGGCCGCCGTGGCCGCTGCTGCTGCTGCTGCTGCTGCTGCTGGCGCTGTGCGGCTG CCCGGCCCCCGCCGCGGCCTCGCCGCTCCTGCTATTTGCCAACCGCCGGGACGTACGGCTGGTGGACGCCGGCGGAGTCAAGCTGGAGTC CACCATCGTGGTCAGCGGCCTGGAGGATGCGGCCGCAGTGGACTTCCAGTTTTCCAAGGGAGCCGTGTACTGGACAGACGTGAGCGAGGA GGCCATCAAGCAGACCTACCTGAACCAGACGGGGGCCGCCGTGCAGAACGTGGTCATCTCCGGCCTGGTCTCTCCCGACGGCCTCGCCTG CGACTGGGTGGGCAAGAAGCTGTACTGGACGGACTCAGAGACCAACCGCATCGAGGTGGCCAACCTCAATGGCACATCCCGGAAGGTGCT CTTCTGGCAGGACCTTGACCAGCCGAGGGCCATCGCCTTGGACCCCGCTCACGGGTACATGTACTGGACAGACTGGGGTGAGACGCCCCG GATTGAGCGGGCAGGGATGGATGGCAGCACCCGGAAGATCATTGTGGACTCGGACATTTACTGGCCCAATGGACTGACCATCGACCTGGA GGAGCAGAAGCTCTACTGGGCTGACGCCAAGCTCAGCTTCATCCACCGTGCCAACCTGGACGGCTCGTTCCGGCAGAAGGTGGTGGAGGG CAGCCTGACGCACCCCTTCGCCCTGACGCTCTCCGGGGACACTCTGTACTGGACAGACTGGCAGACCCGCTCCATCCATGCCTGCAACAA GCGCACTGGGGGGAAGAGGAAGGAGATCCTGAGTGCCCTCTACTCACCCATGGACATCCAGGTGCTGAGCCAGGAGCGGCAGCCTTTCTT CCACACTCGCTGTGAGGAGGACAATGGCGGCTGCTCCCACCTGTGCCTGCTGTCCCCAAGCGAGCCTTTCTACACATGCGCCTGCCCCAC GGGTGTGCAGCTGCAGGACAACGGCAGGACGTGTAAGGCAGGAGCCGAGGAGGTGCTGCTGCTGGCCCGGCGGACGGACCTACGGAGGAT CTCGCTGGACACGCCGGACTTCACCGACATCGTGCTGCAGGTGGACGACATCCGGCACGCCATTGCCATCGACTACGACCCGCTAGAGGG CTATGTCTACTGGACAGATGACGAGGTGCGGGCCATCCGCAGGGCGTACCTGGACGGGTCTGGGGCGCAGACGCTGGTCAACACCGAGAT CAACGACCCCGATGGCATCGCGGTCGACTGGGTGGCCCGAAACCTCTACTGGACCGACACGGGCACGGACCGCATCGAGGTGACGCGCCT CAACGGCACCTCCCGCAAGATCCTGGTGTCGGAGGACCTGGACGAGCCCCGAGCCATCGCACTGCACCCCGTGATGGGCCTCATGTACTG GACAGACTGGGGAGAGAACCCTAAAATCGAGTGTGCCAACTTGGATGGGCAGGAGCGGCGTGTGCTGGTCAATGCCTCCCTCGGGTGGCC CAACGGCCTGGCCCTGGACCTGCAGGAGGGGAAGCTCTACTGGGGAGACGCCAAGACAGACAAGATCGAGGTGATCAATGTTGATGGGAC GAAGAGGCGGACCCTCCTGGAGGACAAGCTCCCGCACATTTTTGGGTTCACGCTGCTGGGGGACTTCATCTACTGGACTGACTGGCAGCG CCGCAGCATCGAGCGGGTGCACAAGGTCAAGGCCAGCCGGGACGTCATCATTGACCAGCTGCCCGACCTGATGGGGCTCAAAGCTGTGAA TGTGGCCAAGGTCGTCGGAACCAACCCGTGTGCGGACAGGAACGGGGGGTGCAGCCACCTGTGCTTCTTCACACCCCACGCAACCCGGTG TGGCTGCCCCATCGGCCTGGAGCTGCTGAGTGACATGAAGACCTGCATCGTGCCTGAGGCCTTCTTGGTCTTCACCAGCAGAGCCGCCAT CCACAGGATCTCCCTCGAGACCAATAACAACGACGTGGCCATCCCGCTCACGGGCGTCAAGGAGGCCTCAGCCCTGGACTTTGATGTGTC CAACAACCACATCTACTGGACAGACGTCAGCCTGAAGACCATCAGCCGCGCCTTCATGAACGGGAGCTCGGTGGAGCACGTGGTGGAGTT TGGCCTTGACTACCCCGAGGGCATGGCCGTTGACTGGATGGGCAAGAACCTCTACTGGGCCGACACTGGGACCAACAGAATCGAAGTGGC GCGGCTGGACGGGCAGTTCCGGCAAGTCCTCGTGTGGAGGGACTTGGACAACCCGAGGTCGCTGGCCCTGGATCCCACCAAGGGCTACAT CTACTGGACCGAGTGGGGCGGCAAGCCGAGGATCGTGCGGGCCTTCATGGACGGGACCAACTGCATGACGCTGGTGGACAAGGTGGGCCG GGCCAACGACCTCACCATTGACTACGCTGACCAGCGCCTCTACTGGACCGACCTGGACACCAACATGATCGAGTCGTCCAACATGCTGGG TCAGGAGCGGGTCGTGATTGCCGACGATCTCCCGCACCCGTTCGGTCTGACGCAGTACAGCGATTATATCTACTGGACAGACTGGAATCT GCACAGCATTGAGCGGGCCGACAAGACTAGCGGCCGGAACCGCACCCTCATCCAGGGCCACCTGGACTTCGTGATGGACATCCTGGTGTT CCACTCCTCCCGCCAGGATGGCCTCAATGACTGTATGCACAACAACGGGCAGTGTGGGCAGCTGTGCCTTGCCATCCCCGGCGGCCACCG CTGCGGCTGCGCCTCACACTACACCCTGGACCCCAGCAGCCGCAACTGCAGCCCGCCCACCACCTTCTTGCTGTTCAGCCAGAAATCTGC CATCAGTCGGATGATCCCGGACGACCAGCACAGCCCGGATCTCATCCTGCCCCTGCATGGACTGAGGAACGTCAAAGCCATCGACTATGA CCCACTGGACAAGTTCATCTACTGGGTGGATGGGCGCCAGAACATCAAGCGAGCCAAGGACGACGGGACCCAGCCCTTTGTTTTGACCTC TCTGAGCCAAGGCCAAAACCCAGACAGGCAGCCCCACGACCTCAGCATCGACATCTACAGCCGGACACTGTTCTGGACGTGCGAGGCCAC CAATACCATCAACGTCCACAGGCTGAGCGGGGAAGCCATGGGGGTGGTGCTGCGTGGGGACCGCGACAAGCCCAGGGCCATCGTCGTCAA CGCGGAGCGAGGGTACCTGTACTTCACCAACATGCAGGACCGGGCAGCCAAGATCGAACGCGCAGCCCTGGACGGCACCGAGCGCGAGGT CCTCTTCACCACCGGCCTCATCCGCCCTGTGGCCCTGGTGGTGGACAACACACTGGGCAAGCTGTTCTGGGTGGACGCGGACCTGAAGCG CATTGAGAGCTGTGACCTGTCAGGGGCCAACCGCCTGACCCTGGAGGACGCCAACATCGTGCAGCCTCTGGGCCTGACCATCCTTGGCAA GCATCTCTACTGGATCGACCGCCAGCAGCAGATGATCGAGCGTGTGGAGAAGACCACCGGGGACAAGCGGACTCGCATCCAGGGCCGTGT CGCCCACCTCACTGGCATCCATGCAGTGGAGGAAGTCAGCCTGGAGGAGTTCTCAGCCCACCCATGTGCCCGTGACAATGGTGGCTGCTC CCACATCTGTATTGCCAAGGGTGATGGGACACCACGGTGCTCATGCCCAGTCCACCTCGTGCTCCTGCAGAACCTGCTGACCTGTGGAGA GCCGCCCACCTGCTCCCCGGACCAGTTTGCATGTGCCACAGGGGAGATCGACTGTATCCCCGGGGCCTGGCGCTGTGACGGCTTTCCCGA GTGCGATGACCAGAGCGACGAGGAGGGCTGCCCCGTGTGCTCCGCCGCCCAGTTCCCCTGCGCGCGGGGTCAGTGTGTGGACCTGCGCCT GCGCTGCGACGGCGAGGCAGACTGTCAGGACCGCTCAGACGAGGCGGACTGTGACGCCATCTGCCTGCCCAACCAGTTCCGGTGTGCGAG CGGCCAGTGTGTCCTCATCAAACAGCAGTGCGACTCCTTCCCCGACTGTATCGACGGCTCCGACGAGCTCATGTGTGAAATCACCAAGCC GCCCTCAGACGACAGCCCGGCCCACAGCAGTGCCATCGGGCCCGTCATTGGCATCATCCTCTCTCTCTTCGTCATGGGTGGTGTCTATTT TGTGTGCCAGCGCGTGGTGTGCCAGCGCTATGCGGGGGCCAACGGGCCCTTCCCGCACGAGTATGTCAGCGGGACCCCGCACGTGCCCCT CAATTTCATAGCCCCGGGCGGTTCCCAGCATGGCCCCTTCACAGGCATCGCATGCGGAAAGTCCATGATGAGCTCCGTGAGCCTGATGGG GGGCCGGGGCGGGGTGCCCCTCTACGACCGGAACCACGTCACAGGGGCCTCGTCCAGCAGCTCGTCCAGCACGAAGGCCACGCTGTACCC GCCGATCCTGAACCCGCCGCCCTCCCCGGCCACGGACCCCTCCCTGTACAACATGGACATGTTCTACTCTTCAAACATTCCGGCCACTGC GAGACCGTACAGCAACCGTTCCTTCAGAGTTGATACAGCGAAGCTCCAGCACAGGTACCAGCAACTGCAGCGTCTTGTCCACCCAGATTT CTTCAGCCAGAGGTCTCAGACTGAAAAGGACTTCTCAGAGAAGCATTCGACCCTGGTGAATGATGCCTATAAGACCCTCCTGGCCCCCCT GAGCAGAGGACTGTACCTTGTAAGCTAAACAGAAAGAATTTACTGACAATGTGAGCAGTGCTTTTGAACAAGATGACTTTGAAGAAGCCA AGGAAATTTTGACAAAGATGAGATACTTTTCAAATATAGAAGAAAAGATCAAGTTAAAGAAGATTCCCCTTTAATTGTGGATAGTTTAAA GTTTAAAAAATAAAGTTCTTGCTGGGCACAGTGGCTCACACCTGTAATCCCAGCACTTTGGGAGGCTGAGGTGGGTGGATGACAAGGTCA GGAGTTCAAGACCAGCTTGGCCAACATAGTGAAACCCCGTCTCTGCTGAAAATACAAAAATTAGCCGGGCATGGTGGCGCGTGCCTGTAA TCCCAGCTACTTGGTAGGCCGAGGCAGGAGAATCGCTTAAACCCGTGAGGTGGAGGTTGCAGTGAGCAGAGATCACGCAACTGCACTCCA >49797_49797_2_LRP5-HSCB_LRP5_chr11_68214001_ENST00000294304_HSCB_chr22_29139870_ENST00000398941_length(amino acids)=1593AA_BP=1526 MEAAPPGPPWPLLLLLLLLLALCGCPAPAAASPLLLFANRRDVRLVDAGGVKLESTIVVSGLEDAAAVDFQFSKGAVYWTDVSEEAIKQT YLNQTGAAVQNVVISGLVSPDGLACDWVGKKLYWTDSETNRIEVANLNGTSRKVLFWQDLDQPRAIALDPAHGYMYWTDWGETPRIERAG MDGSTRKIIVDSDIYWPNGLTIDLEEQKLYWADAKLSFIHRANLDGSFRQKVVEGSLTHPFALTLSGDTLYWTDWQTRSIHACNKRTGGK RKEILSALYSPMDIQVLSQERQPFFHTRCEEDNGGCSHLCLLSPSEPFYTCACPTGVQLQDNGRTCKAGAEEVLLLARRTDLRRISLDTP DFTDIVLQVDDIRHAIAIDYDPLEGYVYWTDDEVRAIRRAYLDGSGAQTLVNTEINDPDGIAVDWVARNLYWTDTGTDRIEVTRLNGTSR KILVSEDLDEPRAIALHPVMGLMYWTDWGENPKIECANLDGQERRVLVNASLGWPNGLALDLQEGKLYWGDAKTDKIEVINVDGTKRRTL LEDKLPHIFGFTLLGDFIYWTDWQRRSIERVHKVKASRDVIIDQLPDLMGLKAVNVAKVVGTNPCADRNGGCSHLCFFTPHATRCGCPIG LELLSDMKTCIVPEAFLVFTSRAAIHRISLETNNNDVAIPLTGVKEASALDFDVSNNHIYWTDVSLKTISRAFMNGSSVEHVVEFGLDYP EGMAVDWMGKNLYWADTGTNRIEVARLDGQFRQVLVWRDLDNPRSLALDPTKGYIYWTEWGGKPRIVRAFMDGTNCMTLVDKVGRANDLT IDYADQRLYWTDLDTNMIESSNMLGQERVVIADDLPHPFGLTQYSDYIYWTDWNLHSIERADKTSGRNRTLIQGHLDFVMDILVFHSSRQ DGLNDCMHNNGQCGQLCLAIPGGHRCGCASHYTLDPSSRNCSPPTTFLLFSQKSAISRMIPDDQHSPDLILPLHGLRNVKAIDYDPLDKF IYWVDGRQNIKRAKDDGTQPFVLTSLSQGQNPDRQPHDLSIDIYSRTLFWTCEATNTINVHRLSGEAMGVVLRGDRDKPRAIVVNAERGY LYFTNMQDRAAKIERAALDGTEREVLFTTGLIRPVALVVDNTLGKLFWVDADLKRIESCDLSGANRLTLEDANIVQPLGLTILGKHLYWI DRQQQMIERVEKTTGDKRTRIQGRVAHLTGIHAVEEVSLEEFSAHPCARDNGGCSHICIAKGDGTPRCSCPVHLVLLQNLLTCGEPPTCS PDQFACATGEIDCIPGAWRCDGFPECDDQSDEEGCPVCSAAQFPCARGQCVDLRLRCDGEADCQDRSDEADCDAICLPNQFRCASGQCVL IKQQCDSFPDCIDGSDELMCEITKPPSDDSPAHSSAIGPVIGIILSLFVMGGVYFVCQRVVCQRYAGANGPFPHEYVSGTPHVPLNFIAP GGSQHGPFTGIACGKSMMSSVSLMGGRGGVPLYDRNHVTGASSSSSSSTKATLYPPILNPPPSPATDPSLYNMDMFYSSNIPATARPYSN -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for LRP5-HSCB |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for LRP5-HSCB |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for LRP5-HSCB |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
