
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:LRP5-LRP4 (FusionGDB2 ID:49803) |
Fusion Gene Summary for LRP5-LRP4 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: LRP5-LRP4 | Fusion gene ID: 49803 | Hgene | Tgene | Gene symbol | LRP5 | LRP4 | Gene ID | 4041 | 4038 |
| Gene name | LDL receptor related protein 5 | LDL receptor related protein 4 | |
| Synonyms | BMND1|EVR1|EVR4|HBM|LR3|LRP-5|LRP-7|LRP7|OPPG|OPS|OPTA1|PCLD4|VBCH2 | CLSS|CMS17|LRP-4|LRP10|MEGF7|SOST2 | |
| Cytomap | 11q13.2 | 11p11.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | low-density lipoprotein receptor-related protein 5low density lipoprotein receptor-related protein 5low density lipoprotein receptor-related protein 7 | low-density lipoprotein receptor-related protein 4multiple epidermal growth factor-like domains 7 | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | O75197 | O75096 | |
| Ensembl transtripts involved in fusion gene | ENST00000294304, ENST00000529481, | ENST00000378623, | |
| Fusion gene scores | * DoF score | 36 X 25 X 12=10800 | 5 X 6 X 6=180 |
| # samples | 41 | 6 | |
| ** MAII score | log2(41/10800*10)=-4.71926359243275 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(6/180*10)=-1.58496250072116 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: LRP5 [Title/Abstract] AND LRP4 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | LRP5(68115711)-LRP4(46897517), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | LRP5 | GO:0008284 | positive regulation of cell proliferation | 9790987 |
| Hgene | LRP5 | GO:0045840 | positive regulation of mitotic nuclear division | 9790987 |
| Hgene | LRP5 | GO:0045893 | positive regulation of transcription, DNA-templated | 15035989|17955262 |
| Hgene | LRP5 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 12857724 |
| Hgene | LRP5 | GO:0060070 | canonical Wnt signaling pathway | 11029007|12121999|12857724|15908424|24706814|25920554 |
| Tgene | LRP4 | GO:0001822 | kidney development | 20381006 |
| Tgene | LRP4 | GO:0060173 | limb development | 20381006 |
| Tgene | LRP4 | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 20093106 |
 Fusion gene breakpoints across LRP5 (5'-gene) Fusion gene breakpoints across LRP5 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
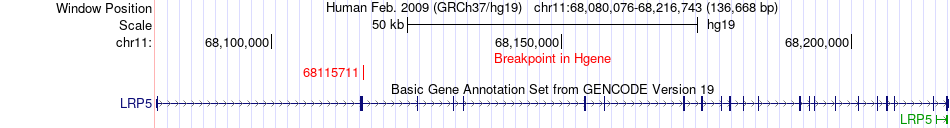 |
 Fusion gene breakpoints across LRP4 (3'-gene) Fusion gene breakpoints across LRP4 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
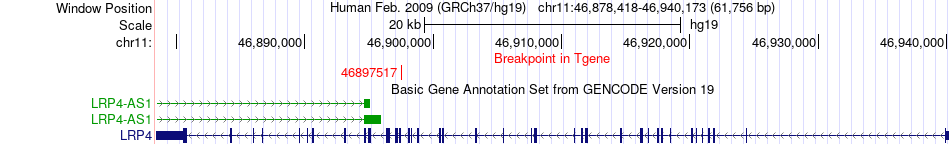 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | Non-Cancer | 61N | LRP5 | chr11 | 68115711 | + | LRP4 | chr11 | 46897517 | - |
Top |
Fusion Gene ORF analysis for LRP5-LRP4 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000294304 | ENST00000378623 | LRP5 | chr11 | 68115711 | + | LRP4 | chr11 | 46897517 | - |
| intron-3CDS | ENST00000529481 | ENST00000378623 | LRP5 | chr11 | 68115711 | + | LRP4 | chr11 | 46897517 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000294304 | LRP5 | chr11 | 68115711 | + | ENST00000378623 | LRP4 | chr11 | 46897517 | - | 4891 | 594 | 106 | 2775 | 889 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000294304 | ENST00000378623 | LRP5 | chr11 | 68115711 | + | LRP4 | chr11 | 46897517 | - | 0.000974972 | 0.999025 |
Top |
Fusion Genomic Features for LRP5-LRP4 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
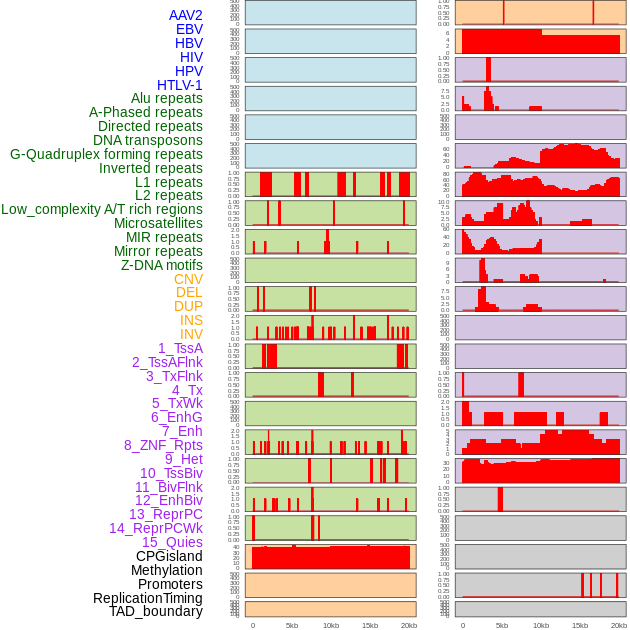 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for LRP5-LRP4 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr11:68115711/chr11:46897517) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| LRP5 | LRP4 |
| FUNCTION: Acts as a coreceptor with members of the frizzled family of seven-transmembrane spanning receptors to transduce signal by Wnt proteins (PubMed:11336703, PubMed:11448771, PubMed:15778503, PubMed:11719191, PubMed:15908424, PubMed:16252235). Activates the canonical Wnt signaling pathway that controls cell fate determination and self-renewal during embryonic development and adult tissue regeneration (PubMed:11336703, PubMed:11719191). In particular, may play an important role in the development of the posterior patterning of the epiblast during gastrulation (By similarity). During bone development, regulates osteoblast proliferation and differentiation thus determining bone mass (PubMed:11719191). Mechanistically, the formation of the signaling complex between Wnt ligand, frizzled receptor and LRP5 coreceptor promotes the recruitment of AXIN1 to LRP5, stabilizing beta-catenin/CTNNB1 and activating TCF/LEF-mediated transcriptional programs (PubMed:11336703, PubMed:25920554, PubMed:24706814, PubMed:14731402). Acts as a coreceptor for non-Wnt proteins, such as norrin/NDP. Binding of norrin/NDP to frizzled 4/FZD4-LRP5 receptor complex triggers beta-catenin/CTNNB1-dependent signaling known to be required for retinal vascular development (PubMed:27228167, PubMed:16252235). Plays a role in controlling postnatal vascular regression in retina via macrophage-induced endothelial cell apoptosis (By similarity). {ECO:0000250|UniProtKB:Q91VN0, ECO:0000269|PubMed:11336703, ECO:0000269|PubMed:11448771, ECO:0000269|PubMed:11719191, ECO:0000269|PubMed:14731402, ECO:0000269|PubMed:15778503, ECO:0000269|PubMed:15908424, ECO:0000269|PubMed:16252235, ECO:0000269|PubMed:24706814, ECO:0000269|PubMed:25920554, ECO:0000269|PubMed:27228167}. | FUNCTION: Mediates SOST-dependent inhibition of bone formation. Functions as a specific facilitator of SOST-mediated inhibition of Wnt signaling. Plays a key role in the formation and the maintenance of the neuromuscular junction (NMJ), the synapse between motor neuron and skeletal muscle. Directly binds AGRIN and recruits it to the MUSK signaling complex. Mediates the AGRIN-induced phosphorylation of MUSK, the kinase of the complex. The activation of MUSK in myotubes induces the formation of NMJ by regulating different processes including the transcription of specific genes and the clustering of AChR in the postsynaptic membrane. Alternatively, may be involved in the negative regulation of the canonical Wnt signaling pathway, being able to antagonize the LRP6-mediated activation of this pathway. More generally, has been proposed to function as a cell surface endocytic receptor binding and internalizing extracellular ligands for degradation by lysosomes. May play an essential role in the process of digit differentiation (By similarity). {ECO:0000250|UniProtKB:Q8VI56, ECO:0000269|PubMed:20381006, ECO:0000269|PubMed:21471202}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 120_162 | 162 | 1616.0 | Repeat | Note=LDL-receptor class B 2 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 123_126 | 162 | 1616.0 | Repeat | Note=YWTD 2 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 75_119 | 162 | 1616.0 | Repeat | Note=LDL-receptor class B 1 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 78_81 | 162 | 1616.0 | Repeat | Note=YWTD 1 |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 1766_1769 | 1178 | 1906.0 | Motif | Endocytosis signal | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 1179_1222 | 1178 | 1906.0 | Repeat | Note=LDL-receptor class B 13 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 1223_1263 | 1178 | 1906.0 | Repeat | Note=LDL-receptor class B 14 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 1264_1306 | 1178 | 1906.0 | Repeat | Note=LDL-receptor class B 15 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 1397_1439 | 1178 | 1906.0 | Repeat | Note=LDL-receptor class B 16 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 1440_1482 | 1178 | 1906.0 | Repeat | Note=LDL-receptor class B 17 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 1483_1526 | 1178 | 1906.0 | Repeat | Note=LDL-receptor class B 18 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 1527_1568 | 1178 | 1906.0 | Repeat | Note=LDL-receptor class B 19 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 1569_1610 | 1178 | 1906.0 | Repeat | Note=LDL-receptor class B 20 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 1747_1905 | 1178 | 1906.0 | Topological domain | Cytoplasmic | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 1726_1746 | 1178 | 1906.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 1495_1610 | 162 | 1616.0 | Compositional bias | Note=Pro-rich |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 1213_1254 | 162 | 1616.0 | Domain | Note=EGF-like 4 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 1258_1296 | 162 | 1616.0 | Domain | LDL-receptor class A 1 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 1297_1333 | 162 | 1616.0 | Domain | LDL-receptor class A 2 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 1335_1371 | 162 | 1616.0 | Domain | LDL-receptor class A 3 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 295_337 | 162 | 1616.0 | Domain | Note=EGF-like 1 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 601_641 | 162 | 1616.0 | Domain | Note=EGF-like 2 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 902_942 | 162 | 1616.0 | Domain | Note=EGF-like 3 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 1500_1506 | 162 | 1616.0 | Motif | Note=PPPSP motif A |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 1538_1545 | 162 | 1616.0 | Motif | Note=PPPSP motif B |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 1574_1581 | 162 | 1616.0 | Motif | Note=PPPSP motif C |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 1591_1596 | 162 | 1616.0 | Motif | Note=PPPSP motif D |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 1605_1612 | 162 | 1616.0 | Motif | Note=PPPSP motif E |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 32_288 | 162 | 1616.0 | Region | Note=Beta-propeller 1 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 341_602 | 162 | 1616.0 | Region | Note=Beta-propeller 2 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 644_903 | 162 | 1616.0 | Region | Note=Beta-propeller 3 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 945_1212 | 162 | 1616.0 | Region | Note=Beta-propeller 4 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 1036_1078 | 162 | 1616.0 | Repeat | Note=LDL-receptor class B 17 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 1079_1123 | 162 | 1616.0 | Repeat | Note=LDL-receptor class B 18 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 1124_1164 | 162 | 1616.0 | Repeat | Note=LDL-receptor class B 19 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 1165_1207 | 162 | 1616.0 | Repeat | Note=LDL-receptor class B 20 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 163_206 | 162 | 1616.0 | Repeat | Note=LDL-receptor class B 3 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 166_169 | 162 | 1616.0 | Repeat | Note=YWTD 3 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 207_247 | 162 | 1616.0 | Repeat | Note=LDL-receptor class B 4 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 248_290 | 162 | 1616.0 | Repeat | Note=LDL-receptor class B 5 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 251_254 | 162 | 1616.0 | Repeat | Note=YWTD 4 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 385_427 | 162 | 1616.0 | Repeat | Note=LDL-receptor class B 6 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 388_391 | 162 | 1616.0 | Repeat | Note=YWTD 5 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 428_470 | 162 | 1616.0 | Repeat | Note=LDL-receptor class B 7 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 431_434 | 162 | 1616.0 | Repeat | Note=YWTD 6 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 471_514 | 162 | 1616.0 | Repeat | Note=LDL-receptor class B 8 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 474_477 | 162 | 1616.0 | Repeat | Note=YWTD 7 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 515_557 | 162 | 1616.0 | Repeat | Note=LDL-receptor class B 9 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 558_600 | 162 | 1616.0 | Repeat | Note=LDL-receptor class B 10 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 559_562 | 162 | 1616.0 | Repeat | Note=YWTD 8 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 687_729 | 162 | 1616.0 | Repeat | Note=LDL-receptor class B 11 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 690_693 | 162 | 1616.0 | Repeat | Note=YWTD 9 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 730_772 | 162 | 1616.0 | Repeat | Note=LDL-receptor class B 12 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 773_815 | 162 | 1616.0 | Repeat | Note=LDL-receptor class B 13 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 816_855 | 162 | 1616.0 | Repeat | Note=LDL-receptor class B 14 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 819_822 | 162 | 1616.0 | Repeat | Note=YWTD 10 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 856_898 | 162 | 1616.0 | Repeat | Note=LDL-receptor class B 15 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 859_862 | 162 | 1616.0 | Repeat | Note=YWTD 11 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 989_1035 | 162 | 1616.0 | Repeat | Note=LDL-receptor class B 16 |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 1408_1615 | 162 | 1616.0 | Topological domain | Cytoplasmic |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 32_1384 | 162 | 1616.0 | Topological domain | Extracellular |
| Hgene | LRP5 | chr11:68115711 | chr11:46897517 | ENST00000294304 | + | 2 | 23 | 1385_1407 | 162 | 1616.0 | Transmembrane | Helical |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 109_144 | 1178 | 1906.0 | Domain | LDL-receptor class A 3 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 147_183 | 1178 | 1906.0 | Domain | LDL-receptor class A 4 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 190_226 | 1178 | 1906.0 | Domain | LDL-receptor class A 5 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 230_266 | 1178 | 1906.0 | Domain | LDL-receptor class A 6 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 269_305 | 1178 | 1906.0 | Domain | LDL-receptor class A 7 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 26_67 | 1178 | 1906.0 | Domain | LDL-receptor class A 1 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 311_350 | 1178 | 1906.0 | Domain | LDL-receptor class A 8 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 354_394 | 1178 | 1906.0 | Domain | EGF-like 1%3B calcium-binding | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 395_434 | 1178 | 1906.0 | Domain | EGF-like 2%3B calcium-binding | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 698_737 | 1178 | 1906.0 | Domain | Note=EGF-like 3 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 70_106 | 1178 | 1906.0 | Domain | LDL-receptor class A 2 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 1093_1135 | 1178 | 1906.0 | Repeat | Note=LDL-receptor class B 11 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 1136_1178 | 1178 | 1906.0 | Repeat | Note=LDL-receptor class B 12 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 480_522 | 1178 | 1906.0 | Repeat | Note=LDL-receptor class B 1 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 523_565 | 1178 | 1906.0 | Repeat | Note=LDL-receptor class B 2 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 566_609 | 1178 | 1906.0 | Repeat | Note=LDL-receptor class B 3 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 610_652 | 1178 | 1906.0 | Repeat | Note=LDL-receptor class B 4 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 653_693 | 1178 | 1906.0 | Repeat | Note=LDL-receptor class B 5 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 785_827 | 1178 | 1906.0 | Repeat | Note=LDL-receptor class B 6 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 828_870 | 1178 | 1906.0 | Repeat | Note=LDL-receptor class B 7 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 871_914 | 1178 | 1906.0 | Repeat | Note=LDL-receptor class B 8 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 915_956 | 1178 | 1906.0 | Repeat | Note=LDL-receptor class B 9 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 957_998 | 1178 | 1906.0 | Repeat | Note=LDL-receptor class B 10 | |
| Tgene | LRP4 | chr11:68115711 | chr11:46897517 | ENST00000378623 | 24 | 38 | 21_1725 | 1178 | 1906.0 | Topological domain | Extracellular |
Top |
Fusion Gene Sequence for LRP5-LRP4 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >49803_49803_1_LRP5-LRP4_LRP5_chr11_68115711_ENST00000294304_LRP4_chr11_46897517_ENST00000378623_length(transcript)=4891nt_BP=594nt GCCCGAGGGGGGAGGCGGAGGCGCCGGGAGCCGCGCGAGGAGCCGCCGCCGCCGCGCCATGGAGCCCGAGTGAGCGCGGCGCGGGCCCGT CCGGCCGCCGGACAACATGGAGGCAGCGCCGCCCGGGCCGCCGTGGCCGCTGCTGCTGCTGCTGCTGCTGCTGCTGGCGCTGTGCGGCTG CCCGGCCCCCGCCGCGGCCTCGCCGCTCCTGCTATTTGCCAACCGCCGGGACGTACGGCTGGTGGACGCCGGCGGAGTCAAGCTGGAGTC CACCATCGTGGTCAGCGGCCTGGAGGATGCGGCCGCAGTGGACTTCCAGTTTTCCAAGGGAGCCGTGTACTGGACAGACGTGAGCGAGGA GGCCATCAAGCAGACCTACCTGAACCAGACGGGGGCCGCCGTGCAGAACGTGGTCATCTCCGGCCTGGTCTCTCCCGACGGCCTCGCCTG CGACTGGGTGGGCAAGAAGCTGTACTGGACGGACTCAGAGACCAACCGCATCGAGGTGGCCAACCTCAATGGCACATCCCGGAAGGTGCT CTTCTGGCAGGACCTTGACCAGCCGAGGGCCATCGCCTTGGACCCCGCTCACGGGTTTATGTACTGGACAGACTGGGGGGAGAATGCCAA GTTAGAGCGGTCCGGAATGGATGGCTCAGACCGCGCGGTGCTCATCAACAACAACCTAGGATGGCCCAATGGACTGACTGTGGACAAGGC CAGCTCCCAACTGCTATGGGCCGATGCCCACACCGAGCGAATTGAGGCTGCTGACCTGAATGGTGCCAATCGGCATACATTGGTGTCACC GGTGCAGCACCCATATGGCCTCACCCTGCTCGACTCCTATATCTACTGGACTGACTGGCAGACTCGGAGCATCCACCGTGCTGACAAGGG TACTGGCAGCAATGTCATCCTCGTGAGGTCCAACCTGCCAGGCCTCATGGACATGCAGGCTGTGGACCGGGCACAGCCACTAGGTTTTAA CAAGTGCGGCTCGAGAAATGGCGGCTGCTCCCACCTCTGCTTGCCTCGGCCTTCTGGCTTCTCCTGTGCCTGCCCCACTGGCATCCAGCT GAAGGGAGATGGGAAGACCTGTGATCCCTCTCCTGAGACCTACCTGCTCTTCTCCAGCCGTGGCTCCATCCGGCGTATCTCACTGGACAC CAGTGACCACACCGATGTGCATGTCCCTGTTCCTGAGCTCAACAATGTCATCTCCCTGGACTATGACAGCGTGGATGGAAAGGTCTATTA CACAGATGTGTTCCTGGATGTTATCAGGCGAGCAGACCTGAACGGCAGCAACATGGAGACAGTGATCGGGCGAGGGCTGAAGACCACTGA CGGGCTGGCAGTGGACTGGGTGGCCAGGAACCTGTACTGGACAGACACAGGTCGAAATACCATTGAGGCGTCCAGGCTGGATGGTTCCTG CCGCAAAGTACTGATCAACAATAGCCTGGATGAGCCCCGGGCCATTGCTGTTTTCCCCAGGAAGGGGTACCTCTTCTGGACAGACTGGGG CCACATTGCCAAGATCGAACGGGCAAACTTGGATGGTTCTGAGCGGAAGGTCCTCATCAACACAGACCTGGGTTGGCCCAATGGCCTTAC CCTGGACTATGATACCCGCAGGATCTACTGGGTGGATGCGCATCTGGACCGGATCGAGAGTGCTGACCTCAATGGGAAACTGCGGCAGGT CTTGGTCAGCCATGTGTCCCACCCCTTTGCCCTCACACAGCAAGACAGGTGGATCTACTGGACAGACTGGCAGACCAAGTCAATCCAGCG TGTTGACAAATACTCAGGCCGGAACAAGGAGACAGTGCTGGCAAATGTGGAAGGACTCATGGATATCATCGTGGTTTCCCCTCAGCGGCA GACAGGGACCAATGCCTGTGGTGTGAACAATGGTGGCTGCACCCACCTCTGCTTTGCCAGAGCCTCGGACTTCGTATGTGCCTGTCCTGA CGAACCTGATAGCCGGCCCTGCTCCCTTGTGCCTGGCCTGGTACCACCAGCTCCTAGGGCTACTGGCATGAGTGAAAAGAGCCCAGTGCT ACCCAACACACCACCTACCACCTTGTATTCTTCAACCACCCGGACCCGCACGTCTCTGGAGGAGGTGGAAGGAAGATGCTCTGAAAGGGA TGCCAGGCTGGGCCTCTGTGCACGTTCCAATGACGCTGTTCCTGCTGCTCCAGGGGAAGGACTTCATATCAGCTACGCCATTGGTGGACT CCTCAGTATTCTGCTGATTTTGGTGGTGATTGCAGCTTTGATGCTGTACAGACACAAAAAATCCAAGTTCACTGATCCTGGAATGGGGAA CCTCACCTACAGCAACCCCTCCTACCGAACATCCACACAGGAAGTGAAGATTGAAGCAATCCCCAAACCAGCCATGTACAACCAGCTGTG CTATAAGAAAGAGGGAGGGCCTGACCATAACTACACCAAGGAGAAGATCAAGATCGTAGAGGGAATCTGCCTCCTGTCTGGGGATGATGC TGAGTGGGATGACCTCAAGCAACTGCGAAGCTCACGGGGGGGCCTCCTCCGGGATCATGTATGCATGAAGACAGACACGGTGTCCATCCA GGCCAGCTCTGGCTCCCTGGATGACACAGAGACGGAGCAGCTGTTACAGGAAGAGCAGTCTGAGTGTAGCAGCGTCCATACTGCAGCCAC TCCAGAAAGACGAGGCTCTCTGCCAGACACGGGCTGGAAACATGAACGCAAGCTCTCCTCAGAGAGCCAGGTCTAAATGCCCACATTCTC TTCCCTGCCTGCCTGTTCCTTCTCCTTTATGGACGTCTAGTCCTTGTGCTCGCTTACACCGCAGGCCCCGCTTCTGTGTGCTTGTCCTCC TCCTCCTCCCACCCCATAACTGTTCCTAAGCCTTCACCGGAGCTGTTTACCACGTGAGTCCATAACTACCTGTGCACAAGAAATGATGGC ACATCACGAGAATTTAGACCTGGATTTTACCATGAACCTCACATCTTGTACTCCATCCTGGGCCCCCTGAAACTGCTTATTCGTGATTCC TCACCAGCGTAGAGCTCCACCTCCCCTTTCCCCAGTACCCTCAGTGCCTGCTTCTCAGTGCTGATGCAGCTGATGACCCAGGACTGCGCT CTGCCCCATCACAGCCAGCATGACTGCTTCTCTGAGAGAACTTGCCCATCAGGGGCTGGGACATGGGGGTGTGGGTAAAGACAGGGATGA AGGATAGAGGCTGAGAGAAGAAGGAAGAATCAGCCCAGCAGGTATGGGCATCTGGGAAACCTCCAGCCTCAAGTGTGTTAGTAACATGAA AAAGCTTTGGGGGGTAGTTGGATCTGGGTGTCTGGTCCATTGCTGGCAGTGGACATTATTCTTGCCCTAAGAGACACTGCCTTTTCAGCA GCAGATACTGGTGAGATGGGGGTGGCTCAGGCTGTTCTTCCTCCTCCTAGAATGTCTGGAGCTGTTTCTACATTCAGATAACTGGGTCCC CTATCACAAGGCTACTGGCTAATAGGAATTCCCTCCTGGTGCCACCACTGGCCAGTACCTTTCCTAAGTCTTTGCTCAAATTAACCAGGT TGTGAGCCAGTGGCTTGAGTGAATGTTAGGCCTTGGGGGCTGAGTCTCTGAAAAGTCTAAGAAGCTCTGCCTAGACCAAATATGGTATAC CTCCTGACCCCTCTCTCCCTCATGTCCTGGGATTCTGGGGAAGAGACCTAGAAACAAGCTTTCAAAGAAAAACCAGAAGTTGTCATAAAT GGTCAGAAAGAACGATCAGGTTGGAGACTTGGGAAACCCAGGGCCTAAAGAGAAGTATCCATGAGGGTCAAACTTCCTGTTGAACTTCCT ATGTTCTTTCTCAAGTGCTCAGGGATCTAAGTTAGTGGACAGCAAGCCTGTGGCTACGGGGTGGTGATGTTCCTCTTCCAGCTGTCCCCT CAGCTAAGGGGCTTAGTTTCCATGTGGGATGCCATCACTTGGTTCATGCTCATTCACACAAAGGGCACGTGTCTCAGCCTGGTATCAGGG AAATTGAGACTTATTTTTGCCCTAAAACGTCTCCCTAGCTGTTCTTCGTGGGGTTTTTTTGTTTGTTTTTTTGCCTAATTTGCTTTTTCT GACCAAGCCTTGTGGCACCAGCAATCTCCAAAGTCCTGTGGTGGGAGGGCTGAATAAATAAAAATACAAAGAGGTGGGTAAGGAGTAGGA AGGTAGAGAGCACCACTGATGAGGCCCTCCTAGCCCATGGCAGACCCAGACCTCTTCTCCCCCAGGAATTAGAAGTGGCAGGAGAGAACA ACAGGGGCTGGGAATGGAGGGGAGAATTTCTAGGGGAAGTTTCCTGAGTTGAAACTTCTCCTGTGGTTACTGGTATTGAGAAATCAGCTA CCAAAGTGAAAAAGGACAAGATCAATTCTTTTCTAGTCAGTTCTAAGACTGCTAGAGAGAGATACCAGGCCCTTAGCCTTGCTCTCAGTA GCGTCAGCCCCAGTTCTGAGCCTCCCCACATTACACTTAACAAGCAGTAAAGGAGTGAGCACTTTGGGTCCTTAGACTCACGTCTGGGGA GGAAGAGCAAGTAGAAAAGTGGCATTTTCTTGATTGGAAAGGGGGAAGGATCTTATTGCACTTGGGCTGTTCAGAATGTAGAAAGGACAT ATTTGAGGAAGTATCTATTTGAGCACTGATTTACTCTGTAAAAAGCAAAATCTCTCTGTCCTAAACTAATGGAAGCGATTCTCCCATGCT CATGTGTAATGGTTTTAACGTTACTCACTGGAGAGATTGGACTTTCTGGAGTTATTTAACCACTATGTTCAGTATTTTAGGACTTTATGA >49803_49803_1_LRP5-LRP4_LRP5_chr11_68115711_ENST00000294304_LRP4_chr11_46897517_ENST00000378623_length(amino acids)=889AA_BP=105 MEAAPPGPPWPLLLLLLLLLALCGCPAPAAASPLLLFANRRDVRLVDAGGVKLESTIVVSGLEDAAAVDFQFSKGAVYWTDVSEEAIKQT YLNQTGAAVQNVVISGLVSPDGLACDWVGKKLYWTDSETNRIEVANLNGTSRKVLFWQDLDQPRAIALDPAHGFMYWTDWGENAKLERSG MDGSDRAVLINNNLGWPNGLTVDKASSQLLWADAHTERIEAADLNGANRHTLVSPVQHPYGLTLLDSYIYWTDWQTRSIHRADKGTGSNV ILVRSNLPGLMDMQAVDRAQPLGFNKCGSRNGGCSHLCLPRPSGFSCACPTGIQLKGDGKTCDPSPETYLLFSSRGSIRRISLDTSDHTD VHVPVPELNNVISLDYDSVDGKVYYTDVFLDVIRRADLNGSNMETVIGRGLKTTDGLAVDWVARNLYWTDTGRNTIEASRLDGSCRKVLI NNSLDEPRAIAVFPRKGYLFWTDWGHIAKIERANLDGSERKVLINTDLGWPNGLTLDYDTRRIYWVDAHLDRIESADLNGKLRQVLVSHV SHPFALTQQDRWIYWTDWQTKSIQRVDKYSGRNKETVLANVEGLMDIIVVSPQRQTGTNACGVNNGGCTHLCFARASDFVCACPDEPDSR PCSLVPGLVPPAPRATGMSEKSPVLPNTPPTTLYSSTTRTRTSLEEVEGRCSERDARLGLCARSNDAVPAAPGEGLHISYAIGGLLSILL ILVVIAALMLYRHKKSKFTDPGMGNLTYSNPSYRTSTQEVKIEAIPKPAMYNQLCYKKEGGPDHNYTKEKIKIVEGICLLSGDDAEWDDL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for LRP5-LRP4 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for LRP5-LRP4 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for LRP5-LRP4 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
