
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:LRRC7-RRP15 (FusionGDB2 ID:50039) |
Fusion Gene Summary for LRRC7-RRP15 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: LRRC7-RRP15 | Fusion gene ID: 50039 | Hgene | Tgene | Gene symbol | LRRC7 | RRP15 | Gene ID | 57554 | 51018 |
| Gene name | leucine rich repeat containing 7 | ribosomal RNA processing 15 homolog | |
| Synonyms | DENSIN | CGI-115|KIAA0507 | |
| Cytomap | 1p31.1 | 1q41 | |
| Type of gene | protein-coding | protein-coding | |
| Description | leucine-rich repeat-containing protein 7densin-180 | RRP15-like proteinribosomal RNA-processing protein 15 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q96NW7 | RRP15 | |
| Ensembl transtripts involved in fusion gene | ENST00000370958, ENST00000310961, ENST00000035383, ENST00000415775, ENST00000565615, | ENST00000491428, ENST00000366932, | |
| Fusion gene scores | * DoF score | 3 X 3 X 3=27 | 7 X 5 X 6=210 |
| # samples | 3 | 9 | |
| ** MAII score | log2(3/27*10)=0.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(9/210*10)=-1.22239242133645 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: LRRC7 [Title/Abstract] AND RRP15 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | LRRC7(70034324)-RRP15(218504290), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across LRRC7 (5'-gene) Fusion gene breakpoints across LRRC7 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
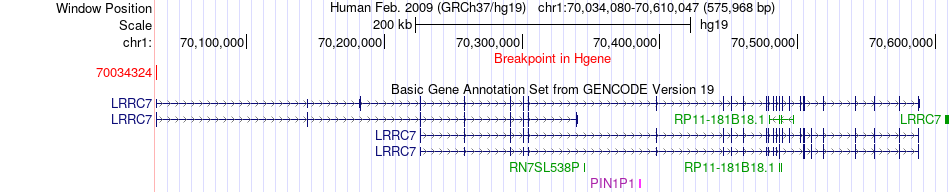 |
 Fusion gene breakpoints across RRP15 (3'-gene) Fusion gene breakpoints across RRP15 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
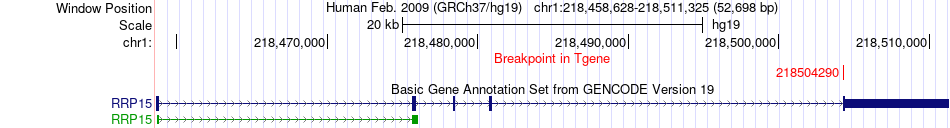 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-HC-8257-01A | LRRC7 | chr1 | 70034324 | - | RRP15 | chr1 | 218504290 | + |
| ChimerDB4 | PRAD | TCGA-HC-8257-01A | LRRC7 | chr1 | 70034324 | + | RRP15 | chr1 | 218504290 | + |
| ChimerDB4 | PRAD | TCGA-HC-8257 | LRRC7 | chr1 | 70034324 | + | RRP15 | chr1 | 218504290 | + |
Top |
Fusion Gene ORF analysis for LRRC7-RRP15 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000370958 | ENST00000491428 | LRRC7 | chr1 | 70034324 | + | RRP15 | chr1 | 218504290 | + |
| 5UTR-3CDS | ENST00000310961 | ENST00000366932 | LRRC7 | chr1 | 70034324 | + | RRP15 | chr1 | 218504290 | + |
| 5UTR-intron | ENST00000310961 | ENST00000491428 | LRRC7 | chr1 | 70034324 | + | RRP15 | chr1 | 218504290 | + |
| In-frame | ENST00000370958 | ENST00000366932 | LRRC7 | chr1 | 70034324 | + | RRP15 | chr1 | 218504290 | + |
| intron-3CDS | ENST00000035383 | ENST00000366932 | LRRC7 | chr1 | 70034324 | + | RRP15 | chr1 | 218504290 | + |
| intron-3CDS | ENST00000415775 | ENST00000366932 | LRRC7 | chr1 | 70034324 | + | RRP15 | chr1 | 218504290 | + |
| intron-3CDS | ENST00000565615 | ENST00000366932 | LRRC7 | chr1 | 70034324 | + | RRP15 | chr1 | 218504290 | + |
| intron-intron | ENST00000035383 | ENST00000491428 | LRRC7 | chr1 | 70034324 | + | RRP15 | chr1 | 218504290 | + |
| intron-intron | ENST00000415775 | ENST00000491428 | LRRC7 | chr1 | 70034324 | + | RRP15 | chr1 | 218504290 | + |
| intron-intron | ENST00000565615 | ENST00000491428 | LRRC7 | chr1 | 70034324 | + | RRP15 | chr1 | 218504290 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000370958 | LRRC7 | chr1 | 70034324 | + | ENST00000366932 | RRP15 | chr1 | 218504290 | + | 7228 | 192 | 5848 | 5450 | 132 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000370958 | ENST00000366932 | LRRC7 | chr1 | 70034324 | + | RRP15 | chr1 | 218504290 | + | 0.8248031 | 0.17519692 |
Top |
Fusion Genomic Features for LRRC7-RRP15 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| LRRC7 | chr1 | 70034324 | + | RRP15 | chr1 | 218504289 | + | 2.72E-07 | 0.99999976 |
| LRRC7 | chr1 | 70034324 | + | RRP15 | chr1 | 218504289 | + | 2.72E-07 | 0.99999976 |
| LRRC7 | chr1 | 70034324 | + | RRP15 | chr1 | 218504289 | + | 2.72E-07 | 0.99999976 |
| LRRC7 | chr1 | 70034324 | + | RRP15 | chr1 | 218504289 | + | 2.72E-07 | 0.99999976 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
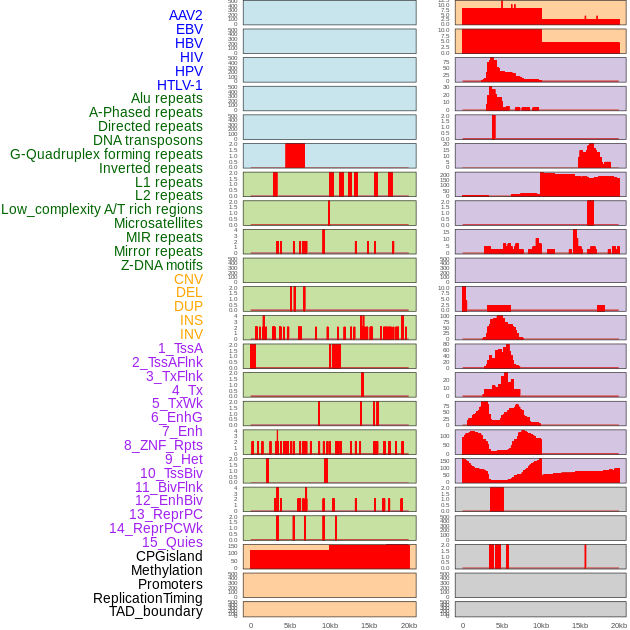 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
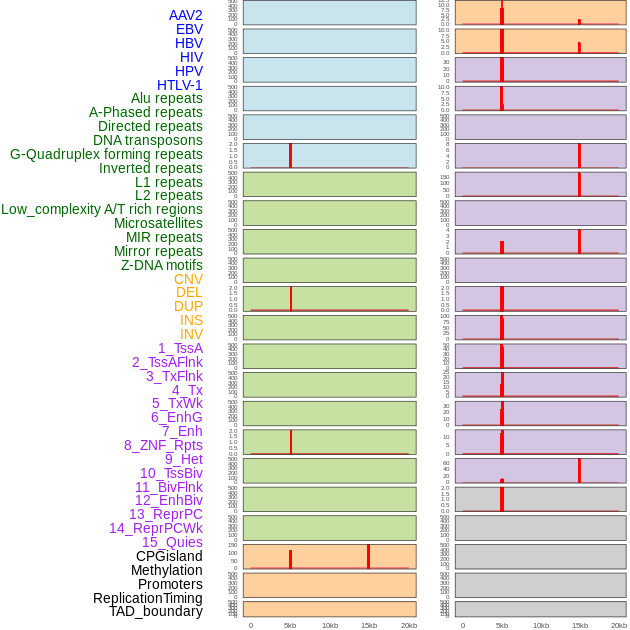 |
Top |
Fusion Protein Features for LRRC7-RRP15 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:70034324/chr1:218504290) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| LRRC7 | RRP15 |
| FUNCTION: Required for normal synaptic spine architecture and function. Necessary for DISC1 and GRM5 localization to postsynaptic density complexes and for both N-methyl D-aspartate receptor-dependent and metabotropic glutamate receptor-dependent long term depression. {ECO:0000269|PubMed:11729199}. | 282 |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LRRC7 | chr1:70034324 | chr1:218504290 | ENST00000035383 | + | 1 | 25 | 1445_1535 | 0 | 1538.0 | Domain | PDZ |
| Hgene | LRRC7 | chr1:70034324 | chr1:218504290 | ENST00000035383 | + | 1 | 25 | 116_137 | 0 | 1538.0 | Repeat | Note=LRR 5 |
| Hgene | LRRC7 | chr1:70034324 | chr1:218504290 | ENST00000035383 | + | 1 | 25 | 139_161 | 0 | 1538.0 | Repeat | Note=LRR 6 |
| Hgene | LRRC7 | chr1:70034324 | chr1:218504290 | ENST00000035383 | + | 1 | 25 | 162_183 | 0 | 1538.0 | Repeat | Note=LRR 7 |
| Hgene | LRRC7 | chr1:70034324 | chr1:218504290 | ENST00000035383 | + | 1 | 25 | 185_206 | 0 | 1538.0 | Repeat | Note=LRR 8 |
| Hgene | LRRC7 | chr1:70034324 | chr1:218504290 | ENST00000035383 | + | 1 | 25 | 208_229 | 0 | 1538.0 | Repeat | Note=LRR 9 |
| Hgene | LRRC7 | chr1:70034324 | chr1:218504290 | ENST00000035383 | + | 1 | 25 | 231_253 | 0 | 1538.0 | Repeat | Note=LRR 10 |
| Hgene | LRRC7 | chr1:70034324 | chr1:218504290 | ENST00000035383 | + | 1 | 25 | 23_44 | 0 | 1538.0 | Repeat | Note=LRR 1 |
| Hgene | LRRC7 | chr1:70034324 | chr1:218504290 | ENST00000035383 | + | 1 | 25 | 254_275 | 0 | 1538.0 | Repeat | Note=LRR 11 |
| Hgene | LRRC7 | chr1:70034324 | chr1:218504290 | ENST00000035383 | + | 1 | 25 | 277_298 | 0 | 1538.0 | Repeat | Note=LRR 12 |
| Hgene | LRRC7 | chr1:70034324 | chr1:218504290 | ENST00000035383 | + | 1 | 25 | 300_321 | 0 | 1538.0 | Repeat | Note=LRR 13 |
| Hgene | LRRC7 | chr1:70034324 | chr1:218504290 | ENST00000035383 | + | 1 | 25 | 323_344 | 0 | 1538.0 | Repeat | Note=LRR 14 |
| Hgene | LRRC7 | chr1:70034324 | chr1:218504290 | ENST00000035383 | + | 1 | 25 | 346_367 | 0 | 1538.0 | Repeat | Note=LRR 15 |
| Hgene | LRRC7 | chr1:70034324 | chr1:218504290 | ENST00000035383 | + | 1 | 25 | 369_391 | 0 | 1538.0 | Repeat | Note=LRR 16 |
| Hgene | LRRC7 | chr1:70034324 | chr1:218504290 | ENST00000035383 | + | 1 | 25 | 392_413 | 0 | 1538.0 | Repeat | Note=LRR 17 |
| Hgene | LRRC7 | chr1:70034324 | chr1:218504290 | ENST00000035383 | + | 1 | 25 | 47_68 | 0 | 1538.0 | Repeat | Note=LRR 2 |
| Hgene | LRRC7 | chr1:70034324 | chr1:218504290 | ENST00000035383 | + | 1 | 25 | 70_91 | 0 | 1538.0 | Repeat | Note=LRR 3 |
| Hgene | LRRC7 | chr1:70034324 | chr1:218504290 | ENST00000035383 | + | 1 | 25 | 93_114 | 0 | 1538.0 | Repeat | Note=LRR 4 |
| Tgene | RRP15 | chr1:70034324 | chr1:218504290 | ENST00000366932 | 3 | 5 | 108_144 | 235 | 283.0 | Coiled coil | Ontology_term=ECO:0000255 |
Top |
Fusion Gene Sequence for LRRC7-RRP15 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >50039_50039_1_LRRC7-RRP15_LRRC7_chr1_70034324_ENST00000370958_RRP15_chr1_218504290_ENST00000366932_length(transcript)=7228nt_BP=192nt GTCTAACGTGGCACCTTCCTGGATTCCCCTCTATCTCCTGTTCTTCCTACCTTCTACTCCTTCCCTCCTCTTCTCCTCCGAAGACCCTGG CGCCCACTCCACTGCGGACCCCTGAACACTTAAGGAATAACCCTTGGCAGCTGCACGACTACGCTCTCCGAAACCTCATGGGCAGTTTCT CCCGCCGGCGATACTGAAGTGAAATCAGAAGAAGGCCCAGGTTGGACGATCCTACGTGATGATTTCATGATGGGAGCATCTATGAAAGAC TGGGACAAGGAAAGTGATGGGCCAGATGACAGCAGACCAGAATCTGCAAGTGACTCTGATACATAAAGCATCATAGGAAATACAATTGCA GTCGTTTTATTTTTTCTAGAAAAATATGTCATCCTCTGATAGTTGGGGAATTATAAGGATACCATTTGTAAGAAAGCCAAAAGACTTTTG CCAGATTTCATATTTCCCCTTTTCATGTACACTTTATATATACTTCATTAAAATTATATTTTAAACCCTTGTATAATTTTAAGCATTGTT CCTCAGAACATTTGTAAAAGGATATATTTCTGCTTGACCAGCGAGATGTGCATTTTGCCAGGATCATATTGGTCATGTCTATTGGTGTAT TATTTCAGTATCACCAATGTTTTCAGAAATACAGTACTAATTCATCATTAAACTCTTTGAAGTTAATATTTTTCTGCCTTCTAACTTATA GACTCAACTATGTATCTGTAGTTTTTGGGAATGGTTGGTGTTTTTTGCTTTGTGTTGGGAAGTTATTGAGAAAACCTATATAATAAAATT TAAAATTATAGTTTTTCAGATTTTGGGTCGTGATTTTAGTTTTAGGTAATCTGGAAGATATGATTTAAGAAACAACAGATTTCAACTTTT TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTGAGACGGAGTCTTGCTCTGTGGCCCCGGATGGAATGCAGTGGCGTGATCTCGGCT CACTGCAAGCACCGCCTCCCAGGTTCACGCCATTCTCCCACCTCAACCTCTCAAGTAGCTGGGACTACAGGTGCCTGCCACCACGCCCAG CTAATTTTTTGTTTTTGTATTTTTAGTAGAGACGGGGTTTCACCATGTTAGCCAGGATGGTCTCAATCTCCTGACCTCGTGATCCACCCG CCTCGGCCTCCCAAAGTGCTGGGATTACAGGCGTGAGCCACTGCGCCCGGCCCAGATTTCAACTTTTATTTTATTTCCTTTCATTTATTA AATAATATAAATAATGACAGATTTATTTTAGGTGTTAGCCTATTTTTGCATGAGAGATTTACAGAGGTGGAAATTCTCATGCTTAACTTA TGCAATGAGACAATTTCTCAGGAAGCAAGTTTGACATGCTTTAGTCTCTCTCTTCTTTTTTTTCCTCCCAAATATTTGAGTTAGCTTAAA TTCTTTTTTATGGACTGCCCAACCAGTGAATTGATGCACTTAGACAACAAGTAATCTGCAGCAGAATATTGCATCCAATTTGAGACTTGA TTCCATATTATTGCCTTAAAGGCTATTAAATTGGGTGACTTTTAGTATCCAGCTGTTAAAAAGAGGAAAGACTGTTTAATATCATTCCAA AATTTTTGTGAAATGTGGCCCAAGATCTAAAGTTTTTAAGGGAATTATTTTTAATAAATATGATCTGTCCCTTTTTGAAAAGGATCCATC CCTTTTCATAATAAAAATAATTCCCTCCTACCACCCAGTTGCACATACTTGCATCTGTTATGTATTGAAGGATAATCCCCCCCAAAAAAA CTATCCAAGAGCAGAGATGTTTATATTTTGAAACCCATTTGAGTGTTACAGTATTATATGACTAAAGCTGAATCAGTCTGTGGCCTGGTA TTTGTTATCATAAAGGAAGATGTACACCTAAGGTATCAAGTGGTTAAAACAGCAAAGTACAAGATGCATTTTTGTATTTATTCTAACTTT TGAACATTAGTGTCTTTCAGTATAAATAAATGTGAACAAATATACCTCAGTAGTAATACAGGAGAACTAAATATCCTGCTATAACTTAAT ACAGATACATATTTCAATATGACTTAACAAACACACAATTTTGAAAAAAAAAAAAAAAAAGTGCAGCAACTTAAACACATTTTTCTCCCC AGAACTAAGATTACACTTAATTTAATTTAATGTGTTTCTGCGTTATACTAGAGTTTTGCAGGAATTTTTCTGAATAGTACATTCACATGA TAGATACGGATAAATACCTTATTTACTAAAATGTTTTGAAACATATTTTATAGTCCCTCATTTGATTTGAAAAAATTTGACAAGATCTTC ACATTGAAAGTAAGGACGAGAATATTTGCAGATTTCCTTCGTTCCAAAATTATTTTATTCCAAAGAAAAATTGAGTTGCCACTATCAGGA AAAAAAAAATGTATCACTTCTGTTAGAAGGCTATGAAGTATATATAAATTATATGACAATTTTTATCATATAGATTTGATATCTTCTATG TATTCGTATTTTTGCATCTTGGGTATTAAGCAAAATACCATATGTATAAATATGAAACCAGGAATGTGCATATTTTAGAAACTAATTGTG TATCTTAAGTGAATGTTTTGAGTAGTTCTGTCACCCCTATTCTAAATATTTTACCTACAACATTTAAATTATTCTGATATCAATCAGATA AATAATCCGAGTGTATTTCATCTTTCTACATTGTCTTTGTGGTTTTTGTTTGTTTGTTTTTGAGACGGTCTCACACTGTCACCCAGGCTG GAGTGCAGTGGTGCAATCTCAGCTCACTGCAACCTCCACCTCCCAGGTTCCAGCTATCCTCCCACCTCAGCCTCCCAAGTAGCTAGGACT ACAGGTGTGCACCACCATGCCCAGCTAATTTTTTGTCTTTTTGATGTAGATGGGGTTACCCCATGTTGCCCAGGCTGGTTGTGAACTCCT GAGCTCAAGGGATCCCCCCGCCTCTTGCCTCCCAAAGTGCTTGGATTACAGGTGTGAGCCACCATACCACCTGTCTTTGAATTTTTTAAC GAGGGGAAAGAAAACTGCAATTCCTGCCTCCCCATTTATTTAAACATAACACCTAGGATTTTATTTTTGGTAACTACTTAGAATAATATA GTAGGTTACTTATCTTCGGTTAGAAAGAACTATGGAAAATAGGAACCAATAATTTGGAAGTATTATTTCCTTATTTCATATATAGAAATA ATTATCTTATTTCCTTATTTCATATATAGAATATTCTAGATACTTGGATGGCTGGGGTTGATGTTCTTAGTATTTATCATAGTTTAGCAT TGGTTGTTATTTTTAAGCATCAGAATTTGAAAGCCAATGATAATACCTGATTAGGACTGAAAATAGAATTATCTTAGTAGTTTAGCACTT TTTATAATGGAGTTGGAGCACTGAATTTATTTTCAAATTACCTAGAATTCTGCATTGCTCAAACATTATTCAATTGATTGCAAAATATAC CTTGGAAACCCTTTGGATAGAACAGTTCAGAGGGTTACCACTAGTACTTAGAGCCTAAAGTCACTCAGTAGGAGGTTCTCAAAGGTCTAT TTGAAGGCAATCCAATTTCTTTAAAATAAAATATTTTTTGCCTTTTAAAACAGATGGGTGAAGAAAGGGACGTATAAATTCTGGGAAACC TCTTATGTTTTCCTCCCATTTCCTTCTATGTCTCCATTAATGTGCTAACTTGAAGGAATTTAACTTCAAGGAAAGTTGACTGATGAAGTT TATGTAGAGCTGTGAATATTATAATTTGTCAATACTTTATTGATAATGTTAACTTTGTTTTTATTTGGAAACATTTTTTCCAGTTGACCA AAGCAGAATTCATTCTGTTCTTCCCTCAAATTCATCTATTAGGATCAATGTGATTGACATTTATCTCCACTCTGCATGTCACTGATATGG TATAGACGCTATAAGCCCTGTAGATATTATGGGTATGAAGACTATTTGAGGTGAATTGGCAGTGAGAGCAAATTTTGGAAACAGGAGAGA GAACTACTTCGTGCCTATAAAAAGCTCTCTCACTCCTTCCCCAAGAAAACAGGAGTCTGAATTTTGGCCAGCTGGGTATTTGAATCTTAC TGACTGTTGCAGCACTGGTAGTCCTGGTAAATTTTTTCAGTGGGCGAGCTTTTCTGAAATTTGATTAAACATATTAGTCAAGGGAATCTG TGAATAGATTTAGAAGTAGAATGTTTGAGGTGGTTGACAAGACATTTAGAATTGTGATTAATGTTTTAATAAAATGCTAAGTTAGAATGA ATAATCATAATTATATAGATGGAAGATATATTCTAACAGTTCTTAAATAATTTTAGATTCTCTAAATTTAGAGTGTTTTCTAGCTTAGAG AGCAATATTAATATTTTTTACCTAGTCTACTGAACATAATTTCAGGGAGTTAGTATTTAGGACTTTTTACCTAGAGATGCGGTAAACCTT TTAAATTTAAATTTGCTTTCTTAAAATACCTGTTCCAACATTGCTCACCTTTCTTCTCCCTCCTTTTTGCCCTCTCCTTCAGTGGGAGGA GCACATATTCAGCTTTCTGTTAAACACTCTTTGGTTTTCAACTGAGTAAGGCAAGCAAGGATATATGTTAGAAGAGTGGGCATAATACTT TATATTAAAATTATAGGGACTAAAAGTCTAAATCATTGCTGTCAATAGGACTTTTTTTGATGATGGAAATAATTTATGTCTGTGCTGTTC AATATGGTAGCCACTCGCAGAGGTGACTACTGAGCATTTCACGTGTGGCTTGTGATACAGAGGAACTAAATTTTTAATTTTACTTAATTA CTTTAAATTTTAAAAGTTACACGTGGCTAGTGGGTCTATCCCAACTATGAAGAGAATAAAGCTACAGAGCATCTATCTAGTCTAATGAAA CAACAGTGAATTTGGGAATTGTAAATCTGTCTTAAGCTTTTTCCATAAACTTTTATTGATATAATTTAGGCATATACAAATACTGGTGAA ATTTTATTGATTGAATTATTGTTGACTTTTTTTTCAGTATTTATATTTTTAAAAAGATATGTTTCTTTATAATTAACTTGTATTGTAGGG TAACCATTGTCCTTAAATGTTAAGCCACTTAGTTAATAACAAATAGCATTTGGTTTTTTAGCATTATTTATCATCTGAGGCTTTCAGAAT TAATGCACCAAACCTTAAATATTTTTGTTGTTCTCTAAAAGAATGGGTTTATGATGCACCAAAAGGACATTTTGAGAACTGCCGGAGCAA CAGATACCACCATGTAAGGAGTGAATTCCCATGTTTTTCACTTATATCTAGAGGCCTACTTATTTAGGAGGTTTCCTAAGTAAGTGTATT TAGTTGTAGGACCTTAGAGTTGCCTAGGTCCATTCTATTCTAGCATATACTCAACATACTGCCAGAGCAAATGTAGGTTGCAGATTCAGG CAAAACACACACACACACACACACAAAACCCTGAAGAGCACAATGAGCATTATTAGCACAGCCAATTTAAAATGATGAGGTGTTAACCAC ATTTGTATAAATAATAGTTTCTCTAACCCTGAATCATTATCCTGCATTTTTAATGCAATCGGTTCATAAACATCAAAGTGAAAACACCAA ATTTGTAAGAGGTCAGGACCAACTGGGGCTCTAAATGGAGAGACAGCTGAGAGCTGTGTCCCTATGCGTGGGTGGCCTTGCAGCTGTTGA TTCACCCTCAGGCTACCAGACAGTTGCCTCTTTGCATTTGCAGAACCAGACTTCAATCTTAGGTAAAAATGTATCCCTCCCATGGACAGG ACATAAACAATAGATTGACATCAGCTCTGAAAACAGTGCAGCAGCCAGACTCAGCAGTTGCCCTGAAATACTGAGAAGTACCAAGCATAA CATCAAGACTGTTTCTACACAATAATATTTGTCTTTGTACAGCAACACTGGGTTCTCTTAAGTATATTATAAATATAAAAGCTCAAAGTC CAAAAGGTTCAATGTGATGATATCCTTCTCTACAAATGCCAGGAACACCAGGGTAGATTGCATGAGACATGACTGCCGTATCTTGGTCCT TACTAGGATTCCCAAATATACTCCTTAATAACCAGTAAAGTTATATTTACACTGGACATTTTAAGAAATACCAGTGTTCGTTGTTAGATG AGAGACAGCATAATTTTGGAATCACAACTGAGTTCAGAGTTCAGTTTTGAGTCCTACTAGTTTTGTTTCCTTGTGTAAGTTTCTTAACCT TTGTAAGGCTAGCATGTAAATCAGATACAATAATATATAGTTCAGGGTGTAAAGATTGGATTAATGTATGCAGTGTCTATCTAGCACAGT GCCTGTTATGTAGTAGCGTGTCAAGAGATGTTCTCTCTTATGCATTTGGAGGATTAGGTGCTGCACCTATAGGTTTCCATTCATTTCTCA TCTATGTAATTCTGAGTCTTCCTTTTCTAGATAGAATTCCCCCCAACAAAAATTTAAAGTCTAACAGGTGGTAGGTGCTTATTGAGGCCC AGCAATTCTACTTCTGGAAACCTTCCTCAGGAAAAGATTCTGATGACAGAAAATGCTGTGTGAACAGTATTATATAGAATTGTTTTTAAT AGCAAGAAATTTTTTAAAACTTACAAGTTCAGTGATATGAGGACATTTACATAAACTATCGAACCACCAGTCTTCTGTCAATTCCTTTTA AAATGATGGTTGTGAAGACTGATGAAAACACAGTGTCAATGATATGTTAACTGAAAAAGTAAGATTAAAAACATTATGGATAGTATAAGT TTCTTAAATGCCAACAATGCATGAGGTGAATAGAATAAGACTTTTTTTTAAAGAATGTAATTTTTGACAGTCTGAAAAGACAGGCCTTAT ATTTATATTGAGAAAAATGATCCTGACATTTAATGAGACACTAAAACAGTTTTTATTTTTTGCTTACTTATTCTGTTCTAGAATCCAAAC TCTACATACTGTTTAATAGAGCATTATTGATATTAGCTGTATTAGATGACTGTATGAAGAGTCCTGTTTGCTAGAATCCTACTTTATCTT TTAAGTTTTGCTAATTACCCTGCAATGAATGTTGAACATAAAAAATTTGACTCCCTTTCCATGTGTCCACTGTTTCCTTGTGCCTTTTGA >50039_50039_1_LRRC7-RRP15_LRRC7_chr1_70034324_ENST00000370958_RRP15_chr1_218504290_ENST00000366932_length(amino acids)=132AA_BP= MSMGGIHFYLRLKSGSANAKRQLSGSLRVNQQLQGHPRIGTQLSAVSPFRAPVGPDLLQIWCFHFDVYEPIALKMQDNDSGLEKLLFIQM -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for LRRC7-RRP15 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for LRRC7-RRP15 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for LRRC7-RRP15 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
