
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:LRRK1-ADAMTS17 (FusionGDB2 ID:50105) |
Fusion Gene Summary for LRRK1-ADAMTS17 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: LRRK1-ADAMTS17 | Fusion gene ID: 50105 | Hgene | Tgene | Gene symbol | LRRK1 | ADAMTS17 | Gene ID | 79705 | 170691 |
| Gene name | leucine rich repeat kinase 1 | ADAM metallopeptidase with thrombospondin type 1 motif 17 | |
| Synonyms | RIPK6|Roco1 | WMS4 | |
| Cytomap | 15q26.3 | 15q26.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | leucine-rich repeat serine/threonine-protein kinase 1 | A disintegrin and metalloproteinase with thrombospondin motifs 17a disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type 1 motif, 17 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q38SD2 | Q8TE56 | |
| Ensembl transtripts involved in fusion gene | ENST00000284395, ENST00000388948, ENST00000532029, ENST00000532145, | ENST00000559976, ENST00000268070, | |
| Fusion gene scores | * DoF score | 5 X 4 X 3=60 | 2 X 2 X 2=8 |
| # samples | 5 | 2 | |
| ** MAII score | log2(5/60*10)=-0.263034405833794 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(2/8*10)=1.32192809488736 | |
| Context | PubMed: LRRK1 [Title/Abstract] AND ADAMTS17 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | LRRK1(101529603)-ADAMTS17(100591936), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | LRRK1-ADAMTS17 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. LRRK1-ADAMTS17 seems lost the major protein functional domain in Hgene partner, which is a kinase due to the frame-shifted ORF. LRRK1-ADAMTS17 seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across LRRK1 (5'-gene) Fusion gene breakpoints across LRRK1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
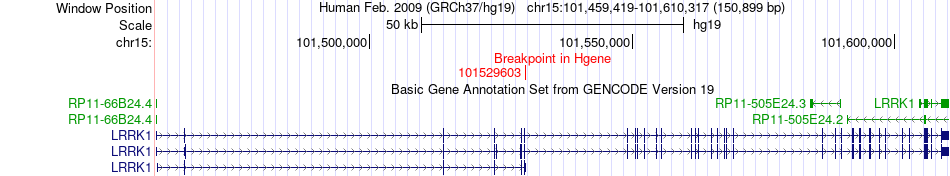 |
 Fusion gene breakpoints across ADAMTS17 (3'-gene) Fusion gene breakpoints across ADAMTS17 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
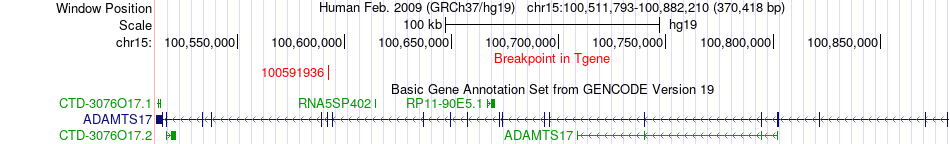 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-A2-A4RW-01A | LRRK1 | chr15 | 101529603 | - | ADAMTS17 | chr15 | 100591936 | - |
| ChimerDB4 | BRCA | TCGA-A2-A4RW-01A | LRRK1 | chr15 | 101529603 | + | ADAMTS17 | chr15 | 100591936 | - |
Top |
Fusion Gene ORF analysis for LRRK1-ADAMTS17 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000284395 | ENST00000559976 | LRRK1 | chr15 | 101529603 | + | ADAMTS17 | chr15 | 100591936 | - |
| 5CDS-intron | ENST00000388948 | ENST00000559976 | LRRK1 | chr15 | 101529603 | + | ADAMTS17 | chr15 | 100591936 | - |
| 5CDS-intron | ENST00000532029 | ENST00000559976 | LRRK1 | chr15 | 101529603 | + | ADAMTS17 | chr15 | 100591936 | - |
| Frame-shift | ENST00000532029 | ENST00000268070 | LRRK1 | chr15 | 101529603 | + | ADAMTS17 | chr15 | 100591936 | - |
| In-frame | ENST00000284395 | ENST00000268070 | LRRK1 | chr15 | 101529603 | + | ADAMTS17 | chr15 | 100591936 | - |
| In-frame | ENST00000388948 | ENST00000268070 | LRRK1 | chr15 | 101529603 | + | ADAMTS17 | chr15 | 100591936 | - |
| intron-3CDS | ENST00000532145 | ENST00000268070 | LRRK1 | chr15 | 101529603 | + | ADAMTS17 | chr15 | 100591936 | - |
| intron-intron | ENST00000532145 | ENST00000559976 | LRRK1 | chr15 | 101529603 | + | ADAMTS17 | chr15 | 100591936 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000388948 | LRRK1 | chr15 | 101529603 | + | ENST00000268070 | ADAMTS17 | chr15 | 100591936 | - | 4927 | 1121 | 344 | 2113 | 589 |
| ENST00000284395 | LRRK1 | chr15 | 101529603 | + | ENST00000268070 | ADAMTS17 | chr15 | 100591936 | - | 4959 | 1153 | 304 | 2145 | 613 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000388948 | ENST00000268070 | LRRK1 | chr15 | 101529603 | + | ADAMTS17 | chr15 | 100591936 | - | 0.016285045 | 0.983715 |
| ENST00000284395 | ENST00000268070 | LRRK1 | chr15 | 101529603 | + | ADAMTS17 | chr15 | 100591936 | - | 0.013593801 | 0.98640615 |
Top |
Fusion Genomic Features for LRRK1-ADAMTS17 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
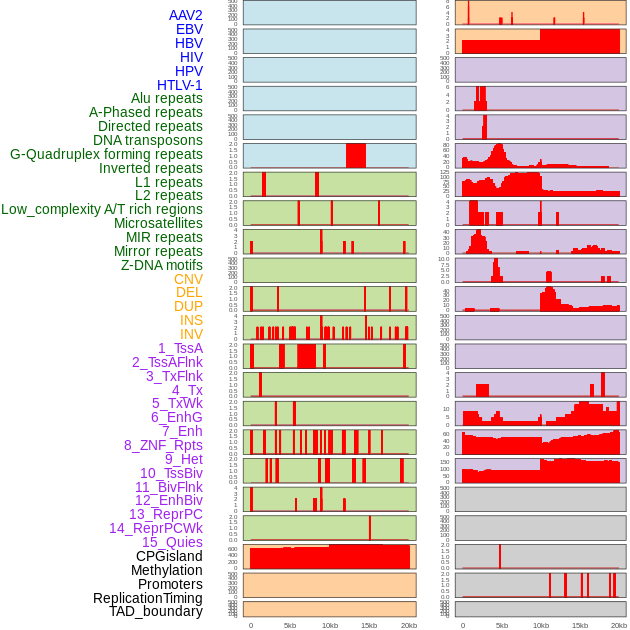 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for LRRK1-ADAMTS17 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr15:101529603/chr15:100591936) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| LRRK1 | ADAMTS17 |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000388948 | + | 6 | 34 | 119_148 | 254 | 2016.0 | Repeat | Note=ANK 2 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000388948 | + | 6 | 34 | 152_182 | 254 | 2016.0 | Repeat | Note=ANK 3 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000388948 | + | 6 | 34 | 193_222 | 254 | 2016.0 | Repeat | Note=ANK 4 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000388948 | + | 6 | 34 | 86_116 | 254 | 2016.0 | Repeat | Note=ANK 1 |
| Tgene | ADAMTS17 | chr15:101529603 | chr15:100591936 | ENST00000268070 | 15 | 22 | 1045_1084 | 765 | 1096.0 | Domain | PLAC | |
| Tgene | ADAMTS17 | chr15:101529603 | chr15:100591936 | ENST00000268070 | 15 | 22 | 800_860 | 765 | 1096.0 | Domain | TSP type-1 2 | |
| Tgene | ADAMTS17 | chr15:101529603 | chr15:100591936 | ENST00000268070 | 15 | 22 | 861_922 | 765 | 1096.0 | Domain | TSP type-1 3 | |
| Tgene | ADAMTS17 | chr15:101529603 | chr15:100591936 | ENST00000268070 | 15 | 22 | 925_968 | 765 | 1096.0 | Domain | TSP type-1 4 | |
| Tgene | ADAMTS17 | chr15:101529603 | chr15:100591936 | ENST00000268070 | 15 | 22 | 972_1029 | 765 | 1096.0 | Domain | TSP type-1 5 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000388948 | + | 6 | 34 | 1242_1525 | 254 | 2016.0 | Domain | Protein kinase |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000388948 | + | 6 | 34 | 632_826 | 254 | 2016.0 | Domain | Roc |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000532029 | + | 1 | 6 | 1242_1525 | 0 | 265.0 | Domain | Protein kinase |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000532029 | + | 1 | 6 | 632_826 | 0 | 265.0 | Domain | Roc |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000388948 | + | 6 | 34 | 1248_1256 | 254 | 2016.0 | Nucleotide binding | ATP |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000532029 | + | 1 | 6 | 1248_1256 | 0 | 265.0 | Nucleotide binding | ATP |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000388948 | + | 6 | 34 | 279_300 | 254 | 2016.0 | Repeat | Note=LRR 1 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000388948 | + | 6 | 34 | 303_324 | 254 | 2016.0 | Repeat | Note=LRR 2 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000388948 | + | 6 | 34 | 330_351 | 254 | 2016.0 | Repeat | Note=LRR 3 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000388948 | + | 6 | 34 | 353_374 | 254 | 2016.0 | Repeat | Note=LRR 4 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000388948 | + | 6 | 34 | 381_402 | 254 | 2016.0 | Repeat | Note=LRR 5 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000388948 | + | 6 | 34 | 405_426 | 254 | 2016.0 | Repeat | Note=LRR 6 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000388948 | + | 6 | 34 | 427_447 | 254 | 2016.0 | Repeat | Note=LRR 7 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000388948 | + | 6 | 34 | 451_472 | 254 | 2016.0 | Repeat | Note=LRR 8 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000388948 | + | 6 | 34 | 474_495 | 254 | 2016.0 | Repeat | Note=LRR 9 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000388948 | + | 6 | 34 | 498_519 | 254 | 2016.0 | Repeat | Note=LRR 10 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000388948 | + | 6 | 34 | 549_570 | 254 | 2016.0 | Repeat | Note=LRR 11 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000388948 | + | 6 | 34 | 572_594 | 254 | 2016.0 | Repeat | Note=LRR 12 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000388948 | + | 6 | 34 | 596_617 | 254 | 2016.0 | Repeat | Note=LRR 13 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000532029 | + | 1 | 6 | 119_148 | 0 | 265.0 | Repeat | Note=ANK 2 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000532029 | + | 1 | 6 | 152_182 | 0 | 265.0 | Repeat | Note=ANK 3 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000532029 | + | 1 | 6 | 193_222 | 0 | 265.0 | Repeat | Note=ANK 4 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000532029 | + | 1 | 6 | 279_300 | 0 | 265.0 | Repeat | Note=LRR 1 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000532029 | + | 1 | 6 | 303_324 | 0 | 265.0 | Repeat | Note=LRR 2 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000532029 | + | 1 | 6 | 330_351 | 0 | 265.0 | Repeat | Note=LRR 3 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000532029 | + | 1 | 6 | 353_374 | 0 | 265.0 | Repeat | Note=LRR 4 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000532029 | + | 1 | 6 | 381_402 | 0 | 265.0 | Repeat | Note=LRR 5 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000532029 | + | 1 | 6 | 405_426 | 0 | 265.0 | Repeat | Note=LRR 6 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000532029 | + | 1 | 6 | 427_447 | 0 | 265.0 | Repeat | Note=LRR 7 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000532029 | + | 1 | 6 | 451_472 | 0 | 265.0 | Repeat | Note=LRR 8 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000532029 | + | 1 | 6 | 474_495 | 0 | 265.0 | Repeat | Note=LRR 9 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000532029 | + | 1 | 6 | 498_519 | 0 | 265.0 | Repeat | Note=LRR 10 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000532029 | + | 1 | 6 | 549_570 | 0 | 265.0 | Repeat | Note=LRR 11 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000532029 | + | 1 | 6 | 572_594 | 0 | 265.0 | Repeat | Note=LRR 12 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000532029 | + | 1 | 6 | 596_617 | 0 | 265.0 | Repeat | Note=LRR 13 |
| Hgene | LRRK1 | chr15:101529603 | chr15:100591936 | ENST00000532029 | + | 1 | 6 | 86_116 | 0 | 265.0 | Repeat | Note=ANK 1 |
| Tgene | ADAMTS17 | chr15:101529603 | chr15:100591936 | ENST00000268070 | 15 | 22 | 599_701 | 765 | 1096.0 | Compositional bias | Note=Cys-rich | |
| Tgene | ADAMTS17 | chr15:101529603 | chr15:100591936 | ENST00000268070 | 15 | 22 | 60_120 | 765 | 1096.0 | Compositional bias | Note=Arg-rich | |
| Tgene | ADAMTS17 | chr15:101529603 | chr15:100591936 | ENST00000268070 | 15 | 22 | 232_452 | 765 | 1096.0 | Domain | Peptidase M12B | |
| Tgene | ADAMTS17 | chr15:101529603 | chr15:100591936 | ENST00000268070 | 15 | 22 | 453_542 | 765 | 1096.0 | Domain | Note=Disintegrin | |
| Tgene | ADAMTS17 | chr15:101529603 | chr15:100591936 | ENST00000268070 | 15 | 22 | 543_598 | 765 | 1096.0 | Domain | TSP type-1 1 | |
| Tgene | ADAMTS17 | chr15:101529603 | chr15:100591936 | ENST00000268070 | 15 | 22 | 199_206 | 765 | 1096.0 | Motif | Cysteine switch | |
| Tgene | ADAMTS17 | chr15:101529603 | chr15:100591936 | ENST00000268070 | 15 | 22 | 702_779 | 765 | 1096.0 | Region | Note=Spacer |
Top |
Fusion Gene Sequence for LRRK1-ADAMTS17 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >50105_50105_1_LRRK1-ADAMTS17_LRRK1_chr15_101529603_ENST00000284395_ADAMTS17_chr15_100591936_ENST00000268070_length(transcript)=4959nt_BP=1153nt GCTGGCGGCGGCCTTGCTCTGCGCGCTCTTCGCGCCGCCCTCCCCGCCCGCCCGCCTCAGGATTGAGGAAGTGCGTCTGGGCCCGGCCCC GGCGCGGGGGGCAGACGGCGGTGGGACGGCCAGGCCCCGGCCCCGCCAGTGTGTCCGCCCGGCCCCGCGTCCCGGAGCGCCCGCACCCGG CCCCGCCGCCGCCTCAGAGCAAGAAAGCTTTCTGCTCAGCCATGGCTACGAGTCCACGCCTTAATGCACCCCACAGCCAGCGGCAGTGGC AGTGACAACAGCGGGACCTGCCTTTGAAGATCGGCTGCTGCAAGGGTTGATGGCTGGCATGTCGCAAAGACCCCCCAGCATGTACTGGTG TGTGGGGCCGGAGGAGTCAGCTGTGTGTCCAGAACGTGCCATGGAGACGCTTAACGGTGCCGGGGACACGGGCGGCAAGCCGTCCACGCG GGGCGGTGACCCTGCAGCGCGGTCCCGCAGGACGGAAGGCATCCGCGCCGCGTACAGGCGGGGAGACCGCGGCGGCGCCCGGGACCTGCT GGAGGAGGCCTGCGACCAGTGCGCGTCCCAGCTGGAAAAGGGCCAGCTTCTGAGCATCCCGGCAGCCTATGGGGATCTGGAGATGGTCCG CTACCTACTCAGCAAGAGACTGGTGGAGCTGCCCACCGAGCCCACGGATGACAACCCAGCCGTGGTGGCAGCGTATTTTGGACACACGGC AGTTGTGCAGGAATTGCTTGAGCCATTGACACCCAAACAAACAGAGTGGGAACAAACAGCACCCTTCAGTGGCACTTTCCCTTTGTCCTG CCATCTCCTGCCAGGTCCCTGCAGTCCCCAGCGGCTTCTGAACTGGATGCTGGCCTTGGCTTGCCAGCGAGGGCACCTGGGGGTTGTGAA GCTCCTGGTCCTGACGCACGGGGCTGACCCGGAGAGCTACGCTGTCAGGAAGAATGAGTTCCCTGTCATCGTGCGCTTGCCCCTGTATGC GGCCATCAAGTCAGGGAATGAAGACATTGCAATATTCCTGCTTCGGCATGGGGCCTATTTCTGTTCCTACATCTTGCTGGATAGTCCTGA CCCCAGCAAACATCTGCTGAGAAAGTACTTCATTGAAGCCAGTCCCTTGCCCAGCAGTTATCCGGGAAAAACAGTGTTGTTATTTCACGA CCAAGATTATGGAATTCATTATGAATACACTGTTCCTGTAAACCGCACTGCGGAAAATCAAAGCGAACCAGAAAAACCGCAGGACTCTTT GTTCATCTGGACCCACAGCGGCTGGGAAGGGTGCAGTGTGCAGTGCGGCGGAGGGGAGCGCAGAACCATCGTCTCGTGTACACGGATTGT CAACAAGACCACAACTCTGGTGAACGACAGTGACTGCCCTCAAGCAAGCCGCCCAGAGCCCCAGGTCCGAAGGTGCAACTTGCACCCCTG CCAGTCACGGTGGGTGGCAGGCCCGTGGAGCCCCTGCTCGGCGACCTGTGAGAAAGGCTTCCAGCACCGGGAGGTGACCTGCGTGTACCA GCTGCAGAACGGCACACACGTCGCTACGCGGCCCCTCTACTGCCCGGGCCCCCGGCCGGCGGCAGTGCAGAGCTGTGAAGGCCAGGACTG CCTGTCCATCTGGGAGGCGTCTGAGTGGTCACAGTGCTCTGCCAGCTGTGGTAAAGGGGTGTGGAAACGGACCGTGGCGTGCACCAACTC ACAAGGGAAATGCGACGCATCCACGAGGCCGAGAGCCGAGGAGGCCTGCGAGGACTACTCAGGCTGCTACGAGTGGAAAACTGGGGACTG GTCTACGTGCTCGTCGACCTGCGGGAAGGGCCTGCAGTCCCGGGTGGTGCAGTGCATGCACAAGGTCACAGGGCGCCACGGCAGCGAGTG CCCCGCCCTCTCGAAGCCTGCCCCCTACAGACAGTGCTACCAGGAGGTCTGCAACGACAGGATCAACGCCAACACCATCACCTCCCCCCG CCTTGCTGCTCTGACCTACAAATGCACACGAGACCAGTGGACGGTATATTGCCGGGTCATCCGAGAAAAGAACCTCTGCCAGGACATGCG GTGGTACCAGCGCTGCTGCCAGACCTGCAGGGACTTCTATGCAAACAAGATGCGCCAGCCACCGCCGAACTCGTGACACGCAGTCCCAAG GGTCGCTCAAAGCTCAGACTCAGGTCTGAAACCCACCCACCCGCAAGCCTACCAGCCTTGTGGCCACGCCCCCACCCGGCTGCCACAAGA ATCCAACTACATAGAACATGAGCGTGGACTTGGCGTTTGCCATTAGTGCTTCCGTACTTAATATATTGTTAACAGCCACTGGCTGGCTTT CTACAGTGAGGAGAAAGTAGGCATGAGTCACAAAGTAACTTCAATTTCTAGGATTTCAGGTACCTCGAAGGGAAGCACCTCTGGCAGACA ACCGTCAAGAGAGAGACATCATTTAGTGTTCTTGTCTTGACTCGCTTTTGACATTTGAATTTCCAGTGCTTGGTATATCATGGAGGAAAC ATCCCCAAAACGAGACATGCTAGAAAAGGCTTTATTCTGAAAGCCGGCGACACCATGGAGGGAGGGGCAGGTGTTGGTGAGCCTCTGCCC GTGGCTTCTCTGGGGAGGGCCGGGCTGCTTAGCACACGTTTCTCTCCATCTACCTTCTTGACCACATGAGAACCAGGACGTTGCCTCCAT GCCCGTCTCTGAGAACATAGTCTCTAAATCCTAGGTGTTGCCTTGGAAGTCTCGTGCGTGGAGTGTAAATCTATATATGCCAGCGAGGAC AGCAGTGCCACGCAGTTCATACCACCCGCATGGGAAGAATGTTCCAAGAGAGTCTGGGTTTGGGGAAGCATCTAATTTTCAGAGCTCTGC TGTCCACCGTGTAGGGAAACAGAAGGGCCTCTCTTCAAGGTGCTGTGACATAAGAAACGGTAATTGCGGTGATGGGGTTGCTTCCTAAGG CAAAGGTAAGCTTGGGCCAGCTTCACTGGGGCGGATGGGCACCTGCCCCGCCTTCCGGGAACATCCACTCTGGCCTGCACTTCCTAAAGC TTTGTACCTTAGAGATGCTGTACCACATCCCAGTGGCTTTCTACCGACCGTGGCCATTTATCTGAAGGTAAGACGACATTTGGGACCTCT GAGGACACAAGCCCAGGCCTAGGATCTGTAGAGCAAGGCCTGACTGCTCTATCCTGGCACGGAGCAGCCTGATGTGCCAGGACCAGGGGA GGAACGCCATCTGGCTGGCACTGCTGCACACCCGCCGAGCCTTCCTGTAGCCCCAGACTTTGTGGTACCCATTATCATCACGCCTGTCAT CATTGACCCATCTTCTTGGTGGGGCAAGGATGATGCATGATGAAGGAAGGTCCTTCCCCCCTGCAGCCCCCTTACGCCTGGCAGCAGACA AGCAGAGTGGCCTCTTTGAGAGCACAGAGGATGGTAGCACCCTACCTGCAAGGAGGCCGGGCAGGGACCCTAGATGCCAGGAGGCCTGTT TTGCTCACCAACTTGGTGGGCATTTCATGGGTGCTTATGTTCTAGGACTTTACCGTAAATAACACCTCCTCCCTGATTTCAGACGGAAGG TCTCACTTGGACTTCCATGGGATCATCTCCCTGTGCTTCTTGATTTATTGGTGCTGTGTTTCTGTGTTTTGTTTTGTTACATGTCACAAC CGTAGAGTTAGCTTAAATCAGAAAGAAGCCTCTCTGCCTTCTCCACCCTGTCTTACGAGCTGTGTTTTTGTTTTTACTACCCTAGAGGCA GAGAAGCGGTAGGGATGTCAGGGAATTTACTCACTTCCACTTGAATCAACGAGAAGTGTTGAGAAACTTCCGTGGGTGCTCTGTGGAAAG AACCGAGGGTGTCAGGATGGAGCGGCCCACCCTCGCCCCGCGGCCTGCGCAGACTGCTGTCCTCCCCTTCAGGCCTGGCCACCAGCAGAC TCCCATGAATTCCCATTTTCACAAAGCCCTCAAAATTTTGGGGGCGCTGAGGTTCAATTTTCTTCATCCCCAAATGAACTGTTAATTCAC TGAAAGACATTTAGACTTTTGCAGCCAAGGAGCAGAGCCGGGCAAGCTCACTAAGGAAGCCTTAGCTTGGTCTCCTGCATGGGCCCCGGG GCCTCCAGATATGCCCACTGTCACCCTGGCAAGGGCCCCACACCCTCAGAATGAAAAGGATTGATTGGGTTATTGTCAGATACCCCCATT AGTTCCCCTAAAGTCTGCATCTGCAGAGCTCTTTTAGCTCATCTTTTTGTTGTCGGTGTTGTTTTTTGAGACAGAGTCTCCCTCTGTTGC CCAGGCTGGAGTGCAGTGGCGCGATCTCAGCTCATTGAAAGCTCCGCCTCCCGGGTTCAGGCCATTCTTCTGCCTCACCCTCCCCAGTAG CTGGGACTACAGGCGCCCACCACCACGCCCACCTACTTTTTTGTATTTTTACTAGAGACGGGGTTTCACCATGTTAGCCAGGATGGTCTC GATCTCCTGACCTCATGATCCACCCGCCTCAGCCTTCCAAAGTGCTGGGATTACAGGAGTGAGCCACCGCGCCCGGCCTTTTAGCTTATC TTGAAATCCACCAGCCTACCTTCTGTTCATGGCACGAGCAGTGATTTCAGTGAAACCATCATAAACATTCTTGTTCCACTAGATGCTGAT CACAGCAGTGATGTTTCTGTGTTAATACCCTGTACTGTCCCTCGAGTACTGTACCTTTGTGCTTGCACACGTGTCACTAACGCACGTGTT ACCAGAGTCTCACGACCCTACGGTGCTATTTTTATTGGTGCAAAATTAAACCCTCTCAGCTGACCGTTGCTGGATAGGATCAGAATGCAG GAAAAGAGCCTCTATGGCAATACGATTGTCCTTTTTTCGTTTCAGAGTCAAATGTATCTATTTTCCATTCGTTAACTTGCATTACATACT >50105_50105_1_LRRK1-ADAMTS17_LRRK1_chr15_101529603_ENST00000284395_ADAMTS17_chr15_100591936_ENST00000268070_length(amino acids)=613AA_BP=283 MLQGLMAGMSQRPPSMYWCVGPEESAVCPERAMETLNGAGDTGGKPSTRGGDPAARSRRTEGIRAAYRRGDRGGARDLLEEACDQCASQL EKGQLLSIPAAYGDLEMVRYLLSKRLVELPTEPTDDNPAVVAAYFGHTAVVQELLEPLTPKQTEWEQTAPFSGTFPLSCHLLPGPCSPQR LLNWMLALACQRGHLGVVKLLVLTHGADPESYAVRKNEFPVIVRLPLYAAIKSGNEDIAIFLLRHGAYFCSYILLDSPDPSKHLLRKYFI EASPLPSSYPGKTVLLFHDQDYGIHYEYTVPVNRTAENQSEPEKPQDSLFIWTHSGWEGCSVQCGGGERRTIVSCTRIVNKTTTLVNDSD CPQASRPEPQVRRCNLHPCQSRWVAGPWSPCSATCEKGFQHREVTCVYQLQNGTHVATRPLYCPGPRPAAVQSCEGQDCLSIWEASEWSQ CSASCGKGVWKRTVACTNSQGKCDASTRPRAEEACEDYSGCYEWKTGDWSTCSSTCGKGLQSRVVQCMHKVTGRHGSECPALSKPAPYRQ -------------------------------------------------------------- >50105_50105_2_LRRK1-ADAMTS17_LRRK1_chr15_101529603_ENST00000388948_ADAMTS17_chr15_100591936_ENST00000268070_length(transcript)=4927nt_BP=1121nt GCGGCCGCCGAGGGGACGCCTCGCGACGGTTCCTGGGAGAGCTGGCGGCGGCCTTGCTCTGCGCGCTCTTCGCGCCGCCCTCCCCGCCCG CCCGCCTCAGGATTGAGGAAGTGCGTCTGGGCCCGGCCCCGGCGCGGGGGGCAGACGGCGGTGGGACGGCCAGGCCCCGGCCCCGCCAGT GTGTCCGCCCGGCCCCGCGTCCCGGAGCGCCCGCACCCGGCCCCGCCGCCGCCTCAGAGCAAGAAAGCTTTCTGCTCAGCCATGGCTACG AGTCCACGCCTTAATGCACCCCACAGCCAGCGGCAGTGGCAGTGACAACAGCGGGACCTGCCTTTGAAGATCGGCTGCTGCAAGGGTTGA TGGCTGGCATGTCGCAAAGACCCCCCAGCATGTACTGGTGTGTGGGGCCGGAGGAGTCAGCTGTGTGTCCAGAACGTGCCATGGAGACGC TTAACGGTGCCGGGGACACGGGCGGCAAGCCGTCCACGCGGGGCGGTGACCCTGCAGCGCGGTCCCGCAGGACGGAAGGCATCCGCGCCG CGTACAGGCGGGGAGACCGCGGCGGCGCCCGGGACCTGCTGGAGGAGGCCTGCGACCAGTGCGCGTCCCAGCTGGAAAAGGGCCAGCTTC TGAGCATCCCGGCAGCCTATGGGGATCTGGAGATGGTCCGCTACCTACTCAGCAAGAGACTGGTGGAGCTGCCCACCGAGCCCACGGATG ACAACCCAGCCGTGGTGGCAGCGTATTTTGGACACACGGCAGTTGTGCAGGAATTGCTTGAGTCCTTACCAGGTCCCTGCAGTCCCCAGC GGCTTCTGAACTGGATGCTGGCCTTGGCTTGCCAGCGAGGGCACCTGGGGGTTGTGAAGCTCCTGGTCCTGACGCACGGGGCTGACCCGG AGAGCTACGCTGTCAGGAAGAATGAGTTCCCTGTCATCGTGCGCTTGCCCCTGTATGCGGCCATCAAGTCAGGGAATGAAGACATTGCAA TATTCCTGCTTCGGCATGGGGCCTATTTCTGTTCCTACATCTTGCTGGATAGTCCTGACCCCAGCAAACATCTGCTGAGAAAGTACTTCA TTGAAGCCAGTCCCTTGCCCAGCAGTTATCCGGGAAAAACAGTGTTGTTATTTCACGACCAAGATTATGGAATTCATTATGAATACACTG TTCCTGTAAACCGCACTGCGGAAAATCAAAGCGAACCAGAAAAACCGCAGGACTCTTTGTTCATCTGGACCCACAGCGGCTGGGAAGGGT GCAGTGTGCAGTGCGGCGGAGGGGAGCGCAGAACCATCGTCTCGTGTACACGGATTGTCAACAAGACCACAACTCTGGTGAACGACAGTG ACTGCCCTCAAGCAAGCCGCCCAGAGCCCCAGGTCCGAAGGTGCAACTTGCACCCCTGCCAGTCACGGTGGGTGGCAGGCCCGTGGAGCC CCTGCTCGGCGACCTGTGAGAAAGGCTTCCAGCACCGGGAGGTGACCTGCGTGTACCAGCTGCAGAACGGCACACACGTCGCTACGCGGC CCCTCTACTGCCCGGGCCCCCGGCCGGCGGCAGTGCAGAGCTGTGAAGGCCAGGACTGCCTGTCCATCTGGGAGGCGTCTGAGTGGTCAC AGTGCTCTGCCAGCTGTGGTAAAGGGGTGTGGAAACGGACCGTGGCGTGCACCAACTCACAAGGGAAATGCGACGCATCCACGAGGCCGA GAGCCGAGGAGGCCTGCGAGGACTACTCAGGCTGCTACGAGTGGAAAACTGGGGACTGGTCTACGTGCTCGTCGACCTGCGGGAAGGGCC TGCAGTCCCGGGTGGTGCAGTGCATGCACAAGGTCACAGGGCGCCACGGCAGCGAGTGCCCCGCCCTCTCGAAGCCTGCCCCCTACAGAC AGTGCTACCAGGAGGTCTGCAACGACAGGATCAACGCCAACACCATCACCTCCCCCCGCCTTGCTGCTCTGACCTACAAATGCACACGAG ACCAGTGGACGGTATATTGCCGGGTCATCCGAGAAAAGAACCTCTGCCAGGACATGCGGTGGTACCAGCGCTGCTGCCAGACCTGCAGGG ACTTCTATGCAAACAAGATGCGCCAGCCACCGCCGAACTCGTGACACGCAGTCCCAAGGGTCGCTCAAAGCTCAGACTCAGGTCTGAAAC CCACCCACCCGCAAGCCTACCAGCCTTGTGGCCACGCCCCCACCCGGCTGCCACAAGAATCCAACTACATAGAACATGAGCGTGGACTTG GCGTTTGCCATTAGTGCTTCCGTACTTAATATATTGTTAACAGCCACTGGCTGGCTTTCTACAGTGAGGAGAAAGTAGGCATGAGTCACA AAGTAACTTCAATTTCTAGGATTTCAGGTACCTCGAAGGGAAGCACCTCTGGCAGACAACCGTCAAGAGAGAGACATCATTTAGTGTTCT TGTCTTGACTCGCTTTTGACATTTGAATTTCCAGTGCTTGGTATATCATGGAGGAAACATCCCCAAAACGAGACATGCTAGAAAAGGCTT TATTCTGAAAGCCGGCGACACCATGGAGGGAGGGGCAGGTGTTGGTGAGCCTCTGCCCGTGGCTTCTCTGGGGAGGGCCGGGCTGCTTAG CACACGTTTCTCTCCATCTACCTTCTTGACCACATGAGAACCAGGACGTTGCCTCCATGCCCGTCTCTGAGAACATAGTCTCTAAATCCT AGGTGTTGCCTTGGAAGTCTCGTGCGTGGAGTGTAAATCTATATATGCCAGCGAGGACAGCAGTGCCACGCAGTTCATACCACCCGCATG GGAAGAATGTTCCAAGAGAGTCTGGGTTTGGGGAAGCATCTAATTTTCAGAGCTCTGCTGTCCACCGTGTAGGGAAACAGAAGGGCCTCT CTTCAAGGTGCTGTGACATAAGAAACGGTAATTGCGGTGATGGGGTTGCTTCCTAAGGCAAAGGTAAGCTTGGGCCAGCTTCACTGGGGC GGATGGGCACCTGCCCCGCCTTCCGGGAACATCCACTCTGGCCTGCACTTCCTAAAGCTTTGTACCTTAGAGATGCTGTACCACATCCCA GTGGCTTTCTACCGACCGTGGCCATTTATCTGAAGGTAAGACGACATTTGGGACCTCTGAGGACACAAGCCCAGGCCTAGGATCTGTAGA GCAAGGCCTGACTGCTCTATCCTGGCACGGAGCAGCCTGATGTGCCAGGACCAGGGGAGGAACGCCATCTGGCTGGCACTGCTGCACACC CGCCGAGCCTTCCTGTAGCCCCAGACTTTGTGGTACCCATTATCATCACGCCTGTCATCATTGACCCATCTTCTTGGTGGGGCAAGGATG ATGCATGATGAAGGAAGGTCCTTCCCCCCTGCAGCCCCCTTACGCCTGGCAGCAGACAAGCAGAGTGGCCTCTTTGAGAGCACAGAGGAT GGTAGCACCCTACCTGCAAGGAGGCCGGGCAGGGACCCTAGATGCCAGGAGGCCTGTTTTGCTCACCAACTTGGTGGGCATTTCATGGGT GCTTATGTTCTAGGACTTTACCGTAAATAACACCTCCTCCCTGATTTCAGACGGAAGGTCTCACTTGGACTTCCATGGGATCATCTCCCT GTGCTTCTTGATTTATTGGTGCTGTGTTTCTGTGTTTTGTTTTGTTACATGTCACAACCGTAGAGTTAGCTTAAATCAGAAAGAAGCCTC TCTGCCTTCTCCACCCTGTCTTACGAGCTGTGTTTTTGTTTTTACTACCCTAGAGGCAGAGAAGCGGTAGGGATGTCAGGGAATTTACTC ACTTCCACTTGAATCAACGAGAAGTGTTGAGAAACTTCCGTGGGTGCTCTGTGGAAAGAACCGAGGGTGTCAGGATGGAGCGGCCCACCC TCGCCCCGCGGCCTGCGCAGACTGCTGTCCTCCCCTTCAGGCCTGGCCACCAGCAGACTCCCATGAATTCCCATTTTCACAAAGCCCTCA AAATTTTGGGGGCGCTGAGGTTCAATTTTCTTCATCCCCAAATGAACTGTTAATTCACTGAAAGACATTTAGACTTTTGCAGCCAAGGAG CAGAGCCGGGCAAGCTCACTAAGGAAGCCTTAGCTTGGTCTCCTGCATGGGCCCCGGGGCCTCCAGATATGCCCACTGTCACCCTGGCAA GGGCCCCACACCCTCAGAATGAAAAGGATTGATTGGGTTATTGTCAGATACCCCCATTAGTTCCCCTAAAGTCTGCATCTGCAGAGCTCT TTTAGCTCATCTTTTTGTTGTCGGTGTTGTTTTTTGAGACAGAGTCTCCCTCTGTTGCCCAGGCTGGAGTGCAGTGGCGCGATCTCAGCT CATTGAAAGCTCCGCCTCCCGGGTTCAGGCCATTCTTCTGCCTCACCCTCCCCAGTAGCTGGGACTACAGGCGCCCACCACCACGCCCAC CTACTTTTTTGTATTTTTACTAGAGACGGGGTTTCACCATGTTAGCCAGGATGGTCTCGATCTCCTGACCTCATGATCCACCCGCCTCAG CCTTCCAAAGTGCTGGGATTACAGGAGTGAGCCACCGCGCCCGGCCTTTTAGCTTATCTTGAAATCCACCAGCCTACCTTCTGTTCATGG CACGAGCAGTGATTTCAGTGAAACCATCATAAACATTCTTGTTCCACTAGATGCTGATCACAGCAGTGATGTTTCTGTGTTAATACCCTG TACTGTCCCTCGAGTACTGTACCTTTGTGCTTGCACACGTGTCACTAACGCACGTGTTACCAGAGTCTCACGACCCTACGGTGCTATTTT TATTGGTGCAAAATTAAACCCTCTCAGCTGACCGTTGCTGGATAGGATCAGAATGCAGGAAAAGAGCCTCTATGGCAATACGATTGTCCT >50105_50105_2_LRRK1-ADAMTS17_LRRK1_chr15_101529603_ENST00000388948_ADAMTS17_chr15_100591936_ENST00000268070_length(amino acids)=589AA_BP=259 MLQGLMAGMSQRPPSMYWCVGPEESAVCPERAMETLNGAGDTGGKPSTRGGDPAARSRRTEGIRAAYRRGDRGGARDLLEEACDQCASQL EKGQLLSIPAAYGDLEMVRYLLSKRLVELPTEPTDDNPAVVAAYFGHTAVVQELLESLPGPCSPQRLLNWMLALACQRGHLGVVKLLVLT HGADPESYAVRKNEFPVIVRLPLYAAIKSGNEDIAIFLLRHGAYFCSYILLDSPDPSKHLLRKYFIEASPLPSSYPGKTVLLFHDQDYGI HYEYTVPVNRTAENQSEPEKPQDSLFIWTHSGWEGCSVQCGGGERRTIVSCTRIVNKTTTLVNDSDCPQASRPEPQVRRCNLHPCQSRWV AGPWSPCSATCEKGFQHREVTCVYQLQNGTHVATRPLYCPGPRPAAVQSCEGQDCLSIWEASEWSQCSASCGKGVWKRTVACTNSQGKCD ASTRPRAEEACEDYSGCYEWKTGDWSTCSSTCGKGLQSRVVQCMHKVTGRHGSECPALSKPAPYRQCYQEVCNDRINANTITSPRLAALT -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for LRRK1-ADAMTS17 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for LRRK1-ADAMTS17 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for LRRK1-ADAMTS17 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
