
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:LSR-CHMP1A (FusionGDB2 ID:50238) |
Fusion Gene Summary for LSR-CHMP1A |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: LSR-CHMP1A | Fusion gene ID: 50238 | Hgene | Tgene | Gene symbol | LSR | CHMP1A | Gene ID | 51599 | 5119 |
| Gene name | lipolysis stimulated lipoprotein receptor | charged multivesicular body protein 1A | |
| Synonyms | ILDR3|LISCH7 | CHMP1|PCH8|PCOLN3|PRSM1|VPS46-1|VPS46A | |
| Cytomap | 19q13.12 | 16q24.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | lipolysis-stimulated lipoprotein receptorLISCH proteinimmunoglobulin-like domain containing receptor 3lipolysis-stimulated remnantliver-specific bHLH-Zip transcription factor | charged multivesicular body protein 1acharged multivesicular body protein 1/chromatin modifying protein 1chromatin modifying protein 1Aprocollagen (type III) N-endopeptidaseprotease, metallo, 1, 33kDvacuolar protein sorting-associated protein 46-1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q86X29 | Q9HD42 | |
| Ensembl transtripts involved in fusion gene | ENST00000597933, ENST00000347609, ENST00000354900, ENST00000360798, ENST00000361790, ENST00000602122, ENST00000427250, | ENST00000253475, ENST00000397901, ENST00000535997, ENST00000550102, ENST00000547614, | |
| Fusion gene scores | * DoF score | 10 X 6 X 6=360 | 45 X 11 X 21=10395 |
| # samples | 11 | 48 | |
| ** MAII score | log2(11/360*10)=-1.71049338280502 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(48/10395*10)=-4.43671154213721 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: LSR [Title/Abstract] AND CHMP1A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | LSR(35749967)-CHMP1A(89713739), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | CHMP1A | GO:0007076 | mitotic chromosome condensation | 11559747 |
| Tgene | CHMP1A | GO:0016192 | vesicle-mediated transport | 11559748 |
| Tgene | CHMP1A | GO:0016458 | gene silencing | 11559747 |
| Tgene | CHMP1A | GO:0045892 | negative regulation of transcription, DNA-templated | 11559747 |
 Fusion gene breakpoints across LSR (5'-gene) Fusion gene breakpoints across LSR (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
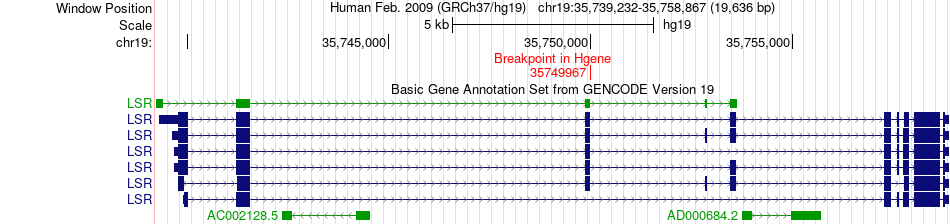 |
 Fusion gene breakpoints across CHMP1A (3'-gene) Fusion gene breakpoints across CHMP1A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
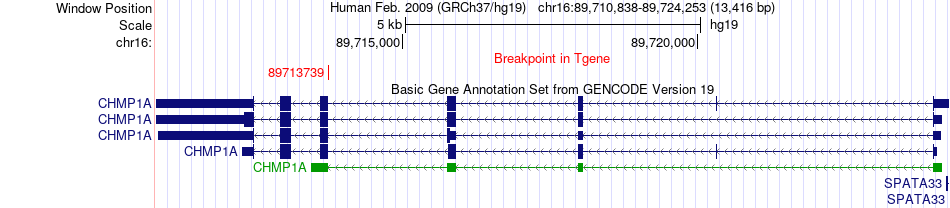 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-G9-A9S0 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
Top |
Fusion Gene ORF analysis for LSR-CHMP1A |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000597933 | ENST00000253475 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| 3UTR-3CDS | ENST00000597933 | ENST00000397901 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| 3UTR-3CDS | ENST00000597933 | ENST00000535997 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| 3UTR-3CDS | ENST00000597933 | ENST00000550102 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| 3UTR-5UTR | ENST00000597933 | ENST00000547614 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| 5CDS-5UTR | ENST00000347609 | ENST00000547614 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| 5CDS-5UTR | ENST00000354900 | ENST00000547614 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| 5CDS-5UTR | ENST00000360798 | ENST00000547614 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| 5CDS-5UTR | ENST00000361790 | ENST00000547614 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| 5CDS-5UTR | ENST00000602122 | ENST00000547614 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| Frame-shift | ENST00000347609 | ENST00000397901 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| Frame-shift | ENST00000347609 | ENST00000535997 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| Frame-shift | ENST00000347609 | ENST00000550102 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| Frame-shift | ENST00000354900 | ENST00000397901 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| Frame-shift | ENST00000354900 | ENST00000535997 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| Frame-shift | ENST00000354900 | ENST00000550102 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| Frame-shift | ENST00000360798 | ENST00000397901 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| Frame-shift | ENST00000360798 | ENST00000535997 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| Frame-shift | ENST00000360798 | ENST00000550102 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| Frame-shift | ENST00000361790 | ENST00000397901 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| Frame-shift | ENST00000361790 | ENST00000535997 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| Frame-shift | ENST00000361790 | ENST00000550102 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| Frame-shift | ENST00000602122 | ENST00000397901 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| Frame-shift | ENST00000602122 | ENST00000535997 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| Frame-shift | ENST00000602122 | ENST00000550102 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| In-frame | ENST00000347609 | ENST00000253475 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| In-frame | ENST00000354900 | ENST00000253475 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| In-frame | ENST00000360798 | ENST00000253475 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| In-frame | ENST00000361790 | ENST00000253475 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| In-frame | ENST00000602122 | ENST00000253475 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| intron-3CDS | ENST00000427250 | ENST00000253475 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| intron-3CDS | ENST00000427250 | ENST00000397901 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| intron-3CDS | ENST00000427250 | ENST00000535997 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| intron-3CDS | ENST00000427250 | ENST00000550102 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
| intron-5UTR | ENST00000427250 | ENST00000547614 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000602122 | LSR | chr19 | 35749967 | + | ENST00000253475 | CHMP1A | chr16 | 89713739 | - | 3172 | 1205 | 136 | 1695 | 519 |
| ENST00000361790 | LSR | chr19 | 35749967 | + | ENST00000253475 | CHMP1A | chr16 | 89713739 | - | 2844 | 877 | 108 | 1367 | 419 |
| ENST00000360798 | LSR | chr19 | 35749967 | + | ENST00000253475 | CHMP1A | chr16 | 89713739 | - | 2799 | 832 | 63 | 1322 | 419 |
| ENST00000354900 | LSR | chr19 | 35749967 | + | ENST00000253475 | CHMP1A | chr16 | 89713739 | - | 2792 | 825 | 56 | 1315 | 419 |
| ENST00000347609 | LSR | chr19 | 35749967 | + | ENST00000253475 | CHMP1A | chr16 | 89713739 | - | 2574 | 607 | 0 | 1097 | 365 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000602122 | ENST00000253475 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - | 0.17064476 | 0.82935524 |
| ENST00000361790 | ENST00000253475 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - | 0.18320368 | 0.8167963 |
| ENST00000360798 | ENST00000253475 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - | 0.17642517 | 0.82357484 |
| ENST00000354900 | ENST00000253475 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - | 0.17758946 | 0.8224105 |
| ENST00000347609 | ENST00000253475 | LSR | chr19 | 35749967 | + | CHMP1A | chr16 | 89713739 | - | 0.1098253 | 0.89017475 |
Top |
Fusion Genomic Features for LSR-CHMP1A |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
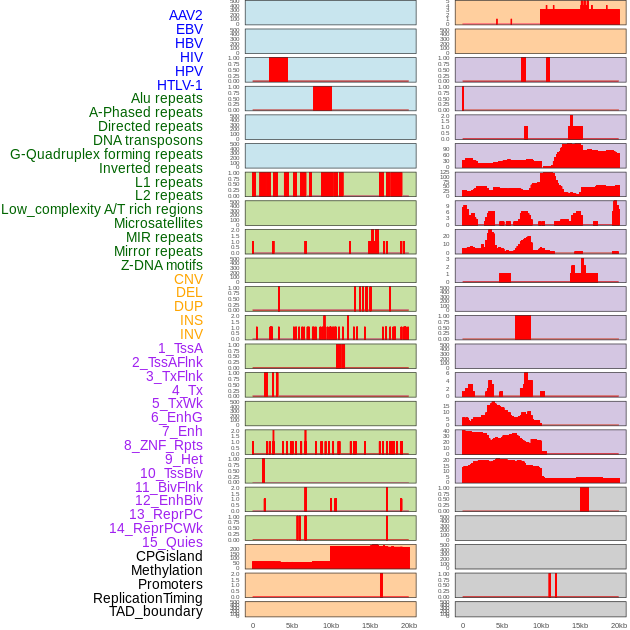 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for LSR-CHMP1A |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:35749967/chr16:89713739) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| LSR | CHMP1A |
| FUNCTION: Probable role in the clearance of triglyceride-rich lipoprotein from blood. Binds chylomicrons, LDL and VLDL in presence of free fatty acids and allows their subsequent uptake in the cells (By similarity). {ECO:0000250}. | FUNCTION: Probable peripherally associated component of the endosomal sorting required for transport complex III (ESCRT-III) which is involved in multivesicular bodies (MVBs) formation and sorting of endosomal cargo proteins into MVBs. MVBs contain intraluminal vesicles (ILVs) that are generated by invagination and scission from the limiting membrane of the endosome and mostly are delivered to lysosomes enabling degradation of membrane proteins, such as stimulated growth factor receptors, lysosomal enzymes and lipids. The MVB pathway appears to require the sequential function of ESCRT-O, -I,-II and -III complexes. ESCRT-III proteins mostly dissociate from the invaginating membrane before the ILV is released. The ESCRT machinery also functions in topologically equivalent membrane fission events, such as the terminal stages of cytokinesis and the budding of enveloped viruses (HIV-1 and other lentiviruses). ESCRT-III proteins are believed to mediate the necessary vesicle extrusion and/or membrane fission activities, possibly in conjunction with the AAA ATPase VPS4. Involved in cytokinesis. Involved in recruiting VPS4A and/or VPS4B to the midbody of dividing cells. May also be involved in chromosome condensation. Targets the Polycomb group (PcG) protein BMI1/PCGF4 to regions of condensed chromatin. May play a role in stable cell cycle progression and in PcG gene silencing. {ECO:0000269|PubMed:11559747, ECO:0000269|PubMed:11559748, ECO:0000269|PubMed:19129479, ECO:0000269|PubMed:23045692}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LSR | chr19:35749967 | chr16:89713739 | ENST00000354900 | + | 3 | 9 | 86_234 | 239 | 631.0 | Domain | Note=Ig-like V-type |
| Hgene | LSR | chr19:35749967 | chr16:89713739 | ENST00000360798 | + | 3 | 8 | 86_234 | 239 | 582.0 | Domain | Note=Ig-like V-type |
| Hgene | LSR | chr19:35749967 | chr16:89713739 | ENST00000361790 | + | 3 | 10 | 86_234 | 239 | 650.0 | Domain | Note=Ig-like V-type |
| Hgene | LSR | chr19:35749967 | chr16:89713739 | ENST00000602122 | + | 3 | 9 | 86_234 | 239 | 630.0 | Domain | Note=Ig-like V-type |
| Tgene | CHMP1A | chr19:35749967 | chr16:89713739 | ENST00000397901 | 3 | 7 | 102_124 | 84 | 197.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | CHMP1A | chr19:35749967 | chr16:89713739 | ENST00000397901 | 3 | 7 | 185_195 | 84 | 197.0 | Motif | Note=MIT-interacting motif |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LSR | chr19:35749967 | chr16:89713739 | ENST00000347609 | + | 3 | 9 | 280_304 | 202 | 592.0 | Compositional bias | Note=Cys-rich |
| Hgene | LSR | chr19:35749967 | chr16:89713739 | ENST00000354900 | + | 3 | 9 | 280_304 | 239 | 631.0 | Compositional bias | Note=Cys-rich |
| Hgene | LSR | chr19:35749967 | chr16:89713739 | ENST00000360798 | + | 3 | 8 | 280_304 | 239 | 582.0 | Compositional bias | Note=Cys-rich |
| Hgene | LSR | chr19:35749967 | chr16:89713739 | ENST00000361790 | + | 3 | 10 | 280_304 | 239 | 650.0 | Compositional bias | Note=Cys-rich |
| Hgene | LSR | chr19:35749967 | chr16:89713739 | ENST00000602122 | + | 3 | 9 | 280_304 | 239 | 630.0 | Compositional bias | Note=Cys-rich |
| Hgene | LSR | chr19:35749967 | chr16:89713739 | ENST00000347609 | + | 3 | 9 | 86_234 | 202 | 592.0 | Domain | Note=Ig-like V-type |
| Hgene | LSR | chr19:35749967 | chr16:89713739 | ENST00000347609 | + | 3 | 9 | 1_259 | 202 | 592.0 | Topological domain | Extracellular |
| Hgene | LSR | chr19:35749967 | chr16:89713739 | ENST00000347609 | + | 3 | 9 | 281_649 | 202 | 592.0 | Topological domain | Cytoplasmic |
| Hgene | LSR | chr19:35749967 | chr16:89713739 | ENST00000354900 | + | 3 | 9 | 1_259 | 239 | 631.0 | Topological domain | Extracellular |
| Hgene | LSR | chr19:35749967 | chr16:89713739 | ENST00000354900 | + | 3 | 9 | 281_649 | 239 | 631.0 | Topological domain | Cytoplasmic |
| Hgene | LSR | chr19:35749967 | chr16:89713739 | ENST00000360798 | + | 3 | 8 | 1_259 | 239 | 582.0 | Topological domain | Extracellular |
| Hgene | LSR | chr19:35749967 | chr16:89713739 | ENST00000360798 | + | 3 | 8 | 281_649 | 239 | 582.0 | Topological domain | Cytoplasmic |
| Hgene | LSR | chr19:35749967 | chr16:89713739 | ENST00000361790 | + | 3 | 10 | 1_259 | 239 | 650.0 | Topological domain | Extracellular |
| Hgene | LSR | chr19:35749967 | chr16:89713739 | ENST00000361790 | + | 3 | 10 | 281_649 | 239 | 650.0 | Topological domain | Cytoplasmic |
| Hgene | LSR | chr19:35749967 | chr16:89713739 | ENST00000602122 | + | 3 | 9 | 1_259 | 239 | 630.0 | Topological domain | Extracellular |
| Hgene | LSR | chr19:35749967 | chr16:89713739 | ENST00000602122 | + | 3 | 9 | 281_649 | 239 | 630.0 | Topological domain | Cytoplasmic |
| Hgene | LSR | chr19:35749967 | chr16:89713739 | ENST00000347609 | + | 3 | 9 | 260_280 | 202 | 592.0 | Transmembrane | Helical |
| Hgene | LSR | chr19:35749967 | chr16:89713739 | ENST00000354900 | + | 3 | 9 | 260_280 | 239 | 631.0 | Transmembrane | Helical |
| Hgene | LSR | chr19:35749967 | chr16:89713739 | ENST00000360798 | + | 3 | 8 | 260_280 | 239 | 582.0 | Transmembrane | Helical |
| Hgene | LSR | chr19:35749967 | chr16:89713739 | ENST00000361790 | + | 3 | 10 | 260_280 | 239 | 650.0 | Transmembrane | Helical |
| Hgene | LSR | chr19:35749967 | chr16:89713739 | ENST00000602122 | + | 3 | 9 | 260_280 | 239 | 630.0 | Transmembrane | Helical |
| Tgene | CHMP1A | chr19:35749967 | chr16:89713739 | ENST00000397901 | 3 | 7 | 5_47 | 84 | 197.0 | Coiled coil | Ontology_term=ECO:0000255 |
Top |
Fusion Gene Sequence for LSR-CHMP1A |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >50238_50238_1_LSR-CHMP1A_LSR_chr19_35749967_ENST00000347609_CHMP1A_chr16_89713739_ENST00000253475_length(transcript)=2574nt_BP=607nt ATGCAACAGGACGGACTTGGAGTAGGGACAAGGAACGGAAGTGGGAAGGGGAGGAGCGTGCACCCCTCCTGGCCTTGGTGCGCGCCGCGC CCCCTAAGGTACTTTGGAAGGGACGCGCGGGCCAGACGCGCCCAGACGGCCGCGATGGCGCTGGCCATCCAGGTGACCGTGTCCAACCCC TACCACGTGGTGATCCTCTTCCAGCCTGTGACCCTGCCCTGTACCTACCAGATGACCTCGACCCCCACGCAACCCATCGTCATCTGGAAG TACAAGTCTTTCTGCCGGGACCGCATCGCCGATGCCTTCTCCCCGGCCAGCGTCGACAACCAGCTCAATGCCCAGCTGGCAGCCGGGAAC CCAGGCTACAACCCCTACGTTGAGTGCCAGGACAGCGTGCGCACCGTCAGGGTCGTGGCCACCAAGCAGGGCAACGCTGTGACCCTGGGA GATTACTACCAGGGCCGGAGGATTACCATCACCGGAAATGCTGACCTGACCTTTGACCAGACGGCGTGGGGGGACAGTGGTGTGTATTAC TGCTCCGTGGTCTCAGCCCAGGACCTCCAGGGGAACAATGAGGCCTACGCAGAGCTCATCGTCCTTGGTGACCAAGAATATGGCCCAGGT GACCAAAGCCCTGGACAAGGCCCTGAGCACCATGGACCTGCAGAAGGTCTCCTCAGTGATGGACAGGTTCGAGCAGCAGGTGCAGAACCT GGACGTCCATACATCGGTGATGGAGGACTCCATGAGCTCGGCCACCACCCTGACCACGCCGCAGGAGCAGGTGGACAGCCTCATCATGCA GATCGCCGAGGAGAATGGCCTGGAGGTGCTGGACCAGCTCAGCCAGCTGCCCGAGGGCGCCTCTGCCGTGGGCGAGAGCTCTGTGCGCAG CCAGGAGGACCAGCTGTCACGGAGGTTGGCCGCCTTGAGGAACTAGCCGTGCCCCGCCGGTGTGCACCGCCTCTGCCCCGTGATGTGCTG GAAGGCTCCTGTCCTCTCCCCACCGCGTCTTGCCTTTGTGCTGACCCCGCGGGGCTGCGGCCGGCAGCCACTCTGCGTCTCTCACCTGCC AGGCCTGCGTGGCCTTAGGGTTGTTCCTGTTCTTTTAGGTTGGGCGGTGGGTCTGTGTCCTGGTGTTGAGTTTCTGCAAATTTCTGGGGG TGATTTCTGTGACTCTGGGCCCACAGCGGGGAGGCCAAGAGGGGCCCTGTGGACTTTCACCCAGCACTGTGGGGGCCTTCAGACTCTGGG GCAGCAGACATGCTGCTTCCCATCAGCCAGAGGGGGTCAGGGCTGCCCTGTTGCCAAACAACTCCCTGAGGCCTCTCCGCACCACCTCAG CGGGCAGGAGGTCCCACCATGTGGACAGACATAGCCCAAGGAGGCACCACAGGTCTATGTGTGCTGGGGGATGTCAGGTGCCACCCAACG CTGTCCTGGTGGTATTTACAATGACATCCTCCTCCTCCATCACTCCAGGGGTGGTGTCTCGGCCGCCCCTACCAGCTGGCTGAGCCCCCT GGCCTCCTGCGCTCCCTCACTTCCCTCAGTTCCCAAAGCTGCCCAGTCCATGGGGACAGAACCGTCACTCAGATCCACATTCAAGTGTGC CCACCCTGCAGTCTTCATCCTCACTCAGCTGCTGCCTCTGGAGGTGCCTTTGGCCACATGTGCTGTGCTGTTTGTCTCCTCGACAGGGAG CCTGTCCACCAGCAGGCTGCGGTCCCAGCGGGTGCGTCTGCAGCTCCTCCCCTTGGGCAGCCTGGTTCTCCCGGAGGACCTTTCCTTGGG GCCCTGCTTCATGACGATGCTGCCTGTGTCACCCTCTACCATCTGTAAACAACTGGGTGCCTTCCCCGACCACACCCCAATGCCTTCCCA GCTTGGAAGCCAAGGCAGCTGATGAAGGGAGCTCAGGAGAGCCGTCTTCAGCTGGGCTGGGGTTGGGGCTGCTGTGAGGAAAACCTGCCA TTGTGGCCCTGGAGAGTCACCAGCAGCTCTTGGGAAGGACTTGCTGGGAGGCTGAGAGAGGCTTTGGGCACAGCCTGCTGTCTTTTCCAT TTCCTAAAGTTTACTTCATTGTCTTGAGGCTTCCAGGTTTTGTTTTTGTTTTTGCCAAAGTAGAAAAGGCAGGTGGTGGGCGGCTGGCAG GGAGTGCGGGTCCCCGCCCCTCTTCAGTCCTGCCCTCCCCTCCTCAGTCCTGCCCACCCCGTGCAGCCCATGCTGAGGCTGCAGTGGTGT CGTGGGTGTTACGTGCAGGAACGTGGAGACCCTGACGTGGGCTCACTGCGTTTGGTTTTCTTTTCAGAACTTGGGAGCCCCCAGGGAGGG GCTAGTGTTGGTAGGTCCTAGACGTGGTTCCCTCCAGCCTCCCCAAAATCAACCCTGGTGTTGAGAGAACGTCCTTCTGTCCATCGTGGG TAACAGCCTTGGGGAGGGTGCAGAGCTCTGCAGAGCCATGGGCCAGGTGGGGCTGCCTCAGTCCTGTCCCCTTGGGCACTGAGGAGAGGG >50238_50238_1_LSR-CHMP1A_LSR_chr19_35749967_ENST00000347609_CHMP1A_chr16_89713739_ENST00000253475_length(amino acids)=365AA_BP=200 MQQDGLGVGTRNGSGKGRSVHPSWPWCAPRPLRYFGRDARARRAQTAAMALAIQVTVSNPYHVVILFQPVTLPCTYQMTSTPTQPIVIWK YKSFCRDRIADAFSPASVDNQLNAQLAAGNPGYNPYVECQDSVRTVRVVATKQGNAVTLGDYYQGRRITITGNADLTFDQTAWGDSGVYY CSVVSAQDLQGNNEAYAELIVLGDQEYGPGDQSPGQGPEHHGPAEGLLSDGQVRAAGAEPGRPYIGDGGLHELGHHPDHAAGAGGQPHHA DRRGEWPGGAGPAQPAARGRLCRGRELCAQPGGPAVTEVGRLEELAVPRRCAPPLPRDVLEGSCPLPTASCLCADPAGLRPAATLRLSPA -------------------------------------------------------------- >50238_50238_2_LSR-CHMP1A_LSR_chr19_35749967_ENST00000354900_CHMP1A_chr16_89713739_ENST00000253475_length(transcript)=2792nt_BP=825nt CGGGTGGAGTGTGGCTCGGAGGACCGCGGCGGGTCAAGCACCTTTCTCCCCCATATCTGAAAGCATGCCCTTTGTCCACGTCGTTTACGC TCATTAAAACTTCCAGAATGCAACAGGACGGACTTGGAGTAGGGACAAGGAACGGAAGTGGGAAGGGGAGGAGCGTGCACCCCTCCTGGC CTTGGTGCGCGCCGCGCCCCCTAAGGTACTTTGGAAGGGACGCGCGGGCCAGACGCGCCCAGACGGCCGCGATGGCGCTGTTGGCCGGCG GGCTCTCCAGAGGGCTGGGCTCCCACCCGGCCGCCGCAGGCCGGGACGCGGTCGTCTTCGTGTGGCTTCTGCTTAGCACCTGGTGCACAG CTCCTGCCAGGGCCATCCAGGTGACCGTGTCCAACCCCTACCACGTGGTGATCCTCTTCCAGCCTGTGACCCTGCCCTGTACCTACCAGA TGACCTCGACCCCCACGCAACCCATCGTCATCTGGAAGTACAAGTCTTTCTGCCGGGACCGCATCGCCGATGCCTTCTCCCCGGCCAGCG TCGACAACCAGCTCAATGCCCAGCTGGCAGCCGGGAACCCAGGCTACAACCCCTACGTTGAGTGCCAGGACAGCGTGCGCACCGTCAGGG TCGTGGCCACCAAGCAGGGCAACGCTGTGACCCTGGGAGATTACTACCAGGGCCGGAGGATTACCATCACCGGAAATGCTGACCTGACCT TTGACCAGACGGCGTGGGGGGACAGTGGTGTGTATTACTGCTCCGTGGTCTCAGCCCAGGACCTCCAGGGGAACAATGAGGCCTACGCAG AGCTCATCGTCCTTGGTGACCAAGAATATGGCCCAGGTGACCAAAGCCCTGGACAAGGCCCTGAGCACCATGGACCTGCAGAAGGTCTCC TCAGTGATGGACAGGTTCGAGCAGCAGGTGCAGAACCTGGACGTCCATACATCGGTGATGGAGGACTCCATGAGCTCGGCCACCACCCTG ACCACGCCGCAGGAGCAGGTGGACAGCCTCATCATGCAGATCGCCGAGGAGAATGGCCTGGAGGTGCTGGACCAGCTCAGCCAGCTGCCC GAGGGCGCCTCTGCCGTGGGCGAGAGCTCTGTGCGCAGCCAGGAGGACCAGCTGTCACGGAGGTTGGCCGCCTTGAGGAACTAGCCGTGC CCCGCCGGTGTGCACCGCCTCTGCCCCGTGATGTGCTGGAAGGCTCCTGTCCTCTCCCCACCGCGTCTTGCCTTTGTGCTGACCCCGCGG GGCTGCGGCCGGCAGCCACTCTGCGTCTCTCACCTGCCAGGCCTGCGTGGCCTTAGGGTTGTTCCTGTTCTTTTAGGTTGGGCGGTGGGT CTGTGTCCTGGTGTTGAGTTTCTGCAAATTTCTGGGGGTGATTTCTGTGACTCTGGGCCCACAGCGGGGAGGCCAAGAGGGGCCCTGTGG ACTTTCACCCAGCACTGTGGGGGCCTTCAGACTCTGGGGCAGCAGACATGCTGCTTCCCATCAGCCAGAGGGGGTCAGGGCTGCCCTGTT GCCAAACAACTCCCTGAGGCCTCTCCGCACCACCTCAGCGGGCAGGAGGTCCCACCATGTGGACAGACATAGCCCAAGGAGGCACCACAG GTCTATGTGTGCTGGGGGATGTCAGGTGCCACCCAACGCTGTCCTGGTGGTATTTACAATGACATCCTCCTCCTCCATCACTCCAGGGGT GGTGTCTCGGCCGCCCCTACCAGCTGGCTGAGCCCCCTGGCCTCCTGCGCTCCCTCACTTCCCTCAGTTCCCAAAGCTGCCCAGTCCATG GGGACAGAACCGTCACTCAGATCCACATTCAAGTGTGCCCACCCTGCAGTCTTCATCCTCACTCAGCTGCTGCCTCTGGAGGTGCCTTTG GCCACATGTGCTGTGCTGTTTGTCTCCTCGACAGGGAGCCTGTCCACCAGCAGGCTGCGGTCCCAGCGGGTGCGTCTGCAGCTCCTCCCC TTGGGCAGCCTGGTTCTCCCGGAGGACCTTTCCTTGGGGCCCTGCTTCATGACGATGCTGCCTGTGTCACCCTCTACCATCTGTAAACAA CTGGGTGCCTTCCCCGACCACACCCCAATGCCTTCCCAGCTTGGAAGCCAAGGCAGCTGATGAAGGGAGCTCAGGAGAGCCGTCTTCAGC TGGGCTGGGGTTGGGGCTGCTGTGAGGAAAACCTGCCATTGTGGCCCTGGAGAGTCACCAGCAGCTCTTGGGAAGGACTTGCTGGGAGGC TGAGAGAGGCTTTGGGCACAGCCTGCTGTCTTTTCCATTTCCTAAAGTTTACTTCATTGTCTTGAGGCTTCCAGGTTTTGTTTTTGTTTT TGCCAAAGTAGAAAAGGCAGGTGGTGGGCGGCTGGCAGGGAGTGCGGGTCCCCGCCCCTCTTCAGTCCTGCCCTCCCCTCCTCAGTCCTG CCCACCCCGTGCAGCCCATGCTGAGGCTGCAGTGGTGTCGTGGGTGTTACGTGCAGGAACGTGGAGACCCTGACGTGGGCTCACTGCGTT TGGTTTTCTTTTCAGAACTTGGGAGCCCCCAGGGAGGGGCTAGTGTTGGTAGGTCCTAGACGTGGTTCCCTCCAGCCTCCCCAAAATCAA CCCTGGTGTTGAGAGAACGTCCTTCTGTCCATCGTGGGTAACAGCCTTGGGGAGGGTGCAGAGCTCTGCAGAGCCATGGGCCAGGTGGGG CTGCCTCAGTCCTGTCCCCTTGGGCACTGAGGAGAGGGGCCCATTCACCTTTCTCCTAGAATGCTGTTGTAAATAAACAAATGGATCCCT >50238_50238_2_LSR-CHMP1A_LSR_chr19_35749967_ENST00000354900_CHMP1A_chr16_89713739_ENST00000253475_length(amino acids)=419AA_BP=254 MKACPLSTSFTLIKTSRMQQDGLGVGTRNGSGKGRSVHPSWPWCAPRPLRYFGRDARARRAQTAAMALLAGGLSRGLGSHPAAAGRDAVV FVWLLLSTWCTAPARAIQVTVSNPYHVVILFQPVTLPCTYQMTSTPTQPIVIWKYKSFCRDRIADAFSPASVDNQLNAQLAAGNPGYNPY VECQDSVRTVRVVATKQGNAVTLGDYYQGRRITITGNADLTFDQTAWGDSGVYYCSVVSAQDLQGNNEAYAELIVLGDQEYGPGDQSPGQ GPEHHGPAEGLLSDGQVRAAGAEPGRPYIGDGGLHELGHHPDHAAGAGGQPHHADRRGEWPGGAGPAQPAARGRLCRGRELCAQPGGPAV -------------------------------------------------------------- >50238_50238_3_LSR-CHMP1A_LSR_chr19_35749967_ENST00000360798_CHMP1A_chr16_89713739_ENST00000253475_length(transcript)=2799nt_BP=832nt CGCGCAGCGGGTGGAGTGTGGCTCGGAGGACCGCGGCGGGTCAAGCACCTTTCTCCCCCATATCTGAAAGCATGCCCTTTGTCCACGTCG TTTACGCTCATTAAAACTTCCAGAATGCAACAGGACGGACTTGGAGTAGGGACAAGGAACGGAAGTGGGAAGGGGAGGAGCGTGCACCCC TCCTGGCCTTGGTGCGCGCCGCGCCCCCTAAGGTACTTTGGAAGGGACGCGCGGGCCAGACGCGCCCAGACGGCCGCGATGGCGCTGTTG GCCGGCGGGCTCTCCAGAGGGCTGGGCTCCCACCCGGCCGCCGCAGGCCGGGACGCGGTCGTCTTCGTGTGGCTTCTGCTTAGCACCTGG TGCACAGCTCCTGCCAGGGCCATCCAGGTGACCGTGTCCAACCCCTACCACGTGGTGATCCTCTTCCAGCCTGTGACCCTGCCCTGTACC TACCAGATGACCTCGACCCCCACGCAACCCATCGTCATCTGGAAGTACAAGTCTTTCTGCCGGGACCGCATCGCCGATGCCTTCTCCCCG GCCAGCGTCGACAACCAGCTCAATGCCCAGCTGGCAGCCGGGAACCCAGGCTACAACCCCTACGTTGAGTGCCAGGACAGCGTGCGCACC GTCAGGGTCGTGGCCACCAAGCAGGGCAACGCTGTGACCCTGGGAGATTACTACCAGGGCCGGAGGATTACCATCACCGGAAATGCTGAC CTGACCTTTGACCAGACGGCGTGGGGGGACAGTGGTGTGTATTACTGCTCCGTGGTCTCAGCCCAGGACCTCCAGGGGAACAATGAGGCC TACGCAGAGCTCATCGTCCTTGGTGACCAAGAATATGGCCCAGGTGACCAAAGCCCTGGACAAGGCCCTGAGCACCATGGACCTGCAGAA GGTCTCCTCAGTGATGGACAGGTTCGAGCAGCAGGTGCAGAACCTGGACGTCCATACATCGGTGATGGAGGACTCCATGAGCTCGGCCAC CACCCTGACCACGCCGCAGGAGCAGGTGGACAGCCTCATCATGCAGATCGCCGAGGAGAATGGCCTGGAGGTGCTGGACCAGCTCAGCCA GCTGCCCGAGGGCGCCTCTGCCGTGGGCGAGAGCTCTGTGCGCAGCCAGGAGGACCAGCTGTCACGGAGGTTGGCCGCCTTGAGGAACTA GCCGTGCCCCGCCGGTGTGCACCGCCTCTGCCCCGTGATGTGCTGGAAGGCTCCTGTCCTCTCCCCACCGCGTCTTGCCTTTGTGCTGAC CCCGCGGGGCTGCGGCCGGCAGCCACTCTGCGTCTCTCACCTGCCAGGCCTGCGTGGCCTTAGGGTTGTTCCTGTTCTTTTAGGTTGGGC GGTGGGTCTGTGTCCTGGTGTTGAGTTTCTGCAAATTTCTGGGGGTGATTTCTGTGACTCTGGGCCCACAGCGGGGAGGCCAAGAGGGGC CCTGTGGACTTTCACCCAGCACTGTGGGGGCCTTCAGACTCTGGGGCAGCAGACATGCTGCTTCCCATCAGCCAGAGGGGGTCAGGGCTG CCCTGTTGCCAAACAACTCCCTGAGGCCTCTCCGCACCACCTCAGCGGGCAGGAGGTCCCACCATGTGGACAGACATAGCCCAAGGAGGC ACCACAGGTCTATGTGTGCTGGGGGATGTCAGGTGCCACCCAACGCTGTCCTGGTGGTATTTACAATGACATCCTCCTCCTCCATCACTC CAGGGGTGGTGTCTCGGCCGCCCCTACCAGCTGGCTGAGCCCCCTGGCCTCCTGCGCTCCCTCACTTCCCTCAGTTCCCAAAGCTGCCCA GTCCATGGGGACAGAACCGTCACTCAGATCCACATTCAAGTGTGCCCACCCTGCAGTCTTCATCCTCACTCAGCTGCTGCCTCTGGAGGT GCCTTTGGCCACATGTGCTGTGCTGTTTGTCTCCTCGACAGGGAGCCTGTCCACCAGCAGGCTGCGGTCCCAGCGGGTGCGTCTGCAGCT CCTCCCCTTGGGCAGCCTGGTTCTCCCGGAGGACCTTTCCTTGGGGCCCTGCTTCATGACGATGCTGCCTGTGTCACCCTCTACCATCTG TAAACAACTGGGTGCCTTCCCCGACCACACCCCAATGCCTTCCCAGCTTGGAAGCCAAGGCAGCTGATGAAGGGAGCTCAGGAGAGCCGT CTTCAGCTGGGCTGGGGTTGGGGCTGCTGTGAGGAAAACCTGCCATTGTGGCCCTGGAGAGTCACCAGCAGCTCTTGGGAAGGACTTGCT GGGAGGCTGAGAGAGGCTTTGGGCACAGCCTGCTGTCTTTTCCATTTCCTAAAGTTTACTTCATTGTCTTGAGGCTTCCAGGTTTTGTTT TTGTTTTTGCCAAAGTAGAAAAGGCAGGTGGTGGGCGGCTGGCAGGGAGTGCGGGTCCCCGCCCCTCTTCAGTCCTGCCCTCCCCTCCTC AGTCCTGCCCACCCCGTGCAGCCCATGCTGAGGCTGCAGTGGTGTCGTGGGTGTTACGTGCAGGAACGTGGAGACCCTGACGTGGGCTCA CTGCGTTTGGTTTTCTTTTCAGAACTTGGGAGCCCCCAGGGAGGGGCTAGTGTTGGTAGGTCCTAGACGTGGTTCCCTCCAGCCTCCCCA AAATCAACCCTGGTGTTGAGAGAACGTCCTTCTGTCCATCGTGGGTAACAGCCTTGGGGAGGGTGCAGAGCTCTGCAGAGCCATGGGCCA GGTGGGGCTGCCTCAGTCCTGTCCCCTTGGGCACTGAGGAGAGGGGCCCATTCACCTTTCTCCTAGAATGCTGTTGTAAATAAACAAATG >50238_50238_3_LSR-CHMP1A_LSR_chr19_35749967_ENST00000360798_CHMP1A_chr16_89713739_ENST00000253475_length(amino acids)=419AA_BP=254 MKACPLSTSFTLIKTSRMQQDGLGVGTRNGSGKGRSVHPSWPWCAPRPLRYFGRDARARRAQTAAMALLAGGLSRGLGSHPAAAGRDAVV FVWLLLSTWCTAPARAIQVTVSNPYHVVILFQPVTLPCTYQMTSTPTQPIVIWKYKSFCRDRIADAFSPASVDNQLNAQLAAGNPGYNPY VECQDSVRTVRVVATKQGNAVTLGDYYQGRRITITGNADLTFDQTAWGDSGVYYCSVVSAQDLQGNNEAYAELIVLGDQEYGPGDQSPGQ GPEHHGPAEGLLSDGQVRAAGAEPGRPYIGDGGLHELGHHPDHAAGAGGQPHHADRRGEWPGGAGPAQPAARGRLCRGRELCAQPGGPAV -------------------------------------------------------------- >50238_50238_4_LSR-CHMP1A_LSR_chr19_35749967_ENST00000361790_CHMP1A_chr16_89713739_ENST00000253475_length(transcript)=2844nt_BP=877nt CTCGGCTCCCACCCTTGTGTACCTGGGCCGAACCATTCACCGGAGCGCGCAGCGGGTGGAGTGTGGCTCGGAGGACCGCGGCGGGTCAAG CACCTTTCTCCCCCATATCTGAAAGCATGCCCTTTGTCCACGTCGTTTACGCTCATTAAAACTTCCAGAATGCAACAGGACGGACTTGGA GTAGGGACAAGGAACGGAAGTGGGAAGGGGAGGAGCGTGCACCCCTCCTGGCCTTGGTGCGCGCCGCGCCCCCTAAGGTACTTTGGAAGG GACGCGCGGGCCAGACGCGCCCAGACGGCCGCGATGGCGCTGTTGGCCGGCGGGCTCTCCAGAGGGCTGGGCTCCCACCCGGCCGCCGCA GGCCGGGACGCGGTCGTCTTCGTGTGGCTTCTGCTTAGCACCTGGTGCACAGCTCCTGCCAGGGCCATCCAGGTGACCGTGTCCAACCCC TACCACGTGGTGATCCTCTTCCAGCCTGTGACCCTGCCCTGTACCTACCAGATGACCTCGACCCCCACGCAACCCATCGTCATCTGGAAG TACAAGTCTTTCTGCCGGGACCGCATCGCCGATGCCTTCTCCCCGGCCAGCGTCGACAACCAGCTCAATGCCCAGCTGGCAGCCGGGAAC CCAGGCTACAACCCCTACGTTGAGTGCCAGGACAGCGTGCGCACCGTCAGGGTCGTGGCCACCAAGCAGGGCAACGCTGTGACCCTGGGA GATTACTACCAGGGCCGGAGGATTACCATCACCGGAAATGCTGACCTGACCTTTGACCAGACGGCGTGGGGGGACAGTGGTGTGTATTAC TGCTCCGTGGTCTCAGCCCAGGACCTCCAGGGGAACAATGAGGCCTACGCAGAGCTCATCGTCCTTGGTGACCAAGAATATGGCCCAGGT GACCAAAGCCCTGGACAAGGCCCTGAGCACCATGGACCTGCAGAAGGTCTCCTCAGTGATGGACAGGTTCGAGCAGCAGGTGCAGAACCT GGACGTCCATACATCGGTGATGGAGGACTCCATGAGCTCGGCCACCACCCTGACCACGCCGCAGGAGCAGGTGGACAGCCTCATCATGCA GATCGCCGAGGAGAATGGCCTGGAGGTGCTGGACCAGCTCAGCCAGCTGCCCGAGGGCGCCTCTGCCGTGGGCGAGAGCTCTGTGCGCAG CCAGGAGGACCAGCTGTCACGGAGGTTGGCCGCCTTGAGGAACTAGCCGTGCCCCGCCGGTGTGCACCGCCTCTGCCCCGTGATGTGCTG GAAGGCTCCTGTCCTCTCCCCACCGCGTCTTGCCTTTGTGCTGACCCCGCGGGGCTGCGGCCGGCAGCCACTCTGCGTCTCTCACCTGCC AGGCCTGCGTGGCCTTAGGGTTGTTCCTGTTCTTTTAGGTTGGGCGGTGGGTCTGTGTCCTGGTGTTGAGTTTCTGCAAATTTCTGGGGG TGATTTCTGTGACTCTGGGCCCACAGCGGGGAGGCCAAGAGGGGCCCTGTGGACTTTCACCCAGCACTGTGGGGGCCTTCAGACTCTGGG GCAGCAGACATGCTGCTTCCCATCAGCCAGAGGGGGTCAGGGCTGCCCTGTTGCCAAACAACTCCCTGAGGCCTCTCCGCACCACCTCAG CGGGCAGGAGGTCCCACCATGTGGACAGACATAGCCCAAGGAGGCACCACAGGTCTATGTGTGCTGGGGGATGTCAGGTGCCACCCAACG CTGTCCTGGTGGTATTTACAATGACATCCTCCTCCTCCATCACTCCAGGGGTGGTGTCTCGGCCGCCCCTACCAGCTGGCTGAGCCCCCT GGCCTCCTGCGCTCCCTCACTTCCCTCAGTTCCCAAAGCTGCCCAGTCCATGGGGACAGAACCGTCACTCAGATCCACATTCAAGTGTGC CCACCCTGCAGTCTTCATCCTCACTCAGCTGCTGCCTCTGGAGGTGCCTTTGGCCACATGTGCTGTGCTGTTTGTCTCCTCGACAGGGAG CCTGTCCACCAGCAGGCTGCGGTCCCAGCGGGTGCGTCTGCAGCTCCTCCCCTTGGGCAGCCTGGTTCTCCCGGAGGACCTTTCCTTGGG GCCCTGCTTCATGACGATGCTGCCTGTGTCACCCTCTACCATCTGTAAACAACTGGGTGCCTTCCCCGACCACACCCCAATGCCTTCCCA GCTTGGAAGCCAAGGCAGCTGATGAAGGGAGCTCAGGAGAGCCGTCTTCAGCTGGGCTGGGGTTGGGGCTGCTGTGAGGAAAACCTGCCA TTGTGGCCCTGGAGAGTCACCAGCAGCTCTTGGGAAGGACTTGCTGGGAGGCTGAGAGAGGCTTTGGGCACAGCCTGCTGTCTTTTCCAT TTCCTAAAGTTTACTTCATTGTCTTGAGGCTTCCAGGTTTTGTTTTTGTTTTTGCCAAAGTAGAAAAGGCAGGTGGTGGGCGGCTGGCAG GGAGTGCGGGTCCCCGCCCCTCTTCAGTCCTGCCCTCCCCTCCTCAGTCCTGCCCACCCCGTGCAGCCCATGCTGAGGCTGCAGTGGTGT CGTGGGTGTTACGTGCAGGAACGTGGAGACCCTGACGTGGGCTCACTGCGTTTGGTTTTCTTTTCAGAACTTGGGAGCCCCCAGGGAGGG GCTAGTGTTGGTAGGTCCTAGACGTGGTTCCCTCCAGCCTCCCCAAAATCAACCCTGGTGTTGAGAGAACGTCCTTCTGTCCATCGTGGG TAACAGCCTTGGGGAGGGTGCAGAGCTCTGCAGAGCCATGGGCCAGGTGGGGCTGCCTCAGTCCTGTCCCCTTGGGCACTGAGGAGAGGG >50238_50238_4_LSR-CHMP1A_LSR_chr19_35749967_ENST00000361790_CHMP1A_chr16_89713739_ENST00000253475_length(amino acids)=419AA_BP=254 MKACPLSTSFTLIKTSRMQQDGLGVGTRNGSGKGRSVHPSWPWCAPRPLRYFGRDARARRAQTAAMALLAGGLSRGLGSHPAAAGRDAVV FVWLLLSTWCTAPARAIQVTVSNPYHVVILFQPVTLPCTYQMTSTPTQPIVIWKYKSFCRDRIADAFSPASVDNQLNAQLAAGNPGYNPY VECQDSVRTVRVVATKQGNAVTLGDYYQGRRITITGNADLTFDQTAWGDSGVYYCSVVSAQDLQGNNEAYAELIVLGDQEYGPGDQSPGQ GPEHHGPAEGLLSDGQVRAAGAEPGRPYIGDGGLHELGHHPDHAAGAGGQPHHADRRGEWPGGAGPAQPAARGRLCRGRELCAQPGGPAV -------------------------------------------------------------- >50238_50238_5_LSR-CHMP1A_LSR_chr19_35749967_ENST00000602122_CHMP1A_chr16_89713739_ENST00000253475_length(transcript)=3172nt_BP=1205nt AGACTCGCTCCTCCCGGCAGGGCGCACCTAGGCGGGTCCATCGCCAGCCGGGGAGAGGGGTTTGGGCAGGGAGGGAACAGGTGCGCGGCG GGACCCGCCCTATCTCAACAGGTGAATCGCTCCAAGTGGGTCTCGGTTGCATGGATCTCGGTGCGCTTGGTTTGGCCGGAGCAGATGGGG GCCGGAAGGGACCTGTGGTCCGCAGGCGCCCTCCCAGCGGGCCAGTCACTTGGTTCGGGCCCTGGGGGACGGAGCGCACCTGGGTCAGCC CACTTCCGGGGAGGGAGGCAGAGGAACCCCTCCCCGCCGCTCACCCCTAAGCCCAGCCCTCGGCTCCCACCCTTGTGTACCTGGGCCGAA CCATTCACCGGAGCGCGCAGCGGGTGGAGTGTGGCTCGGAGGACCGCGGCGGGTCAAGCACCTTTCTCCCCCATATCTGAAAGCATGCCC TTTGTCCACGTCGTTTACGCTCATTAAAACTTCCAGAATGCAACAGGACGGACTTGGAGTAGGGACAAGGAACGGAAGTGGGAAGGGGAG GAGCGTGCACCCCTCCTGGCCTTGGTGCGCGCCGCGCCCCCTAAGGTACTTTGGAAGGGACGCGCGGGCCAGACGCGCCCAGACGGCCGC GATGGCGCTGTTGGCCGGCGGGCTCTCCAGAGGGCTGGGCTCCCACCCGGCCGCCGCAGGCCGGGACGCGGTCGTCTTCGTGTGGCTTCT GCTTAGCACCTGGTGCACAGCTCCTGCCAGGGCCATCCAGGTGACCGTGTCCAACCCCTACCACGTGGTGATCCTCTTCCAGCCTGTGAC CCTGCCCTGTACCTACCAGATGACCTCGACCCCCACGCAACCCATCGTCATCTGGAAGTACAAGTCTTTCTGCCGGGACCGCATCGCCGA TGCCTTCTCCCCGGCCAGCGTCGACAACCAGCTCAATGCCCAGCTGGCAGCCGGGAACCCAGGCTACAACCCCTACGTTGAGTGCCAGGA CAGCGTGCGCACCGTCAGGGTCGTGGCCACCAAGCAGGGCAACGCTGTGACCCTGGGAGATTACTACCAGGGCCGGAGGATTACCATCAC CGGAAATGCTGACCTGACCTTTGACCAGACGGCGTGGGGGGACAGTGGTGTGTATTACTGCTCCGTGGTCTCAGCCCAGGACCTCCAGGG GAACAATGAGGCCTACGCAGAGCTCATCGTCCTTGGTGACCAAGAATATGGCCCAGGTGACCAAAGCCCTGGACAAGGCCCTGAGCACCA TGGACCTGCAGAAGGTCTCCTCAGTGATGGACAGGTTCGAGCAGCAGGTGCAGAACCTGGACGTCCATACATCGGTGATGGAGGACTCCA TGAGCTCGGCCACCACCCTGACCACGCCGCAGGAGCAGGTGGACAGCCTCATCATGCAGATCGCCGAGGAGAATGGCCTGGAGGTGCTGG ACCAGCTCAGCCAGCTGCCCGAGGGCGCCTCTGCCGTGGGCGAGAGCTCTGTGCGCAGCCAGGAGGACCAGCTGTCACGGAGGTTGGCCG CCTTGAGGAACTAGCCGTGCCCCGCCGGTGTGCACCGCCTCTGCCCCGTGATGTGCTGGAAGGCTCCTGTCCTCTCCCCACCGCGTCTTG CCTTTGTGCTGACCCCGCGGGGCTGCGGCCGGCAGCCACTCTGCGTCTCTCACCTGCCAGGCCTGCGTGGCCTTAGGGTTGTTCCTGTTC TTTTAGGTTGGGCGGTGGGTCTGTGTCCTGGTGTTGAGTTTCTGCAAATTTCTGGGGGTGATTTCTGTGACTCTGGGCCCACAGCGGGGA GGCCAAGAGGGGCCCTGTGGACTTTCACCCAGCACTGTGGGGGCCTTCAGACTCTGGGGCAGCAGACATGCTGCTTCCCATCAGCCAGAG GGGGTCAGGGCTGCCCTGTTGCCAAACAACTCCCTGAGGCCTCTCCGCACCACCTCAGCGGGCAGGAGGTCCCACCATGTGGACAGACAT AGCCCAAGGAGGCACCACAGGTCTATGTGTGCTGGGGGATGTCAGGTGCCACCCAACGCTGTCCTGGTGGTATTTACAATGACATCCTCC TCCTCCATCACTCCAGGGGTGGTGTCTCGGCCGCCCCTACCAGCTGGCTGAGCCCCCTGGCCTCCTGCGCTCCCTCACTTCCCTCAGTTC CCAAAGCTGCCCAGTCCATGGGGACAGAACCGTCACTCAGATCCACATTCAAGTGTGCCCACCCTGCAGTCTTCATCCTCACTCAGCTGC TGCCTCTGGAGGTGCCTTTGGCCACATGTGCTGTGCTGTTTGTCTCCTCGACAGGGAGCCTGTCCACCAGCAGGCTGCGGTCCCAGCGGG TGCGTCTGCAGCTCCTCCCCTTGGGCAGCCTGGTTCTCCCGGAGGACCTTTCCTTGGGGCCCTGCTTCATGACGATGCTGCCTGTGTCAC CCTCTACCATCTGTAAACAACTGGGTGCCTTCCCCGACCACACCCCAATGCCTTCCCAGCTTGGAAGCCAAGGCAGCTGATGAAGGGAGC TCAGGAGAGCCGTCTTCAGCTGGGCTGGGGTTGGGGCTGCTGTGAGGAAAACCTGCCATTGTGGCCCTGGAGAGTCACCAGCAGCTCTTG GGAAGGACTTGCTGGGAGGCTGAGAGAGGCTTTGGGCACAGCCTGCTGTCTTTTCCATTTCCTAAAGTTTACTTCATTGTCTTGAGGCTT CCAGGTTTTGTTTTTGTTTTTGCCAAAGTAGAAAAGGCAGGTGGTGGGCGGCTGGCAGGGAGTGCGGGTCCCCGCCCCTCTTCAGTCCTG CCCTCCCCTCCTCAGTCCTGCCCACCCCGTGCAGCCCATGCTGAGGCTGCAGTGGTGTCGTGGGTGTTACGTGCAGGAACGTGGAGACCC TGACGTGGGCTCACTGCGTTTGGTTTTCTTTTCAGAACTTGGGAGCCCCCAGGGAGGGGCTAGTGTTGGTAGGTCCTAGACGTGGTTCCC TCCAGCCTCCCCAAAATCAACCCTGGTGTTGAGAGAACGTCCTTCTGTCCATCGTGGGTAACAGCCTTGGGGAGGGTGCAGAGCTCTGCA GAGCCATGGGCCAGGTGGGGCTGCCTCAGTCCTGTCCCCTTGGGCACTGAGGAGAGGGGCCCATTCACCTTTCTCCTAGAATGCTGTTGT >50238_50238_5_LSR-CHMP1A_LSR_chr19_35749967_ENST00000602122_CHMP1A_chr16_89713739_ENST00000253475_length(amino acids)=519AA_BP=354 MHGSRCAWFGRSRWGPEGTCGPQAPSQRASHLVRALGDGAHLGQPTSGEGGRGTPPRRSPLSPALGSHPCVPGPNHSPERAAGGVWLGGP RRVKHLSPPYLKACPLSTSFTLIKTSRMQQDGLGVGTRNGSGKGRSVHPSWPWCAPRPLRYFGRDARARRAQTAAMALLAGGLSRGLGSH PAAAGRDAVVFVWLLLSTWCTAPARAIQVTVSNPYHVVILFQPVTLPCTYQMTSTPTQPIVIWKYKSFCRDRIADAFSPASVDNQLNAQL AAGNPGYNPYVECQDSVRTVRVVATKQGNAVTLGDYYQGRRITITGNADLTFDQTAWGDSGVYYCSVVSAQDLQGNNEAYAELIVLGDQE YGPGDQSPGQGPEHHGPAEGLLSDGQVRAAGAEPGRPYIGDGGLHELGHHPDHAAGAGGQPHHADRRGEWPGGAGPAQPAARGRLCRGRE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for LSR-CHMP1A |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for LSR-CHMP1A |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for LSR-CHMP1A |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
