
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:LTBP2-NEK9 (FusionGDB2 ID:50282) |
Fusion Gene Summary for LTBP2-NEK9 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: LTBP2-NEK9 | Fusion gene ID: 50282 | Hgene | Tgene | Gene symbol | LTBP2 | NEK9 | Gene ID | 4054 | 91754 |
| Gene name | latent transforming growth factor beta binding protein 3 | NIMA related kinase 9 | |
| Synonyms | DASS|GPHYSD3|LTBP-3|LTBP2|STHAG6|pp6425 | APUG|LCCS10|NC|NERCC|NERCC1 | |
| Cytomap | 11q13.1 | 14q24.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | latent-transforming growth factor beta-binding protein 3latent TGF beta binding protein 3 | serine/threonine-protein kinase Nek9NIMA (never in mitosis gene a)- related kinase 9nercc1 kinasenimA-related protein kinase 9 | |
| Modification date | 20200322 | 20200320 | |
| UniProtAcc | Q14767 | Q8TD19 | |
| Ensembl transtripts involved in fusion gene | ENST00000261978, ENST00000556690, ENST00000557425, | ENST00000555763, ENST00000238616, | |
| Fusion gene scores | * DoF score | 5 X 5 X 2=50 | 4 X 7 X 3=84 |
| # samples | 5 | 7 | |
| ** MAII score | log2(5/50*10)=0 | log2(7/84*10)=-0.263034405833794 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: LTBP2 [Title/Abstract] AND NEK9 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | LTBP2(74999129)-NEK9(75558181), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across LTBP2 (5'-gene) Fusion gene breakpoints across LTBP2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
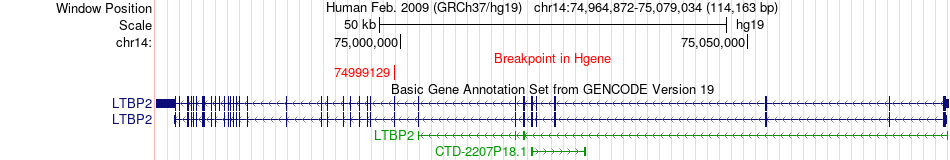 |
 Fusion gene breakpoints across NEK9 (3'-gene) Fusion gene breakpoints across NEK9 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
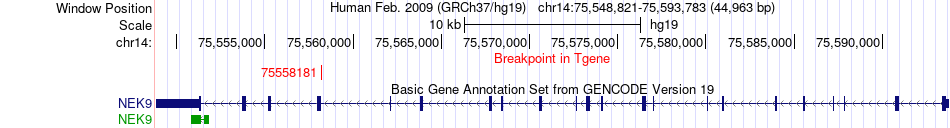 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-25-1315-01A | LTBP2 | chr14 | 74999129 | - | NEK9 | chr14 | 75558181 | - |
Top |
Fusion Gene ORF analysis for LTBP2-NEK9 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000261978 | ENST00000555763 | LTBP2 | chr14 | 74999129 | - | NEK9 | chr14 | 75558181 | - |
| 5CDS-intron | ENST00000556690 | ENST00000555763 | LTBP2 | chr14 | 74999129 | - | NEK9 | chr14 | 75558181 | - |
| In-frame | ENST00000261978 | ENST00000238616 | LTBP2 | chr14 | 74999129 | - | NEK9 | chr14 | 75558181 | - |
| In-frame | ENST00000556690 | ENST00000238616 | LTBP2 | chr14 | 74999129 | - | NEK9 | chr14 | 75558181 | - |
| intron-3CDS | ENST00000557425 | ENST00000238616 | LTBP2 | chr14 | 74999129 | - | NEK9 | chr14 | 75558181 | - |
| intron-intron | ENST00000557425 | ENST00000555763 | LTBP2 | chr14 | 74999129 | - | NEK9 | chr14 | 75558181 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000261978 | LTBP2 | chr14 | 74999129 | - | ENST00000238616 | NEK9 | chr14 | 75558181 | - | 5526 | 2374 | 33 | 3080 | 1015 |
| ENST00000556690 | LTBP2 | chr14 | 74999129 | - | ENST00000238616 | NEK9 | chr14 | 75558181 | - | 5267 | 2115 | 23 | 2821 | 932 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000261978 | ENST00000238616 | LTBP2 | chr14 | 74999129 | - | NEK9 | chr14 | 75558181 | - | 0.011272243 | 0.9887278 |
| ENST00000556690 | ENST00000238616 | LTBP2 | chr14 | 74999129 | - | NEK9 | chr14 | 75558181 | - | 0.011673998 | 0.988326 |
Top |
Fusion Genomic Features for LTBP2-NEK9 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
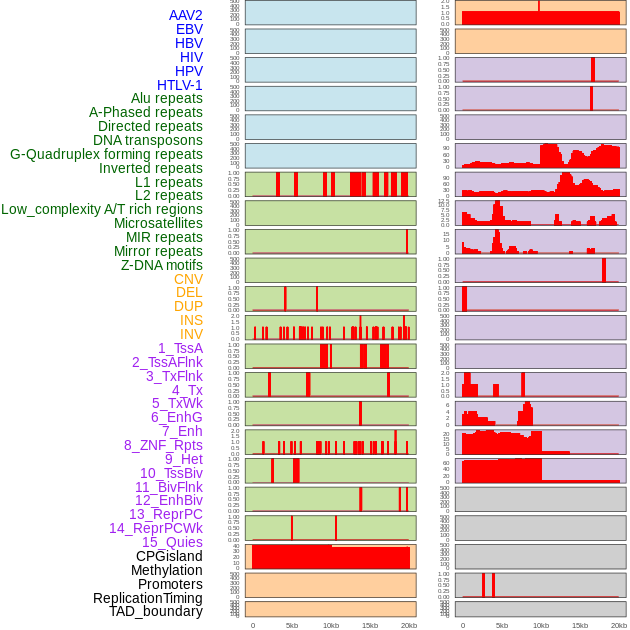 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for LTBP2-NEK9 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr14:74999129/chr14:75558181) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| LTBP2 | NEK9 |
| FUNCTION: May play an integral structural role in elastic-fiber architectural organization and/or assembly. {ECO:0000303|PubMed:10743502, ECO:0000303|PubMed:11104663}. | FUNCTION: Pleiotropic regulator of mitotic progression, participating in the control of spindle dynamics and chromosome separation. Phosphorylates different histones, myelin basic protein, beta-casein, and BICD2. Phosphorylates histone H3 on serine and threonine residues and beta-casein on serine residues. Important for G1/S transition and S phase progression. Phosphorylates NEK6 and NEK7 and stimulates their activity by releasing the autoinhibitory functions of Tyr-108 and Tyr-97 respectively. {ECO:0000269|PubMed:12840024, ECO:0000269|PubMed:14660563, ECO:0000269|PubMed:19941817}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 187_219 | 662 | 1822.0 | Domain | EGF-like 1 |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 396_428 | 662 | 1822.0 | Domain | EGF-like 2 |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 552_604 | 662 | 1822.0 | Domain | TB 1 |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 622_662 | 662 | 1822.0 | Domain | EGF-like 3%3B calcium-binding |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 375_377 | 662 | 1822.0 | Motif | Cell attachment site |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 232_249 | 662 | 1822.0 | Region | Note=Heparin-binding |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 344_354 | 662 | 1822.0 | Region | Note=Heparin-binding |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 94_115 | 662 | 1822.0 | Region | Note=Heparin-binding |
| Tgene | NEK9 | chr14:74999129 | chr14:75558181 | ENST00000238616 | 17 | 22 | 892_939 | 744 | 980.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | NEK9 | chr14:74999129 | chr14:75558181 | ENST00000238616 | 17 | 22 | 752_760 | 744 | 980.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | NEK9 | chr14:74999129 | chr14:75558181 | ENST00000238616 | 17 | 22 | 765_888 | 744 | 980.0 | Compositional bias | Note=Pro/Ser/Thr-rich |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 856_1372 | 662 | 1822.0 | Compositional bias | Note=Cys-rich |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 1010_1050 | 662 | 1822.0 | Domain | EGF-like 8%3B calcium-binding |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 1051_1092 | 662 | 1822.0 | Domain | EGF-like 9%3B calcium-binding |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 1093_1134 | 662 | 1822.0 | Domain | EGF-like 10%3B calcium-binding |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 1135_1175 | 662 | 1822.0 | Domain | EGF-like 11%3B calcium-binding |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 1176_1217 | 662 | 1822.0 | Domain | EGF-like 12%3B calcium-binding |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 1218_1258 | 662 | 1822.0 | Domain | EGF-like 13%3B calcium-binding |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 1259_1302 | 662 | 1822.0 | Domain | EGF-like 14%3B calcium-binding |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 1303_1344 | 662 | 1822.0 | Domain | EGF-like 15%3B calcium-binding |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 1345_1387 | 662 | 1822.0 | Domain | EGF-like 16%3B calcium-binding |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 1411_1463 | 662 | 1822.0 | Domain | TB 3 |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 1485_1527 | 662 | 1822.0 | Domain | EGF-like 17%3B calcium-binding |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 1528_1567 | 662 | 1822.0 | Domain | EGF-like 18%3B calcium-binding |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 1584_1636 | 662 | 1822.0 | Domain | TB 4 |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 1733_1773 | 662 | 1822.0 | Domain | EGF-like 19%3B calcium-binding |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 1774_1818 | 662 | 1822.0 | Domain | EGF-like 20%3B calcium-binding |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 672_724 | 662 | 1822.0 | Domain | TB 2 |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 844_886 | 662 | 1822.0 | Domain | EGF-like 4 |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 887_929 | 662 | 1822.0 | Domain | EGF-like 5%3B calcium-binding |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 930_969 | 662 | 1822.0 | Domain | EGF-like 6%3B calcium-binding |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 970_1009 | 662 | 1822.0 | Domain | EGF-like 7%3B calcium-binding |
| Hgene | LTBP2 | chr14:74999129 | chr14:75558181 | ENST00000261978 | - | 10 | 36 | 1639_1821 | 662 | 1822.0 | Region | C-terminal domain |
| Tgene | NEK9 | chr14:74999129 | chr14:75558181 | ENST00000238616 | 17 | 22 | 52_308 | 744 | 980.0 | Domain | Protein kinase | |
| Tgene | NEK9 | chr14:74999129 | chr14:75558181 | ENST00000238616 | 17 | 22 | 58_66 | 744 | 980.0 | Nucleotide binding | ATP | |
| Tgene | NEK9 | chr14:74999129 | chr14:75558181 | ENST00000238616 | 17 | 22 | 388_444 | 744 | 980.0 | Repeat | Note=RCC1 1 | |
| Tgene | NEK9 | chr14:74999129 | chr14:75558181 | ENST00000238616 | 17 | 22 | 445_498 | 744 | 980.0 | Repeat | Note=RCC1 2 | |
| Tgene | NEK9 | chr14:74999129 | chr14:75558181 | ENST00000238616 | 17 | 22 | 499_550 | 744 | 980.0 | Repeat | Note=RCC1 3 | |
| Tgene | NEK9 | chr14:74999129 | chr14:75558181 | ENST00000238616 | 17 | 22 | 551_615 | 744 | 980.0 | Repeat | Note=RCC1 4 | |
| Tgene | NEK9 | chr14:74999129 | chr14:75558181 | ENST00000238616 | 17 | 22 | 616_668 | 744 | 980.0 | Repeat | Note=RCC1 5 | |
| Tgene | NEK9 | chr14:74999129 | chr14:75558181 | ENST00000238616 | 17 | 22 | 669_726 | 744 | 980.0 | Repeat | Note=RCC1 6 |
Top |
Fusion Gene Sequence for LTBP2-NEK9 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >50282_50282_1_LTBP2-NEK9_LTBP2_chr14_74999129_ENST00000261978_NEK9_chr14_75558181_ENST00000238616_length(transcript)=5526nt_BP=2374nt CCTCGCTCCCTCTCCGGTAATGAGGGGGCTGAGCTGTCCCTCCGAGGAGGGGGCCTGGTGTGGATAAAAGAGACGAAAAAGCCGGGGGAG GTTTCCAAAAATAAAACCGTCCGGGTCCCCTTCAGACGGCTGCAGGCACAGGGAGGAGGCGCGAAGGTGCAGCAGCCGTGCGAGCCCAGC TGGAGTAGGAGCGCGGACTCGAGGCTCGGGGCGCGCAGCCCTCGTTCCGCCGAGAGCCGGGCCCCCAGTCGGCCGCTTCAGGGCCCCCTA GACTCAGAGAAGCTGGCCGCCGGGCGGGGCCGGGAGAACAGCCCGCGGGCGTCCAGCGTGCCGACCACAAAGCTCTTCGCGGTGCCCGCG CGCACCACTCTCCAGCCGCCCCGCGCCATGAGGCCGCGGACCAAAGCCCGCAGCCCGGGGCGCGCCCTGCGGAACCCCTGGAGAGGCTTC CTGCCGCTCACCCTGGCTCTCTTCGTGGGCGCGGGTCATGCCCAAAGGGACCCCGTAGGGAGATACGAGCCGGCTGGTGGAGACGCGAAT CGACTGCGGCGCCCTGGGGGCAGCTACCCGGCAGCGGCTGCAGCCAAGGTGTACAGTCTGTTCCGGGAGCAGGACGCGCCTGTCGCGGGC TTGCAGCCCGTGGAGCGGGCCCAGCCGGGCTGGGGGAGCCCCAGGAGGCCCACCGAGGCGGAGGCCAGGAGGCCGTCCCGCGCGCAGCAG TCGCGGCGTGTCCAGCCACCTGCGCAGACCCGGAGAAGCACTCCCCTGGGCCAGCAGCAACCAGCACCCCGGACCCGGGCCGCGCCGGCT CTCCCACGCCTGGGGACCCCACAGCGGTCTGGGGCTGCGCCCCCAACCCCGCCGCGAGGGCGGCTCACGGGGAGGAACGTCTGCGGGGGA CAGTGCTGCCCAGGATGGACAACAGCAAACAGCACCAACCACTGTATCAAACCCGTTTGCGAGCCGCCGTGCCAGAACCGGGGCTCCTGC AGCCGCCCGCAGCTCTGTGTCTGCCGCTCTGGTTTCCGTGGAGCCCGCTGCGAGGAGGTCATTCCCGATGAGGAATTTGACCCCCAGAAC TCCAGGCTGGCACCTCGACGCTGGGCCGAGCGTTCACCCAACCTGCGCAGGAGCAGTGCGGCTGGAGAGGGCACCTTGGCCAGAGCACAG CCGCCAGCACCACAGTCGCCGCCCGCACCACAGTCGCCACCAGCTGGGACCCTGAGTGGCCTCAGCCAGACCCACCCTTCCCAGCAGCAC GTGGGGTTGTCCCGCACTGTCCGACTTCACCCGACTGCCACGGCCAGTAGCCAGCTCTCTTCCAACGCCCTGCCCCCGGGACCAGGCCTT GAGCAGAGAGATGGCACCCAACAGGCGGTACCTCTGGAGCACCCCTCATCCCCCTGGGGGCTGAACCTCACGGAGAAAATCAAGAAGATC AAGATCGTCTTCACTCCCACCATCTGCAAGCAGACCTGTGCCCGTGGACACTGTGCCAACAGCTGTGAGAGGGGCGACACCACCACCCTG TACAGCCAGGGCGGCCATGGGCACGATCCCAAGTCTGGCTTCCGCATCTATTTCTGCCAGATCCCCTGCCTGAACGGAGGCCGCTGCATC GGCAGGGACGAATGCTGGTGCCCCGCCAACTCCACCGGGAAGTTCTGCCACCTGCCTATCCCGCAGCCGGACAGGGAGCCTCCAGGGAGG GGGTCCCGCCCCAGGGCCTTGCTGGAAGCCCCACTGAAGCAGTCCACTTTCACACTGCCGCTCTCCAACCAGCTGGCCTCCGTGAACCCC TCCCTGGTGAAGGTGCACATTCACCACCCACCCGAGGCCTCAGTGCAGATCCACCAGGTGGCCCAGGTGCGGGGCGGGGTGGAGGAGGCC CTAGTGGAGAACAGCGTGGAGACCAGACCCCCGCCCTGGCTGCCTGCCAGCCCTGGCCACAGCCTCTGGGACAGCAACAACATCCCTGCT CGGTCTGGAGAGCCCCCTCGGCCACTGCCCCCAGCAGCACCCAGGCCTCGAGGACTGCTGGGCCGGTGTTACCTGAACACTGTGAACGGA CAGTGTGCCAACCCTCTGCTGGAGCTGACTACCCAGGAGGACTGCTGTGGCAGTGTGGGAGCCTTCTGGGGGGTGACTTTGTGTGCCCCA TGCCCACCCAGACCAGCCTCCCCGGTGATTGAGAATGGCCAGCTGGAGTGTCCTCAGGGGTACAAGAGACTGAACCTCACTCACTGCCAA GATATCAACGAGTGCTTGACCCTGGGCCTGTGCAAGGACGCGGAGTGTGTGAATACCAGGGGCAGCTACCTGTGCACATGCAGACCTGGC CTCATGCTGGATCCATCGCGGAGCCGCTGTGTGTTGTTTCAGAGCTCTAGCCCGGGAGGAGGCGGCGGGGGCGGCGGTGGTGAAGAAGAG GACAGTCAGCAGGAATCTGAAACTCCTGACCCAAGTGGAGGCTTCCGAGGAACAATGGAAGCAGACCGAGGAATGGAAGGTTTAATCAGT CCCACAGAGGCCATGGGGAACAGTAATGGGGCCAGCAGCTCCTGTCCTGGCTGGCTTCGAAAGGAGCTGGAAAATGCAGAATTTATCCCC ATGCCTGACAGCCCATCTCCTCTCAGTGCAGCGTTTTCAGAATCTGAGAAAGATACCCTGCCCTATGAAGAGCTGCAAGGACTCAAAGTG GCCTCTGAAGCTCCTTTGGAACACAAACCCCAAGTAGAAGCCTCGTCACCTCGGCTGAATCCTGCAGTAACCTGTGCTGGGAAGGGAACA CCACTGACTCCTCCTGCGTGTGCGTGCAGCTCTCTGCAGGTGGAGGTTGAGAGATTGCAGGGTCTGGTGTTAAAGTGTCTGGCTGAACAA CAGAAGCTACAGCAAGAAAACCTCCAGATTTTTACCCAACTGCAGAAGTTGAACAAGAAATTAGAAGGAGGGCAGCAGGTGGGGATGCAT TCCAAAGGAACTCAGACAGCAAAGGAAGAGATGGAAATGGATCCAAAGCCTGACTTAGATTCAGATTCCTGGTGCCTCCTGGGAACAGAC TCCTGTAGACCCAGCCTCTAGTCTCCTGAGCCTGTAGAGCCCCCAGGAGACTGGGACCCAAAGAACTTCACAGCACACTTACCGAATGCA GAGAGCAGCTTTCCTGGCTTTGTTCACTTGCAGAAAAGGAGCGCAAGGCAGAGGCTCTGAAGCACTTTCCTTGTACATTTGGAGAGTGGC ATTGCCTTTTAGATAGGATCTAGGAGTGATTTTATTGTTTTGGAGAATGGAAGGGCCCCCATGGCCCTGGCTTTGTCATCAGTGACTGCC ATAGCAACAGCAGCTCTGTACCTCATCTGTTGATCCCACCTTTGAAGAGGAGACACAGTGCTCACCTTAATTGCGCTGGTAGCAGCTTAT ATCCCATGTATCATTTTCACCATTGATTGGAAGCTGCCTTGGGAATTCAGTACCAGGCATTACCCCTCTGGGTGGGAGAGGGAGAAGTGT AAAGTTGGAGTGGGCTGGAATCAGGTGTGGCCCGCCCAGTGTCCTCTGCAGAGTGGTGAAGTAGTCTGGCCCTCTTGGGAGCCCTGAGTC CAGGAAAATATGTCTGATGGAGTCAATCTAGGGCTTGTTTCGAAAAAGTTCAGTTACTCTGTGCAGCTAAATGCTTTAGGAGGAAAGGTA GGCTTAGGTTGCTTTTCCTCTGAGGGTTGATTGAAATTTCTTCAGTGAGGAATAGAGAAAGGGCAGGACCCTCCTCATCACACAGCTGGT GTTTCAGGCTGTGACCAATGCAGGGTGGGATTTCCTAACTGTGGATGAGGGGATGAGGTGTCTCTGAGGGATGAGGTGTCTCAGAGAATT GAGTCCATGGGGCAGTCAGAATAGCCTTAAGAGAAAATCATGAAGGAGAAGAGGTCCTCCTTTAGCTGCCTCTACTTGGTATCTTAGAGA GGGCTTAGAGGGCTCTCAGTCTTCTGCCCATGAAAAGACTTCTTTGAGCCTCTGCCTTCATGGCTCTTAGGGTTCTGATCTTGATATCAG CAGCCCCAACCACTTTCTTTCTGAATGTCTAGTCAGTATTTTTCCCCTTTTGGTGTTTTATGAAGCCATGTGGTAACGAATGAATCTGTA TCATTTTTCCTACCTGAGTGGCCCAAAGCCAGCACCAAACCCTGGGAGTCCCTGAAGCCTAACAGAACAGGTAGAACTTGCAAAAGGAAT TTGGCTGAGAGCTCACTTCTAATCCTGTACTCACTGTGTCTTTGTAGATAGAAACAAGCTAGCTTTACCAAAGAGAAATAGTGTCATAGA AGAACAAAACTTCATATAGAAAGTTCTAGGCAAGTATTTGATGGTTTCCTTAAGGATGTGAGCTTTGTATTTCCACCTAGCCTTGTAAAA TGTTCCTGTGGTATTTTGTGTCACACATCCTACCTTTGATGAGTCTCACATCCCACCTTCCTATAAACAGCTAAATTAATTTTGTTTCAT CTTCCCCAGACCAAAATGTTTGATAATCTTATCAACATTGTGGGAGGTCTTGTGCAATGGAAATTTTGCCATTTCTCCAAAACTGGTGGC ATAAAGGCTGATGCTTGGGGAGAACCCCATTGCTCGGGACAGGCAACTCTGTTCAATGGGATCTTCTTTGGTTTGATGTTCCCATTGTTT TCTCAGTTCTGGGAAGCCTAGTACATTAGTACTAATGTAATCACTGAAACCTTTTCTTGAAATAAGGGAAGCAGCCAAACTTTGATTAAA GTTGCAAGTTCTGGGGACTTGCGGGGGTTGTCATAAACTGTAACAGTGGGTTTTGGTTCAGCATGTAAATGCAACTTTGATTTTCTTGAG GACCGATTGACCTGTCATGTCCCTGTATCCTCATGCTCATCATCTCAGCAGGCCTGAGAGGCTGGGTCAGTTTGGGTGTTCATCATGAGG ATTGCTTCTGCCATGGAGCTGATGGACATGGGCAGGTTGCTGAGAAGGTGGGGTGAAAGTGAGTGCCGGGGGTGGGTGAGTGCCCTGGTC TTGTTCATAGGGGAGCCTTTCCCTAGCAGTGGAACGCTGTGGTCATTTTCTCTAGCATATTCCCTTGGGAAGTCTAGATTTGCTATTAAT CTGGCTGAGAATCTAAGTTCTGTGCCTTAGAGACAGTTTGCACTTTCCCATATTGTGCCTGGGACAGCCATATGATTTTTTTTCCCACCA AACAAGTATGCAAACAGAAACCAGTTCAAAGGGGGATGGAGTAAAAGATGAGGCAGTAGAAATGCCTTTGAATGGTTTTCTGTAGCTAAT TCTCTTTAAATTTTGTCCTGCTTTTTTTCTTTATGCAGTGCTAGGTGTTTTAAGTTTTCTAGTAGTATTGCTTTTGAGTTACAGTATAAC CTGAGTTACTCCTCTGCTCTAACATTGTTGCAGAAGAACTAGTTTATTGTTAGCCAGGTTGCTTGAAAGGTTGAGAGTGGAGTGGTTTGG >50282_50282_1_LTBP2-NEK9_LTBP2_chr14_74999129_ENST00000261978_NEK9_chr14_75558181_ENST00000238616_length(amino acids)=1015AA_BP=781 MSLRGGGLVWIKETKKPGEVSKNKTVRVPFRRLQAQGGGAKVQQPCEPSWSRSADSRLGARSPRSAESRAPSRPLQGPLDSEKLAAGRGR ENSPRASSVPTTKLFAVPARTTLQPPRAMRPRTKARSPGRALRNPWRGFLPLTLALFVGAGHAQRDPVGRYEPAGGDANRLRRPGGSYPA AAAAKVYSLFREQDAPVAGLQPVERAQPGWGSPRRPTEAEARRPSRAQQSRRVQPPAQTRRSTPLGQQQPAPRTRAAPALPRLGTPQRSG AAPPTPPRGRLTGRNVCGGQCCPGWTTANSTNHCIKPVCEPPCQNRGSCSRPQLCVCRSGFRGARCEEVIPDEEFDPQNSRLAPRRWAER SPNLRRSSAAGEGTLARAQPPAPQSPPAPQSPPAGTLSGLSQTHPSQQHVGLSRTVRLHPTATASSQLSSNALPPGPGLEQRDGTQQAVP LEHPSSPWGLNLTEKIKKIKIVFTPTICKQTCARGHCANSCERGDTTTLYSQGGHGHDPKSGFRIYFCQIPCLNGGRCIGRDECWCPANS TGKFCHLPIPQPDREPPGRGSRPRALLEAPLKQSTFTLPLSNQLASVNPSLVKVHIHHPPEASVQIHQVAQVRGGVEEALVENSVETRPP PWLPASPGHSLWDSNNIPARSGEPPRPLPPAAPRPRGLLGRCYLNTVNGQCANPLLELTTQEDCCGSVGAFWGVTLCAPCPPRPASPVIE NGQLECPQGYKRLNLTHCQDINECLTLGLCKDAECVNTRGSYLCTCRPGLMLDPSRSRCVLFQSSSPGGGGGGGGGEEEDSQQESETPDP SGGFRGTMEADRGMEGLISPTEAMGNSNGASSSCPGWLRKELENAEFIPMPDSPSPLSAAFSESEKDTLPYEELQGLKVASEAPLEHKPQ VEASSPRLNPAVTCAGKGTPLTPPACACSSLQVEVERLQGLVLKCLAEQQKLQQENLQIFTQLQKLNKKLEGGQQVGMHSKGTQTAKEEM -------------------------------------------------------------- >50282_50282_2_LTBP2-NEK9_LTBP2_chr14_74999129_ENST00000556690_NEK9_chr14_75558181_ENST00000238616_length(transcript)=5267nt_BP=2115nt AGGGCCCCCTAGACTCAGAGAAGCTGGCCGCCGGGCGGGGCCGGGAGAACAGCCCGCGGGCGTCCAGCGTGCCGACCACAAAGCTCTTCG CGGTGCCCGCGCGCACCACTCTCCAGCCGCCCCGCGCCATGAGGCCGCGGACCAAAGCCCGCAGCCCGGGGCGCGCCCTGCGGAACCCCT GGAGAGGCTTCCTGCCGCTCACCCTGGCTCTCTTCGTGGGCGCGGGTCATGCCCAAAGGGACCCCGTAGGGAGATACGAGCCGGCTGGTG GAGACGCGAATCGACTGCGGCGCCCTGGGGGCAGCTACCCGGCAGCGGCTGCAGCCAAGGTGTACAGTCTGTTCCGGGAGCAGGACGCGC CTGTCGCGGGCTTGCAGCCCGTGGAGCGGGCCCAGCCGGGCTGGGGGAGCCCCAGGAGGCCCACCGAGGCGGAGGCCAGGAGGCCGTCCC GCGCGCAGCAGTCGCGGCGTGTCCAGCCACCTGCGCAGACCCGGAGAAGCACTCCCCTGGGCCAGCAGCAACCAGCACCCCGGACCCGGG CCGCGCCGGCTCTCCCACGCCTGGGGACCCCACAGCGGTCTGGGGCTGCGCCCCCAACCCCGCCGCGAGGGCGGCTCACGGGGAGGAACG TCTGCGGGGGACAGTGCTGCCCAGGATGGACAACAGCAAACAGCACCAACCACTGTATCAAACCCGTTTGCGAGCCGCCGTGCCAGAACC GGGGCTCCTGCAGCCGCCCGCAGCTCTGTGTCTGCCGCTCTGGTTTCCGTGGAGCCCGCTGCGAGGAGGTCATTCCCGATGAGGAATTTG ACCCCCAGAACTCCAGGCTGGCACCTCGACGCTGGGCCGAGCGTTCACCCAACCTGCGCAGGAGCAGTGCGGCTGGAGAGGGCACCTTGG CCAGAGCACAGCCGCCAGCACCACAGTCGCCGCCCGCACCACAGTCGCCACCAGCTGGGACCCTGAGTGGCCTCAGCCAGACCCACCCTT CCCAGCAGCACGTGGGGTTGTCCCGCACTGTCCGACTTCACCCGACTGCCACGGCCAGTAGCCAGCTCTCTTCCAACGCCCTGCCCCCGG GACCAGGCCTTGAGCAGAGAGATGGCACCCAACAGGCGGTACCTCTGGAGCACCCCTCATCCCCCTGGGGGCTGAACCTCACGGAGAAAA TCAAGAAGATCAAGATCGTCTTCACTCCCACCATCTGCAAGCAGACCTGTGCCCGTGGACACTGTGCCAACAGCTGTGAGAGGGGCGACA CCACCACCCTGTACAGCCAGGGCGGCCATGGGCACGATCCCAAGTCTGGCTTCCGCATCTATTTCTGCCAGATCCCCTGCCTGAACGGAG GCCGCTGCATCGGCAGGGACGAATGCTGGTGCCCCGCCAACTCCACCGGGAAGTTCTGCCACCTGCCTATCCCGCAGCCGGACAGGGAGC CTCCAGGGAGGGGGTCCCGCCCCAGGGCCTTGCTGGAAGCCCCACTGAAGCAGTCCACTTTCACACTGCCGCTCTCCAACCAGCTGGCCT CCGTGAACCCCTCCCTGGTGAAGGTGCACATTCACCACCCACCCGAGGCCTCAGTGCAGATCCACCAGGTGGCCCAGGTGCGGGGCGGGG TGGAGGAGGCCCTAGTGGAGAACAGCGTGGAGACCAGACCCCCGCCCTGGCTGCCTGCCAGCCCTGGCCACAGCCTCTGGGACAGCAACA ACATCCCTGCTCGGTCTGGAGAGCCCCCTCGGCCACTGCCCCCAGCAGCACCCAGGCCTCGAGGACTGCTGGGCCGGTGTTACCTGAACA CTGTGAACGGACAGTGTGCCAACCCTCTGCTGGAGCTGACTACCCAGGAGGACTGCTGTGGCAGTGTGGGAGCCTTCTGGGGGGTGACTT TGTGTGCCCCATGCCCACCCAGACCAGCCTCCCCGGTGATTGAGAATGGCCAGCTGGAGTGTCCTCAGGGGTACAAGAGACTGAACCTCA CTCACTGCCAAGATATCAACGAGTGCTTGACCCTGGGCCTGTGCAAGGACGCGGAGTGTGTGAATACCAGGGGCAGCTACCTGTGCACAT GCAGACCTGGCCTCATGCTGGATCCATCGCGGAGCCGCTGTGTGTTGTTTCAGAGCTCTAGCCCGGGAGGAGGCGGCGGGGGCGGCGGTG GTGAAGAAGAGGACAGTCAGCAGGAATCTGAAACTCCTGACCCAAGTGGAGGCTTCCGAGGAACAATGGAAGCAGACCGAGGAATGGAAG GTTTAATCAGTCCCACAGAGGCCATGGGGAACAGTAATGGGGCCAGCAGCTCCTGTCCTGGCTGGCTTCGAAAGGAGCTGGAAAATGCAG AATTTATCCCCATGCCTGACAGCCCATCTCCTCTCAGTGCAGCGTTTTCAGAATCTGAGAAAGATACCCTGCCCTATGAAGAGCTGCAAG GACTCAAAGTGGCCTCTGAAGCTCCTTTGGAACACAAACCCCAAGTAGAAGCCTCGTCACCTCGGCTGAATCCTGCAGTAACCTGTGCTG GGAAGGGAACACCACTGACTCCTCCTGCGTGTGCGTGCAGCTCTCTGCAGGTGGAGGTTGAGAGATTGCAGGGTCTGGTGTTAAAGTGTC TGGCTGAACAACAGAAGCTACAGCAAGAAAACCTCCAGATTTTTACCCAACTGCAGAAGTTGAACAAGAAATTAGAAGGAGGGCAGCAGG TGGGGATGCATTCCAAAGGAACTCAGACAGCAAAGGAAGAGATGGAAATGGATCCAAAGCCTGACTTAGATTCAGATTCCTGGTGCCTCC TGGGAACAGACTCCTGTAGACCCAGCCTCTAGTCTCCTGAGCCTGTAGAGCCCCCAGGAGACTGGGACCCAAAGAACTTCACAGCACACT TACCGAATGCAGAGAGCAGCTTTCCTGGCTTTGTTCACTTGCAGAAAAGGAGCGCAAGGCAGAGGCTCTGAAGCACTTTCCTTGTACATT TGGAGAGTGGCATTGCCTTTTAGATAGGATCTAGGAGTGATTTTATTGTTTTGGAGAATGGAAGGGCCCCCATGGCCCTGGCTTTGTCAT CAGTGACTGCCATAGCAACAGCAGCTCTGTACCTCATCTGTTGATCCCACCTTTGAAGAGGAGACACAGTGCTCACCTTAATTGCGCTGG TAGCAGCTTATATCCCATGTATCATTTTCACCATTGATTGGAAGCTGCCTTGGGAATTCAGTACCAGGCATTACCCCTCTGGGTGGGAGA GGGAGAAGTGTAAAGTTGGAGTGGGCTGGAATCAGGTGTGGCCCGCCCAGTGTCCTCTGCAGAGTGGTGAAGTAGTCTGGCCCTCTTGGG AGCCCTGAGTCCAGGAAAATATGTCTGATGGAGTCAATCTAGGGCTTGTTTCGAAAAAGTTCAGTTACTCTGTGCAGCTAAATGCTTTAG GAGGAAAGGTAGGCTTAGGTTGCTTTTCCTCTGAGGGTTGATTGAAATTTCTTCAGTGAGGAATAGAGAAAGGGCAGGACCCTCCTCATC ACACAGCTGGTGTTTCAGGCTGTGACCAATGCAGGGTGGGATTTCCTAACTGTGGATGAGGGGATGAGGTGTCTCTGAGGGATGAGGTGT CTCAGAGAATTGAGTCCATGGGGCAGTCAGAATAGCCTTAAGAGAAAATCATGAAGGAGAAGAGGTCCTCCTTTAGCTGCCTCTACTTGG TATCTTAGAGAGGGCTTAGAGGGCTCTCAGTCTTCTGCCCATGAAAAGACTTCTTTGAGCCTCTGCCTTCATGGCTCTTAGGGTTCTGAT CTTGATATCAGCAGCCCCAACCACTTTCTTTCTGAATGTCTAGTCAGTATTTTTCCCCTTTTGGTGTTTTATGAAGCCATGTGGTAACGA ATGAATCTGTATCATTTTTCCTACCTGAGTGGCCCAAAGCCAGCACCAAACCCTGGGAGTCCCTGAAGCCTAACAGAACAGGTAGAACTT GCAAAAGGAATTTGGCTGAGAGCTCACTTCTAATCCTGTACTCACTGTGTCTTTGTAGATAGAAACAAGCTAGCTTTACCAAAGAGAAAT AGTGTCATAGAAGAACAAAACTTCATATAGAAAGTTCTAGGCAAGTATTTGATGGTTTCCTTAAGGATGTGAGCTTTGTATTTCCACCTA GCCTTGTAAAATGTTCCTGTGGTATTTTGTGTCACACATCCTACCTTTGATGAGTCTCACATCCCACCTTCCTATAAACAGCTAAATTAA TTTTGTTTCATCTTCCCCAGACCAAAATGTTTGATAATCTTATCAACATTGTGGGAGGTCTTGTGCAATGGAAATTTTGCCATTTCTCCA AAACTGGTGGCATAAAGGCTGATGCTTGGGGAGAACCCCATTGCTCGGGACAGGCAACTCTGTTCAATGGGATCTTCTTTGGTTTGATGT TCCCATTGTTTTCTCAGTTCTGGGAAGCCTAGTACATTAGTACTAATGTAATCACTGAAACCTTTTCTTGAAATAAGGGAAGCAGCCAAA CTTTGATTAAAGTTGCAAGTTCTGGGGACTTGCGGGGGTTGTCATAAACTGTAACAGTGGGTTTTGGTTCAGCATGTAAATGCAACTTTG ATTTTCTTGAGGACCGATTGACCTGTCATGTCCCTGTATCCTCATGCTCATCATCTCAGCAGGCCTGAGAGGCTGGGTCAGTTTGGGTGT TCATCATGAGGATTGCTTCTGCCATGGAGCTGATGGACATGGGCAGGTTGCTGAGAAGGTGGGGTGAAAGTGAGTGCCGGGGGTGGGTGA GTGCCCTGGTCTTGTTCATAGGGGAGCCTTTCCCTAGCAGTGGAACGCTGTGGTCATTTTCTCTAGCATATTCCCTTGGGAAGTCTAGAT TTGCTATTAATCTGGCTGAGAATCTAAGTTCTGTGCCTTAGAGACAGTTTGCACTTTCCCATATTGTGCCTGGGACAGCCATATGATTTT TTTTCCCACCAAACAAGTATGCAAACAGAAACCAGTTCAAAGGGGGATGGAGTAAAAGATGAGGCAGTAGAAATGCCTTTGAATGGTTTT CTGTAGCTAATTCTCTTTAAATTTTGTCCTGCTTTTTTTCTTTATGCAGTGCTAGGTGTTTTAAGTTTTCTAGTAGTATTGCTTTTGAGT TACAGTATAACCTGAGTTACTCCTCTGCTCTAACATTGTTGCAGAAGAACTAGTTTATTGTTAGCCAGGTTGCTTGAAAGGTTGAGAGTG >50282_50282_2_LTBP2-NEK9_LTBP2_chr14_74999129_ENST00000556690_NEK9_chr14_75558181_ENST00000238616_length(amino acids)=932AA_BP=698 MAAGRGRENSPRASSVPTTKLFAVPARTTLQPPRAMRPRTKARSPGRALRNPWRGFLPLTLALFVGAGHAQRDPVGRYEPAGGDANRLRR PGGSYPAAAAAKVYSLFREQDAPVAGLQPVERAQPGWGSPRRPTEAEARRPSRAQQSRRVQPPAQTRRSTPLGQQQPAPRTRAAPALPRL GTPQRSGAAPPTPPRGRLTGRNVCGGQCCPGWTTANSTNHCIKPVCEPPCQNRGSCSRPQLCVCRSGFRGARCEEVIPDEEFDPQNSRLA PRRWAERSPNLRRSSAAGEGTLARAQPPAPQSPPAPQSPPAGTLSGLSQTHPSQQHVGLSRTVRLHPTATASSQLSSNALPPGPGLEQRD GTQQAVPLEHPSSPWGLNLTEKIKKIKIVFTPTICKQTCARGHCANSCERGDTTTLYSQGGHGHDPKSGFRIYFCQIPCLNGGRCIGRDE CWCPANSTGKFCHLPIPQPDREPPGRGSRPRALLEAPLKQSTFTLPLSNQLASVNPSLVKVHIHHPPEASVQIHQVAQVRGGVEEALVEN SVETRPPPWLPASPGHSLWDSNNIPARSGEPPRPLPPAAPRPRGLLGRCYLNTVNGQCANPLLELTTQEDCCGSVGAFWGVTLCAPCPPR PASPVIENGQLECPQGYKRLNLTHCQDINECLTLGLCKDAECVNTRGSYLCTCRPGLMLDPSRSRCVLFQSSSPGGGGGGGGGEEEDSQQ ESETPDPSGGFRGTMEADRGMEGLISPTEAMGNSNGASSSCPGWLRKELENAEFIPMPDSPSPLSAAFSESEKDTLPYEELQGLKVASEA PLEHKPQVEASSPRLNPAVTCAGKGTPLTPPACACSSLQVEVERLQGLVLKCLAEQQKLQQENLQIFTQLQKLNKKLEGGQQVGMHSKGT -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for LTBP2-NEK9 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Tgene | NEK9 | chr14:74999129 | chr14:75558181 | ENST00000238616 | 17 | 22 | 732_891 | 744.3333333333334 | 980.0 | NEK6 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for LTBP2-NEK9 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for LTBP2-NEK9 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
