
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:LYN-TGS1 (FusionGDB2 ID:50448) |
Fusion Gene Summary for LYN-TGS1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: LYN-TGS1 | Fusion gene ID: 50448 | Hgene | Tgene | Gene symbol | LYN | TGS1 | Gene ID | 4067 | 286826 |
| Gene name | LYN proto-oncogene, Src family tyrosine kinase | lin-9 DREAM MuvB core complex component | |
| Synonyms | JTK8|p53Lyn|p56Lyn | BARA|BARPsv|Lin-9|TGS|TGS1|TGS2 | |
| Cytomap | 8q12.1 | 1q42.12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | tyrosine-protein kinase Lynlck/Yes-related novel protein tyrosine kinasev-yes-1 Yamaguchi sarcoma viral related oncogene homolog | protein lin-9 homologTUDOR gene similar proteinbeta subunit-associated regulator of apoptosislin-9 homologpRB-associated proteinrb related pathway actortype I interferon receptor beta chain-associated protein | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | P07948 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000420292, ENST00000519728, ENST00000520220, | ENST00000260129, | |
| Fusion gene scores | * DoF score | 11 X 7 X 7=539 | 5 X 6 X 5=150 |
| # samples | 13 | 6 | |
| ** MAII score | log2(13/539*10)=-2.05177364972405 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(6/150*10)=-1.32192809488736 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: LYN [Title/Abstract] AND TGS1 [Title/Abstract] AND fusion [Title/Abstract] | ||
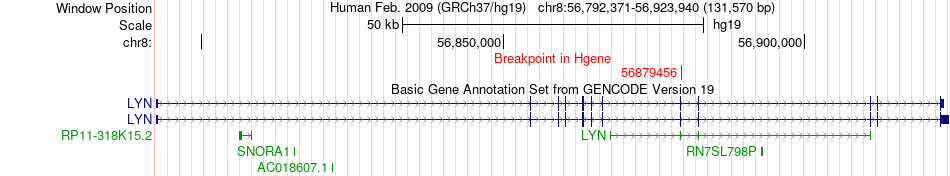
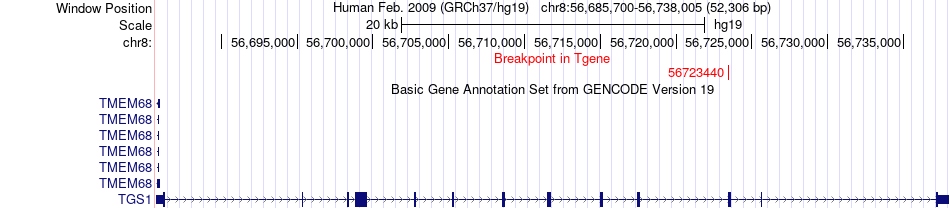
| Most frequent breakpoint | LYN(56879456)-TGS1(56723440), # samples:5 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | LYN | GO:0006468 | protein phosphorylation | 11517336 |
| Hgene | LYN | GO:0006974 | cellular response to DNA damage stimulus | 10891478|11517336 |
| Hgene | LYN | GO:0018108 | peptidyl-tyrosine phosphorylation | 7682714|11782428 |
| Hgene | LYN | GO:0046777 | protein autophosphorylation | 7682714 |
| Hgene | LYN | GO:0051272 | positive regulation of cellular component movement | 16467205 |
| Hgene | LYN | GO:0070304 | positive regulation of stress-activated protein kinase signaling cascade | 10891478 |
 Fusion gene breakpoints across LYN (5'-gene) Fusion gene breakpoints across LYN (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across TGS1 (3'-gene) Fusion gene breakpoints across TGS1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | HNSC | TCGA-CQ-6228 | LYN | chr8 | 56879456 | + | TGS1 | chr8 | 56723440 | + |
| ChimerDB4 | HNSC | TCGA-CV-7242 | LYN | chr8 | 56879456 | + | TGS1 | chr8 | 56723440 | + |
| ChimerDB4 | LGG | TCGA-HT-7602 | LYN | chr8 | 56879456 | + | TGS1 | chr8 | 56723440 | + |
| ChimerDB4 | OV | TCGA-24-1565-01A | LYN | chr8 | 56879456 | + | TGS1 | chr8 | 56723440 | + |
| ChimerDB4 | SKCM | TCGA-FS-A1ZA-06A | LYN | chr8 | 56879456 | + | TGS1 | chr8 | 56723440 | + |
Top |
Fusion Gene ORF analysis for LYN-TGS1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000420292 | ENST00000260129 | LYN | chr8 | 56879456 | + | TGS1 | chr8 | 56723440 | + |
| In-frame | ENST00000519728 | ENST00000260129 | LYN | chr8 | 56879456 | + | TGS1 | chr8 | 56723440 | + |
| In-frame | ENST00000520220 | ENST00000260129 | LYN | chr8 | 56879456 | + | TGS1 | chr8 | 56723440 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000519728 | LYN | chr8 | 56879456 | + | ENST00000260129 | TGS1 | chr8 | 56723440 | + | 2431 | 1269 | 50 | 1687 | 545 |
| ENST00000520220 | LYN | chr8 | 56879456 | + | ENST00000260129 | TGS1 | chr8 | 56723440 | + | 2346 | 1184 | 28 | 1602 | 524 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000519728 | ENST00000260129 | LYN | chr8 | 56879456 | + | TGS1 | chr8 | 56723440 | + | 0.000982315 | 0.9990177 |
| ENST00000520220 | ENST00000260129 | LYN | chr8 | 56879456 | + | TGS1 | chr8 | 56723440 | + | 0.001324736 | 0.9986753 |
Top |
Fusion Genomic Features for LYN-TGS1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| LYN | chr8 | 56879456 | + | TGS1 | chr8 | 56723439 | + | 8.97E-08 | 0.9999999 |
| LYN | chr8 | 56879456 | + | TGS1 | chr8 | 56723439 | + | 8.97E-08 | 0.9999999 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for LYN-TGS1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr8:56879456/chr8:56723440) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| LYN | . |
| FUNCTION: Non-receptor tyrosine-protein kinase that transmits signals from cell surface receptors and plays an important role in the regulation of innate and adaptive immune responses, hematopoiesis, responses to growth factors and cytokines, integrin signaling, but also responses to DNA damage and genotoxic agents. Functions primarily as negative regulator, but can also function as activator, depending on the context. Required for the initiation of the B-cell response, but also for its down-regulation and termination. Plays an important role in the regulation of B-cell differentiation, proliferation, survival and apoptosis, and is important for immune self-tolerance. Acts downstream of several immune receptors, including the B-cell receptor, CD79A, CD79B, CD5, CD19, CD22, FCER1, FCGR2, FCGR1A, TLR2 and TLR4. Plays a role in the inflammatory response to bacterial lipopolysaccharide. Mediates the responses to cytokines and growth factors in hematopoietic progenitors, platelets, erythrocytes, and in mature myeloid cells, such as dendritic cells, neutrophils and eosinophils. Acts downstream of EPOR, KIT, MPL, the chemokine receptor CXCR4, as well as the receptors for IL3, IL5 and CSF2. Plays an important role in integrin signaling. Regulates cell proliferation, survival, differentiation, migration, adhesion, degranulation, and cytokine release. Down-regulates signaling pathways by phosphorylation of immunoreceptor tyrosine-based inhibitory motifs (ITIM), that then serve as binding sites for phosphatases, such as PTPN6/SHP-1, PTPN11/SHP-2 and INPP5D/SHIP-1, that modulate signaling by dephosphorylation of kinases and their substrates. Phosphorylates LIME1 in response to CD22 activation. Phosphorylates BTK, CBL, CD5, CD19, CD72, CD79A, CD79B, CSF2RB, DOK1, HCLS1, LILRB3/PIR-B, MS4A2/FCER1B, SYK and TEC. Promotes phosphorylation of SIRPA, PTPN6/SHP-1, PTPN11/SHP-2 and INPP5D/SHIP-1. Mediates phosphorylation of the BCR-ABL fusion protein. Required for rapid phosphorylation of FER in response to FCER1 activation. Mediates KIT phosphorylation. Acts as an effector of EPOR (erythropoietin receptor) in controlling KIT expression and may play a role in erythroid differentiation during the switch between proliferation and maturation. Depending on the context, activates or inhibits several signaling cascades. Regulates phosphatidylinositol 3-kinase activity and AKT1 activation. Regulates activation of the MAP kinase signaling cascade, including activation of MAP2K1/MEK1, MAPK1/ERK2, MAPK3/ERK1, MAPK8/JNK1 and MAPK9/JNK2. Mediates activation of STAT5A and/or STAT5B. Phosphorylates LPXN on 'Tyr-72'. Kinase activity facilitates TLR4-TLR6 heterodimerization and signal initiation. Phosphorylates SCIMP on 'Tyr-107'; this enhances binding of SCIMP to TLR4, promoting the phosphorylation of TLR4, and a selective cytokine response to lipopolysaccharide in macrophages (By similarity). Phosphorylates CLNK (By similarity). {ECO:0000250|UniProtKB:P25911, ECO:0000269|PubMed:10574931, ECO:0000269|PubMed:10748115, ECO:0000269|PubMed:10891478, ECO:0000269|PubMed:11435302, ECO:0000269|PubMed:11517336, ECO:0000269|PubMed:11825908, ECO:0000269|PubMed:14726379, ECO:0000269|PubMed:15795233, ECO:0000269|PubMed:16467205, ECO:0000269|PubMed:17640867, ECO:0000269|PubMed:17977829, ECO:0000269|PubMed:18056483, ECO:0000269|PubMed:18070987, ECO:0000269|PubMed:18235045, ECO:0000269|PubMed:18577747, ECO:0000269|PubMed:18802065, ECO:0000269|PubMed:19290919, ECO:0000269|PubMed:20037584, ECO:0000269|PubMed:7687428}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LYN | chr8:56879456 | chr8:56723440 | ENST00000519728 | + | 9 | 13 | 129_226 | 324 | 513.0 | Domain | SH2 |
| Hgene | LYN | chr8:56879456 | chr8:56723440 | ENST00000519728 | + | 9 | 13 | 63_123 | 324 | 513.0 | Domain | SH3 |
| Hgene | LYN | chr8:56879456 | chr8:56723440 | ENST00000520220 | + | 9 | 13 | 129_226 | 303 | 492.0 | Domain | SH2 |
| Hgene | LYN | chr8:56879456 | chr8:56723440 | ENST00000520220 | + | 9 | 13 | 63_123 | 303 | 492.0 | Domain | SH3 |
| Hgene | LYN | chr8:56879456 | chr8:56723440 | ENST00000519728 | + | 9 | 13 | 253_261 | 324 | 513.0 | Nucleotide binding | ATP |
| Hgene | LYN | chr8:56879456 | chr8:56723440 | ENST00000520220 | + | 9 | 13 | 253_261 | 303 | 492.0 | Nucleotide binding | ATP |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LYN | chr8:56879456 | chr8:56723440 | ENST00000519728 | + | 9 | 13 | 247_501 | 324 | 513.0 | Domain | Protein kinase |
| Hgene | LYN | chr8:56879456 | chr8:56723440 | ENST00000520220 | + | 9 | 13 | 247_501 | 303 | 492.0 | Domain | Protein kinase |
| Tgene | TGS1 | chr8:56879456 | chr8:56723440 | ENST00000260129 | 9 | 13 | 611_624 | 714 | 854.0 | Compositional bias | Note=Lys-rich | |
| Tgene | TGS1 | chr8:56879456 | chr8:56723440 | ENST00000260129 | 9 | 13 | 631_846 | 714 | 854.0 | Region | Note=Sufficient for catalytic activity |
Top |
Fusion Gene Sequence for LYN-TGS1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >50448_50448_1_LYN-TGS1_LYN_chr8_56879456_ENST00000519728_TGS1_chr8_56723440_ENST00000260129_length(transcript)=2431nt_BP=1269nt TCGGCCGAGCCCAGAGACAGCCAGTTCCTCTCCCGCCGCGCCGGGCCGCGCTGCCGCTCGCTCCCCGGCCGTGGCGCCTCCGGGCCAGAC GCGCTGCAGCCTCCAGCCCGCGGCAAGCGGGGCGGCCGCGCCACCCCCGGCCCCGCGCCAGCAGCCCCTCGCCGCGCGTCCAGCGTTCCC GGCCAGCAGCCTCCCCATACGCAGGTCCTGCTGGGCCGCCCCGTCGCGCCCCCCACTCTGAACTCAAGTCACCGTGGAGCTCCGCCGCCC CGAAACTTTCACCGCGAGCGGGAAATATGGGATGTATAAAATCAAAAGGGAAAGACAGCTTGAGTGACGATGGAGTAGATTTGAAGACTC AACCAGTACGTAATACTGAAAGAACTATTTATGTGAGAGATCCAACGTCCAATAAACAGCAAAGGCCAGTTCCAGAATCTCAGCTTTTAC CTGGACAGAGGTTTCAAACTAAAGATCCAGAGGAACAAGGAGACATTGTGGTAGCCTTGTACCCCTATGATGGCATCCACCCGGACGACT TGTCTTTCAAGAAAGGAGAGAAGATGAAAGTCCTGGAGGAGCATGGAGAATGGTGGAAAGCAAAGTCCCTTTTAACAAAAAAAGAAGGCT TCATCCCCAGCAACTATGTGGCCAAACTCAACACCTTAGAAACAGAAGAGTGGTTTTTCAAGGATATAACCAGGAAGGACGCAGAAAGGC AGCTTTTGGCACCAGGAAATAGCGCTGGAGCTTTCCTTATTAGAGAAAGTGAAACATTAAAAGGAAGCTTCTCTCTGTCTGTCAGAGACT TTGACCCTGTGCATGGTGATGTTATTAAGCACTACAAAATTAGAAGTCTGGATAATGGGGGCTATTACATCTCTCCACGAATCACTTTTC CCTGTATCAGCGACATGATTAAACATTACCAAAAGCAGGCAGATGGCTTGTGCAGAAGATTGGAGAAGGCTTGTATTAGTCCCAAGCCAC AGAAGCCATGGGATAAAGATGCCTGGGAGATCCCCCGGGAGTCCATCAAGTTGGTGAAAAGGCTTGGCGCTGGGCAGTTTGGGGAAGTCT GGATGGGTTACTATAACAACAGTACCAAGGTGGCTGTGAAAACCCTGAAGCCAGGAACTATGTCTGTGCAAGCCTTCCTGGAAGAAGCCA ACCTCATGAAGACCCTGCAGCATGACAAGCTCGTGAGGCTCTACGCTGTGGTCACCAGGGAGGAGCCCATTTACATCATCACCGAGTACA TGGCCAAGGTGATTGCCATTGATATCGATCCTGTTAAGATTGCCCTTGCTCGCAATAATGCAGAAGTTTATGGGATAGCAGATAAGATAG AGTTCATCTGTGGAGATTTCTTGCTGCTGGCTTCTTTTTTAAAGGCTGATGTTGTGTTCCTCAGCCCACCTTGGGGAGGGCCAGACTATG CCACTGCAGAGACCTTTGACATTAGAACAATGATGTCTCCTGATGGCTTTGAAATTTTCAGACTTTCTAAGAAGATCACTAATAATATTG TTTATTTTCTTCCAAGAAATGCTGATATTGACCAGGTGGCATCCTTAGCTGGGCCTGGAGGGCAAGTGGAAATAGAACAGAACTTCCTTA ACAACAAATTGAAGACAATCACTGCATATTTTGGTGACCTAATTCGAAGACCAGCCTCTGAAACCTAACTATGCAGCAGTGCGAGGACAA AAGATCATGGAGTGGTCAAAATATTCAGATGAGACATTTGGCATGTCTTCCTTTATTCACTGATATTTTCTACCCATGGTCTTATATCAC CGTATGAAATGGAAACTTACAGGACTTAAATATCAGTGAAATATTTTGAGATCTTTGAATAATTCCTTTAGAGGAATTATACAAAATTAA TATATATGAGTCCTTTGTAATTTATTTTTTTTTGAGACAGGATCTCACTTTTACCGCCCAGGCTGGAGTGCAGTGCCATGATCACAGCTC ACTGCAGGTTCAGCCTTCTGAGTTCAAGCAATCCTTCTGTCTCAGTCTCCTCAGTAGCTGGGGCTTTAGGTGGGCACTGCCACACCGTAC TAATTTTTGTATTTTTTGTAGAGACGAGGTCCCACCATGTTGCCCAGGCTGGTGTCAAACTCCTGGGCTCAGTCAGTCCCCCCATCTCAC CCTCCCCAAGTGCTGGAATTACAGGCGTGAGCTACTGTGCCCAGCCTTACGGACATCCTTTTGAATTATCTTTTTCACTCATAGAATATG AATACATTTATTTAGACTTTTTCTAGAACTTTCCTGTTTTCATGTCTTTGCTTCATCTGGAATTGGCTTAACACCCTTTTATAAAGTTTG TGTTTGTAAAATTTCCATTGTGACATCAATACGCAATATATTTTGTAATATAGGAGTTTCTATTTTTTTATTAAAATGGCAATGAAAGCA >50448_50448_1_LYN-TGS1_LYN_chr8_56879456_ENST00000519728_TGS1_chr8_56723440_ENST00000260129_length(amino acids)=545AA_BP=406 MPLAPRPWRLRARRAAASSPRQAGRPRHPRPRASSPSPRVQRSRPAASPYAGPAGPPRRAPHSELKSPWSSAAPKLSPRAGNMGCIKSKG KDSLSDDGVDLKTQPVRNTERTIYVRDPTSNKQQRPVPESQLLPGQRFQTKDPEEQGDIVVALYPYDGIHPDDLSFKKGEKMKVLEEHGE WWKAKSLLTKKEGFIPSNYVAKLNTLETEEWFFKDITRKDAERQLLAPGNSAGAFLIRESETLKGSFSLSVRDFDPVHGDVIKHYKIRSL DNGGYYISPRITFPCISDMIKHYQKQADGLCRRLEKACISPKPQKPWDKDAWEIPRESIKLVKRLGAGQFGEVWMGYYNNSTKVAVKTLK PGTMSVQAFLEEANLMKTLQHDKLVRLYAVVTREEPIYIITEYMAKVIAIDIDPVKIALARNNAEVYGIADKIEFICGDFLLLASFLKAD VVFLSPPWGGPDYATAETFDIRTMMSPDGFEIFRLSKKITNNIVYFLPRNADIDQVASLAGPGGQVEIEQNFLNNKLKTITAYFGDLIRR -------------------------------------------------------------- >50448_50448_2_LYN-TGS1_LYN_chr8_56879456_ENST00000520220_TGS1_chr8_56723440_ENST00000260129_length(transcript)=2346nt_BP=1184nt AGTTCCTCTCCCGCCGCGCCGGGCCGCGCTGCCGCTCGCTCCCCGGCCGTGGCGCCTCCGGGCCAGACGCGCTGCAGCCTCCAGCCCGCG GCAAGCGGGGCGGCCGCGCCACCCCCGGCCCCGCGCCAGCAGCCCCTCGCCGCGCGTCCAGCGTTCCCGGCCAGCAGCCTCCCCATACGC AGGTCCTGCTGGGCCGCCCCGTCGCGCCCCCCACTCTGAACTCAAGTCACCGTGGAGCTCCGCCGCCCCGAAACTTTCACCGCGAGCGGG AAATATGGGATGTATAAAATCAAAAGGGAAAGACAGCTTGAGTGACGATGGAGTAGATTTGAAGACTCAACCAGTTCCAGAATCTCAGCT TTTACCTGGACAGAGGTTTCAAACTAAAGATCCAGAGGAACAAGGAGACATTGTGGTAGCCTTGTACCCCTATGATGGCATCCACCCGGA CGACTTGTCTTTCAAGAAAGGAGAGAAGATGAAAGTCCTGGAGGAGCATGGAGAATGGTGGAAAGCAAAGTCCCTTTTAACAAAAAAAGA AGGCTTCATCCCCAGCAACTATGTGGCCAAACTCAACACCTTAGAAACAGAAGAGTGGTTTTTCAAGGATATAACCAGGAAGGACGCAGA AAGGCAGCTTTTGGCACCAGGAAATAGCGCTGGAGCTTTCCTTATTAGAGAAAGTGAAACATTAAAAGGAAGCTTCTCTCTGTCTGTCAG AGACTTTGACCCTGTGCATGGTGATGTTATTAAGCACTACAAAATTAGAAGTCTGGATAATGGGGGCTATTACATCTCTCCACGAATCAC TTTTCCCTGTATCAGCGACATGATTAAACATTACCAAAAGCAGGCAGATGGCTTGTGCAGAAGATTGGAGAAGGCTTGTATTAGTCCCAA GCCACAGAAGCCATGGGATAAAGATGCCTGGGAGATCCCCCGGGAGTCCATCAAGTTGGTGAAAAGGCTTGGCGCTGGGCAGTTTGGGGA AGTCTGGATGGGTTACTATAACAACAGTACCAAGGTGGCTGTGAAAACCCTGAAGCCAGGAACTATGTCTGTGCAAGCCTTCCTGGAAGA AGCCAACCTCATGAAGACCCTGCAGCATGACAAGCTCGTGAGGCTCTACGCTGTGGTCACCAGGGAGGAGCCCATTTACATCATCACCGA GTACATGGCCAAGGTGATTGCCATTGATATCGATCCTGTTAAGATTGCCCTTGCTCGCAATAATGCAGAAGTTTATGGGATAGCAGATAA GATAGAGTTCATCTGTGGAGATTTCTTGCTGCTGGCTTCTTTTTTAAAGGCTGATGTTGTGTTCCTCAGCCCACCTTGGGGAGGGCCAGA CTATGCCACTGCAGAGACCTTTGACATTAGAACAATGATGTCTCCTGATGGCTTTGAAATTTTCAGACTTTCTAAGAAGATCACTAATAA TATTGTTTATTTTCTTCCAAGAAATGCTGATATTGACCAGGTGGCATCCTTAGCTGGGCCTGGAGGGCAAGTGGAAATAGAACAGAACTT CCTTAACAACAAATTGAAGACAATCACTGCATATTTTGGTGACCTAATTCGAAGACCAGCCTCTGAAACCTAACTATGCAGCAGTGCGAG GACAAAAGATCATGGAGTGGTCAAAATATTCAGATGAGACATTTGGCATGTCTTCCTTTATTCACTGATATTTTCTACCCATGGTCTTAT ATCACCGTATGAAATGGAAACTTACAGGACTTAAATATCAGTGAAATATTTTGAGATCTTTGAATAATTCCTTTAGAGGAATTATACAAA ATTAATATATATGAGTCCTTTGTAATTTATTTTTTTTTGAGACAGGATCTCACTTTTACCGCCCAGGCTGGAGTGCAGTGCCATGATCAC AGCTCACTGCAGGTTCAGCCTTCTGAGTTCAAGCAATCCTTCTGTCTCAGTCTCCTCAGTAGCTGGGGCTTTAGGTGGGCACTGCCACAC CGTACTAATTTTTGTATTTTTTGTAGAGACGAGGTCCCACCATGTTGCCCAGGCTGGTGTCAAACTCCTGGGCTCAGTCAGTCCCCCCAT CTCACCCTCCCCAAGTGCTGGAATTACAGGCGTGAGCTACTGTGCCCAGCCTTACGGACATCCTTTTGAATTATCTTTTTCACTCATAGA ATATGAATACATTTATTTAGACTTTTTCTAGAACTTTCCTGTTTTCATGTCTTTGCTTCATCTGGAATTGGCTTAACACCCTTTTATAAA GTTTGTGTTTGTAAAATTTCCATTGTGACATCAATACGCAATATATTTTGTAATATAGGAGTTTCTATTTTTTTATTAAAATGGCAATGA >50448_50448_2_LYN-TGS1_LYN_chr8_56879456_ENST00000520220_TGS1_chr8_56723440_ENST00000260129_length(amino acids)=524AA_BP=385 MPLAPRPWRLRARRAAASSPRQAGRPRHPRPRASSPSPRVQRSRPAASPYAGPAGPPRRAPHSELKSPWSSAAPKLSPRAGNMGCIKSKG KDSLSDDGVDLKTQPVPESQLLPGQRFQTKDPEEQGDIVVALYPYDGIHPDDLSFKKGEKMKVLEEHGEWWKAKSLLTKKEGFIPSNYVA KLNTLETEEWFFKDITRKDAERQLLAPGNSAGAFLIRESETLKGSFSLSVRDFDPVHGDVIKHYKIRSLDNGGYYISPRITFPCISDMIK HYQKQADGLCRRLEKACISPKPQKPWDKDAWEIPRESIKLVKRLGAGQFGEVWMGYYNNSTKVAVKTLKPGTMSVQAFLEEANLMKTLQH DKLVRLYAVVTREEPIYIITEYMAKVIAIDIDPVKIALARNNAEVYGIADKIEFICGDFLLLASFLKADVVFLSPPWGGPDYATAETFDI -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for LYN-TGS1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for LYN-TGS1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for LYN-TGS1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
