
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:MAN2A1-TRIM9 (FusionGDB2 ID:51100) |
Fusion Gene Summary for MAN2A1-TRIM9 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: MAN2A1-TRIM9 | Fusion gene ID: 51100 | Hgene | Tgene | Gene symbol | MAN2A1 | TRIM9 | Gene ID | 4124 | 114088 |
| Gene name | mannosidase alpha class 2A member 1 | tripartite motif containing 9 | |
| Synonyms | AMan II|GOLIM7|MANA2|MANII | RNF91|SPRING | |
| Cytomap | 5q21.3 | 14q22.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | alpha-mannosidase 2Golgi alpha-mannosidase IIgolgi integral membrane protein 7mannosidase, alpha type IImannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase | E3 ubiquitin-protein ligase TRIM9RING finger protein 91RING-type E3 ubiquitin transferase TRIM9SNAP-25-interacting RING finger proteinhomolog of rat RING finger Springtripartite motif-containing protein 9 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q16706 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000261483, ENST00000505313, | ENST00000557456, ENST00000298355, ENST00000338969, ENST00000360392, | |
| Fusion gene scores | * DoF score | 14 X 12 X 8=1344 | 5 X 4 X 5=100 |
| # samples | 14 | 5 | |
| ** MAII score | log2(14/1344*10)=-3.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/100*10)=-1 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: MAN2A1 [Title/Abstract] AND TRIM9 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | MAN2A1(109026253)-TRIM9(51492078), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | TRIM9 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 20085810 |
 Fusion gene breakpoints across MAN2A1 (5'-gene) Fusion gene breakpoints across MAN2A1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
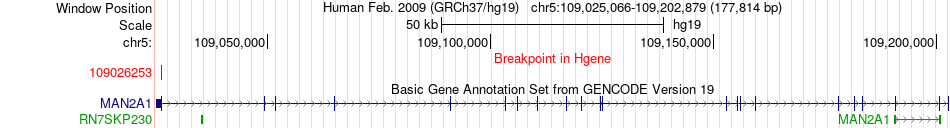 |
 Fusion gene breakpoints across TRIM9 (3'-gene) Fusion gene breakpoints across TRIM9 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
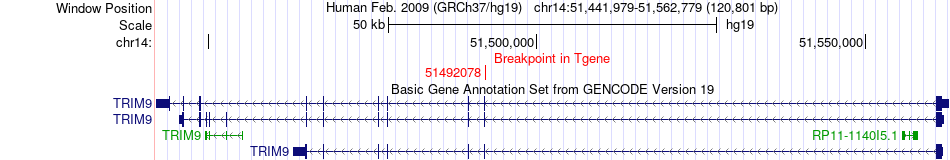 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-DX-A3M1-01A | MAN2A1 | chr5 | 109026253 | - | TRIM9 | chr14 | 51492078 | - |
Top |
Fusion Gene ORF analysis for MAN2A1-TRIM9 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000261483 | ENST00000557456 | MAN2A1 | chr5 | 109026253 | - | TRIM9 | chr14 | 51492078 | - |
| In-frame | ENST00000261483 | ENST00000298355 | MAN2A1 | chr5 | 109026253 | - | TRIM9 | chr14 | 51492078 | - |
| In-frame | ENST00000261483 | ENST00000338969 | MAN2A1 | chr5 | 109026253 | - | TRIM9 | chr14 | 51492078 | - |
| In-frame | ENST00000261483 | ENST00000360392 | MAN2A1 | chr5 | 109026253 | - | TRIM9 | chr14 | 51492078 | - |
| intron-3CDS | ENST00000505313 | ENST00000298355 | MAN2A1 | chr5 | 109026253 | - | TRIM9 | chr14 | 51492078 | - |
| intron-3CDS | ENST00000505313 | ENST00000338969 | MAN2A1 | chr5 | 109026253 | - | TRIM9 | chr14 | 51492078 | - |
| intron-3CDS | ENST00000505313 | ENST00000360392 | MAN2A1 | chr5 | 109026253 | - | TRIM9 | chr14 | 51492078 | - |
| intron-intron | ENST00000505313 | ENST00000557456 | MAN2A1 | chr5 | 109026253 | - | TRIM9 | chr14 | 51492078 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000261483 | MAN2A1 | chr5 | 109026253 | - | ENST00000298355 | TRIM9 | chr14 | 51492078 | - | 4527 | 1187 | 731 | 2497 | 588 |
| ENST00000261483 | MAN2A1 | chr5 | 109026253 | - | ENST00000338969 | TRIM9 | chr14 | 51492078 | - | 3287 | 1187 | 731 | 2773 | 680 |
| ENST00000261483 | MAN2A1 | chr5 | 109026253 | - | ENST00000360392 | TRIM9 | chr14 | 51492078 | - | 3954 | 1187 | 731 | 2017 | 428 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000261483 | ENST00000298355 | MAN2A1 | chr5 | 109026253 | - | TRIM9 | chr14 | 51492078 | - | 0.001130439 | 0.9988695 |
| ENST00000261483 | ENST00000338969 | MAN2A1 | chr5 | 109026253 | - | TRIM9 | chr14 | 51492078 | - | 0.00680826 | 0.9931918 |
| ENST00000261483 | ENST00000360392 | MAN2A1 | chr5 | 109026253 | - | TRIM9 | chr14 | 51492078 | - | 0.002272456 | 0.9977276 |
Top |
Fusion Genomic Features for MAN2A1-TRIM9 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
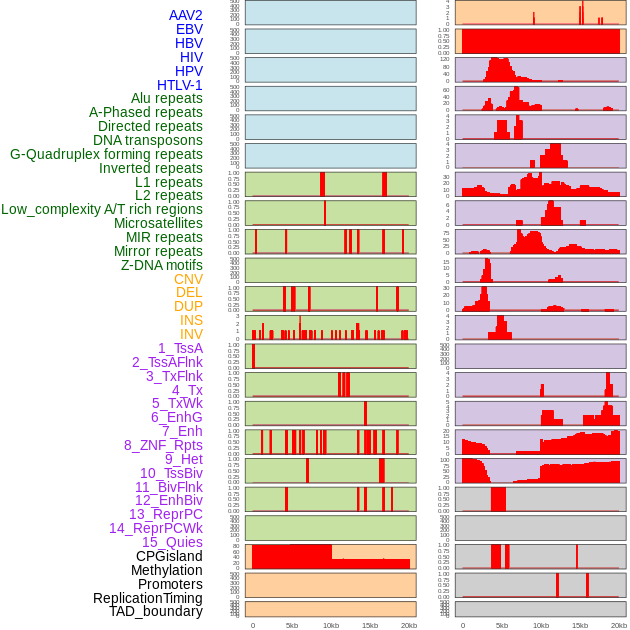 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for MAN2A1-TRIM9 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr5:109026253/chr14:51492078) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MAN2A1 | . |
| FUNCTION: Catalyzes the first committed step in the biosynthesis of complex N-glycans. It controls conversion of high mannose to complex N-glycans; the final hydrolytic step in the N-glycan maturation pathway. {ECO:0000250|UniProtKB:P28494}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MAN2A1 | chr5:109026253 | chr14:51492078 | ENST00000261483 | - | 1 | 22 | 1_5 | 45 | 1145.0 | Topological domain | Cytoplasmic |
| Hgene | MAN2A1 | chr5:109026253 | chr14:51492078 | ENST00000261483 | - | 1 | 22 | 6_26 | 45 | 1145.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Tgene | TRIM9 | chr5:109026253 | chr14:51492078 | ENST00000298355 | 0 | 10 | 273_340 | 274 | 711.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | TRIM9 | chr5:109026253 | chr14:51492078 | ENST00000338969 | 0 | 12 | 273_340 | 274 | 803.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | TRIM9 | chr5:109026253 | chr14:51492078 | ENST00000360392 | 0 | 7 | 273_340 | 274 | 551.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | TRIM9 | chr5:109026253 | chr14:51492078 | ENST00000298355 | 0 | 10 | 374_432 | 274 | 711.0 | Domain | COS | |
| Tgene | TRIM9 | chr5:109026253 | chr14:51492078 | ENST00000298355 | 0 | 10 | 440_535 | 274 | 711.0 | Domain | Fibronectin type-III | |
| Tgene | TRIM9 | chr5:109026253 | chr14:51492078 | ENST00000298355 | 0 | 10 | 533_702 | 274 | 711.0 | Domain | B30.2/SPRY | |
| Tgene | TRIM9 | chr5:109026253 | chr14:51492078 | ENST00000338969 | 0 | 12 | 374_432 | 274 | 803.0 | Domain | COS | |
| Tgene | TRIM9 | chr5:109026253 | chr14:51492078 | ENST00000338969 | 0 | 12 | 440_535 | 274 | 803.0 | Domain | Fibronectin type-III | |
| Tgene | TRIM9 | chr5:109026253 | chr14:51492078 | ENST00000338969 | 0 | 12 | 533_702 | 274 | 803.0 | Domain | B30.2/SPRY | |
| Tgene | TRIM9 | chr5:109026253 | chr14:51492078 | ENST00000360392 | 0 | 7 | 374_432 | 274 | 551.0 | Domain | COS | |
| Tgene | TRIM9 | chr5:109026253 | chr14:51492078 | ENST00000360392 | 0 | 7 | 440_535 | 274 | 551.0 | Domain | Fibronectin type-III | |
| Tgene | TRIM9 | chr5:109026253 | chr14:51492078 | ENST00000360392 | 0 | 7 | 533_702 | 274 | 551.0 | Domain | B30.2/SPRY |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MAN2A1 | chr5:109026253 | chr14:51492078 | ENST00000261483 | - | 1 | 22 | 27_1144 | 45 | 1145.0 | Topological domain | Lumenal |
| Tgene | TRIM9 | chr5:109026253 | chr14:51492078 | ENST00000298355 | 0 | 10 | 10_50 | 274 | 711.0 | Zinc finger | RING-type | |
| Tgene | TRIM9 | chr5:109026253 | chr14:51492078 | ENST00000298355 | 0 | 10 | 163_212 | 274 | 711.0 | Zinc finger | B box-type 1 | |
| Tgene | TRIM9 | chr5:109026253 | chr14:51492078 | ENST00000298355 | 0 | 10 | 224_266 | 274 | 711.0 | Zinc finger | B box-type 2 | |
| Tgene | TRIM9 | chr5:109026253 | chr14:51492078 | ENST00000338969 | 0 | 12 | 10_50 | 274 | 803.0 | Zinc finger | RING-type | |
| Tgene | TRIM9 | chr5:109026253 | chr14:51492078 | ENST00000338969 | 0 | 12 | 163_212 | 274 | 803.0 | Zinc finger | B box-type 1 | |
| Tgene | TRIM9 | chr5:109026253 | chr14:51492078 | ENST00000338969 | 0 | 12 | 224_266 | 274 | 803.0 | Zinc finger | B box-type 2 | |
| Tgene | TRIM9 | chr5:109026253 | chr14:51492078 | ENST00000360392 | 0 | 7 | 10_50 | 274 | 551.0 | Zinc finger | RING-type | |
| Tgene | TRIM9 | chr5:109026253 | chr14:51492078 | ENST00000360392 | 0 | 7 | 163_212 | 274 | 551.0 | Zinc finger | B box-type 1 | |
| Tgene | TRIM9 | chr5:109026253 | chr14:51492078 | ENST00000360392 | 0 | 7 | 224_266 | 274 | 551.0 | Zinc finger | B box-type 2 |
Top |
Fusion Gene Sequence for MAN2A1-TRIM9 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >51100_51100_1_MAN2A1-TRIM9_MAN2A1_chr5_109026253_ENST00000261483_TRIM9_chr14_51492078_ENST00000298355_length(transcript)=4527nt_BP=1187nt GGCGGGGCCAGCCGCCCGCTCGGCTCAGGCGCTGCGGGCGCCTATTGACCCAGCGGCTGCTGCGCCGCCGCTGTCTCCTCCTGCTCGTGG CGGGCGGTGCTGGAGCGCCAAGTGGCGCTGGAGAACCGGCGCTTCCTTTCGCCGCTTCCGCCGCCATCTCCGCGTTTGTGGGGCGGGAAA GAGGGAGGGGGCTAGCGGCTGCAGCTGGAGCGGGCTTCTCTCCGGGGACGGTCCTTTCCTCCCTGCTCTCCTTTTCCTTCTTTCCGCGTT GCCGCCGCCCGCCCCCTGCGCCTCCCCGCGGAGCCTGGGTCCGGGAGGGGGAAGGTAGGGGCGGCGGGGGGCGGGAGAGTCTGGCGAGCG GACGCTAGCTCTGAGGAAACTCATCAATCCGTGAGCCCCGGAGTCCGGGGTGCACATCGGCCCAGCCGCAGCGTCGGCGGCGGCGGCGGC AGCAGCACGAAGGGGCTCAGTCGGGGTAGGCGGGGGCGGTGCCGGTGCCGCGGGGGCGGGCCCGACCGTCCCGCCCAGAAGTTGTAGGGC TTGGCTCCTCGCGATCTTGTTCCTTTCCCCTCCGCTTCTCTGACCTAGCTGCGCGGCCCCGGCCCGGGAGCTGCCGAACCCGCGCCTCCC CTGGGTGAGGAGGACACGCCTGCCCTCGTCGAGAAAACTTTTCCTGCCGACTCAGTTGGGGCGGCGGTGGCAGGAAGTGCGGGCAGCGAC CTCTCCTCCGCCTGCCCCGCGCGCCCTGCCGGAGGTCGGCGCTGAGCTTGCGATCAAGTTTGTGGGGGCCCCCCTTCCCAGTTGCCGGCG AGTCTCGCCTCGAGAGGGGCGCCCGACCCCGGGGAGGGCGGCAGGCCAGGGCGAAGGCCAAGGGCGTGTGGTGGCGCCGGAGACTAGGTG CGGAGCAAGGCGGGGACTCGCACCCGCATCCGAGAGCGCGGAGGTCGCGCAGCCCGGGAGAAGGGAGCCTCCGGCGGCTGCTTCCTAGAG TCCACAGTGCGCTGTCTCCTTTGGCTGAGGAGAGTGTCCTGGCCCCGAGTCTATCGAGGAAAATGAAGTTAAGCCGCCAGTTCACCGTGT TCGGCAGTGCGATCTTCTGTGTGGTGATTTTCTCGCTCTACCTGATGCTGGACCGGGGTCACTTAGACTACCCCAGGAACCCGCGCCGCG AGGGCTCCTTCCCTCAGAGCCAGCTCTCCCAGGCGCTGAACGGACTGTCAGACAGGGCCAAAGAAGCCAAGGAGTTTCTGGTACAGCTGC GCAACATGGTCCAGCAGATCCAGGAGAACAGTGTGGAGTTTGAAGCCTGTCTGGTGGCCCAATGTGATGCCCTCATCGATGCCCTCAACA GAAGAAAAGCCCAGCTGCTGGCCCGCGTCAACAAGGAGCATGAGCACAAGCTGAAGGTGGTTCGAGATCAGATCTCTCACTGCACAGTGA AATTGCGCCAGACCACAGGTCTCATGGAGTACTGCTTGGAGGTGATTAAGGAAAATGATCCTAGTGGTTTTTTGCAGATTTCTGACGCCC TCATAAGAAGAGTGCACCTGACTGAGGATCAGTGGGGTAAAGGCACACTCACTCCAAGGATGACCACGGACTTTGACTTGAGTCTGGACA ACAGCCCTCTGCTGCAATCCATCCACCAGCTGGATTTCGTGCAAGTGAAAGCTTCCTCTCCAGTCCCAGCAACCCCTATCCTACAGCTGG AGGAATGTTGTACCCACAACAACAGCGCTACGTTGTCCTGGAAACAGCCACCTCTGTCCACGGTGCCCGCCGATGGATACATTCTGGAGC TGGATGATGGCAACGGTGGTCAATTCCGGGAGGTGTATGTGGGGAAGGAGACAATGTGCACTGTGGATGGTCTTCACTTCAACAGCACAT ACAACGCTCGGGTCAAGGCCTTCAACAAAACAGGAGTCAGCCCGTACAGCAAGACCCTGGTCCTCCAAACGTCTGAGGTGGCCTGGTTTG CTTTCGACCCTGGCTCGGCGCACTCGGACATCATCCTCTCCAATGACAACCTGACAGTGACCTGTAGTAGCTATGATGACCGGGTGGTGC TAGGGAAGACTGGCTTCTCCAAGGGCATCCACTACTGGGAGCTCACGGTAGATCGCTATGACAACCACCCTGATCCTGCCTTTGGTGTGG CTCGCATGGACGTGATGAAGGATGTGATGTTAGGAAAAGACGACAAAGCTTGGGCAATGTATGTGGACAATAACCGGAGCTGGTTCATGC ACAACAACTCGCACACCAACAGAACTGAGGGAGGGATCACAAAAGGGGCCACAATTGGGGTCCTCCTCGACTTAAATAGAAAAAACTTGA CATTTTTTATCAACGATGAACAACAAGGTCCCATAGCATTTGATAACGTGGAGGGCCTCTTCTTCCCTGCGGTCAGCCTGAACAGGAACG TGCAGGTCACGCTGCACACCGGGCTCCCAGTCCCCGACTTCTACTCCAGCAGAGCATCAATAGCCTAAGGATGTGCCGTGGAGGCGCCAG CTGCCTGTTCTTACCTCCGCCTGCGAGAGCCACAGCAAGGAGCTCAGCCAGCCGTGGTGGGGTGCAGAGTTGGCAGGAGTGGGAGAAGGA GGAGAGAAAAGCTGGTCCTCTGCAGTCTTTACACCCACAGCTCTGCCCTTTTCCCTTTCAACCTCTCCTCCGCTGTCATGCCTGCTTCCG CTTCCATGTCCAACAATTCTAACCAACAAAGGACCTAGACAGCCCACCAAGTCACTTGGTTCCCACTCCCAGATTTTGCTTTTATTTAAC TTAATTTTTTATGTAGGTGAGTTATATTTTCTTTCTTTTCTGATCAGGTTATTGGTGACTTACTGGACTGGCACCGCCAGGAGAAAATTC TCCTGCTAACTTTTTTTCTTAAGCTTTCTGTCAAACAATGAGGTTGTAGGGGGAGGTAGGGAAGAATGAGCTGAATTTGTAGCATACAGC TTACCTGCTAGAATGTTCTTACCCTCTTACCTCTCCTGTAGCGTTAGCTCTGCAGAGCTAAGCTTTGGGAGAATGAATCTATCACTGAGA AGTTTTACTACTCATTGAAGCACAAAAATATCCGCTACACAGGTTCACATCAGGGTAGGATCGTTAGGATGGCTCTATAATTAGGATAGG CCAGAACCCTGGCCACTGTATAAAATTGGAAAGTCTCTAAGACACAGGCAAACCAGGGGACTTAATGATTTGGTTCGTTTATTTAATAAA AGCTTTTAATTGAGAACCTACTATTGACCAGGCACTGTTCAAGACACTAGAACATTATGTTCTAGACACTGTGGGGACACAGTGGTGAAC AGACAGGCATGGTCTCTGCCCTTCGGAGCTTATATTCCAGTAGGAGAAAAAGCAAAAATAAAATAATTATAAATGGTCATATGTACTAGA AAGGAAATGGGCAAGGGTCAAAGAGACTCATGGGCAAGATCTTTCAGGCTTCCTAGAATGATGCTAAAAATCCACACATTCTGGCCATAG CACCAGAAATGTACCAACTCAATGCCTTTTCAGCTGCACTATTTAATTATGGTACCTGCTGCCACTCATCACAGAGCCCTTCTTGGTAGA AATGATGTCAGCATTCTTATTTGCTGATGCTGCTATGATACCACGCCTTAAAAATTGACAGTTGAAAAAAAAAAGAGTGACCAGAGGGCA AAGGACCCATTCTCAGTAATGGGAGATCTTGAGTTGCAGTCCACAGCATTCACATTCTCAGGATAAACATGCTTGTCTCCATATATACTA TCTGTGCCTGTTATGAATGAAAAATCCATCCACTCTGCTGCCATTCACAGCCTAATTTTCTGGAGTAGTCCAAACAATGTTTGGAAAAAC ATGGGACTGTATGTCATTACTATGACTGATAGCAGAATAATAATGCATCTTACTTCTATATAGAGATAGACTTACATAGGTTACCCTTGA AATTCATTAGTTTGTCATAAAGTTTTAGGAAAGGTAGGACCCGGAAAGAAGTTCTAATTAGTTGTCTAAATATTTTTCAGTGAGCCAAGA AATTCACCATGAAAAAACAAGAATAACAAATAGAAGGGAAGAGATAGGATGGGAAAGCTAACAAATTAAAGTTTTGGCAAAAAGGAATAT ATGTAAATAGCTAATTATTTACTTTTGTGCTTACTTTATTTAGATTATTTCTATCAGTTACAATCTTTTTCTAGTTAAGTGTACCTAATT TATGGAATGGGTGCTATCCTGTTTATGTGTGTCTTGGTTTTTCTTGGCTACAGAAAAACTGTTGCAGGGCAACACTAGTTTGATATTTGA TTTACTCTCCAATGAGACTCAATGGCTGGGCCGTGGTAGACTCATAGTTCCTCTTGTTCTTTATTAAATTCATCCTGCTAATTAGATTTC TAGTGACTTGTAACATGTAGTTTACACTGAATTGCAATTACAGATGCATACAACTACTATACTAGAAAAAATTGTCATTTCTGATGTGCC >51100_51100_1_MAN2A1-TRIM9_MAN2A1_chr5_109026253_ENST00000261483_TRIM9_chr14_51492078_ENST00000298355_length(amino acids)=588AA_BP=151 MPRAPCRRSALSLRSSLWGPPFPVAGESRLERGARPRGGRQARAKAKGVWWRRRLGAEQGGDSHPHPRARRSRSPGEGSLRRLLPRVHSA LSPLAEESVLAPSLSRKMKLSRQFTVFGSAIFCVVIFSLYLMLDRGHLDYPRNPRREGSFPQSQLSQALNGLSDRAKEAKEFLVQLRNMV QQIQENSVEFEACLVAQCDALIDALNRRKAQLLARVNKEHEHKLKVVRDQISHCTVKLRQTTGLMEYCLEVIKENDPSGFLQISDALIRR VHLTEDQWGKGTLTPRMTTDFDLSLDNSPLLQSIHQLDFVQVKASSPVPATPILQLEECCTHNNSATLSWKQPPLSTVPADGYILELDDG NGGQFREVYVGKETMCTVDGLHFNSTYNARVKAFNKTGVSPYSKTLVLQTSEVAWFAFDPGSAHSDIILSNDNLTVTCSSYDDRVVLGKT GFSKGIHYWELTVDRYDNHPDPAFGVARMDVMKDVMLGKDDKAWAMYVDNNRSWFMHNNSHTNRTEGGITKGATIGVLLDLNRKNLTFFI -------------------------------------------------------------- >51100_51100_2_MAN2A1-TRIM9_MAN2A1_chr5_109026253_ENST00000261483_TRIM9_chr14_51492078_ENST00000338969_length(transcript)=3287nt_BP=1187nt GGCGGGGCCAGCCGCCCGCTCGGCTCAGGCGCTGCGGGCGCCTATTGACCCAGCGGCTGCTGCGCCGCCGCTGTCTCCTCCTGCTCGTGG CGGGCGGTGCTGGAGCGCCAAGTGGCGCTGGAGAACCGGCGCTTCCTTTCGCCGCTTCCGCCGCCATCTCCGCGTTTGTGGGGCGGGAAA GAGGGAGGGGGCTAGCGGCTGCAGCTGGAGCGGGCTTCTCTCCGGGGACGGTCCTTTCCTCCCTGCTCTCCTTTTCCTTCTTTCCGCGTT GCCGCCGCCCGCCCCCTGCGCCTCCCCGCGGAGCCTGGGTCCGGGAGGGGGAAGGTAGGGGCGGCGGGGGGCGGGAGAGTCTGGCGAGCG GACGCTAGCTCTGAGGAAACTCATCAATCCGTGAGCCCCGGAGTCCGGGGTGCACATCGGCCCAGCCGCAGCGTCGGCGGCGGCGGCGGC AGCAGCACGAAGGGGCTCAGTCGGGGTAGGCGGGGGCGGTGCCGGTGCCGCGGGGGCGGGCCCGACCGTCCCGCCCAGAAGTTGTAGGGC TTGGCTCCTCGCGATCTTGTTCCTTTCCCCTCCGCTTCTCTGACCTAGCTGCGCGGCCCCGGCCCGGGAGCTGCCGAACCCGCGCCTCCC CTGGGTGAGGAGGACACGCCTGCCCTCGTCGAGAAAACTTTTCCTGCCGACTCAGTTGGGGCGGCGGTGGCAGGAAGTGCGGGCAGCGAC CTCTCCTCCGCCTGCCCCGCGCGCCCTGCCGGAGGTCGGCGCTGAGCTTGCGATCAAGTTTGTGGGGGCCCCCCTTCCCAGTTGCCGGCG AGTCTCGCCTCGAGAGGGGCGCCCGACCCCGGGGAGGGCGGCAGGCCAGGGCGAAGGCCAAGGGCGTGTGGTGGCGCCGGAGACTAGGTG CGGAGCAAGGCGGGGACTCGCACCCGCATCCGAGAGCGCGGAGGTCGCGCAGCCCGGGAGAAGGGAGCCTCCGGCGGCTGCTTCCTAGAG TCCACAGTGCGCTGTCTCCTTTGGCTGAGGAGAGTGTCCTGGCCCCGAGTCTATCGAGGAAAATGAAGTTAAGCCGCCAGTTCACCGTGT TCGGCAGTGCGATCTTCTGTGTGGTGATTTTCTCGCTCTACCTGATGCTGGACCGGGGTCACTTAGACTACCCCAGGAACCCGCGCCGCG AGGGCTCCTTCCCTCAGAGCCAGCTCTCCCAGGCGCTGAACGGACTGTCAGACAGGGCCAAAGAAGCCAAGGAGTTTCTGGTACAGCTGC GCAACATGGTCCAGCAGATCCAGGAGAACAGTGTGGAGTTTGAAGCCTGTCTGGTGGCCCAATGTGATGCCCTCATCGATGCCCTCAACA GAAGAAAAGCCCAGCTGCTGGCCCGCGTCAACAAGGAGCATGAGCACAAGCTGAAGGTGGTTCGAGATCAGATCTCTCACTGCACAGTGA AATTGCGCCAGACCACAGGTCTCATGGAGTACTGCTTGGAGGTGATTAAGGAAAATGATCCTAGTGGTTTTTTGCAGATTTCTGACGCCC TCATAAGAAGAGTGCACCTGACTGAGGATCAGTGGGGTAAAGGCACACTCACTCCAAGGATGACCACGGACTTTGACTTGAGTCTGGACA ACAGCCCTCTGCTGCAATCCATCCACCAGCTGGATTTCGTGCAAGTGAAAGTCCCAGCAACCCCTATCCTACAGCTGGAGGAATGTTGTA CCCACAACAACAGCGCTACGTTGTCCTGGAAACAGCCACCTCTGTCCACGGTGCCCGCCGATGGATACATTCTGGAGCTGGATGATGGCA ACGGTGGTCAATTCCGGGAGGTGTATGTGGGGAAGGAGACAATGTGCACTGTGGATGGTCTTCACTTCAACAGCACATACAACGCTCGGG TCAAGGCCTTCAACAAAACAGGAGTCAGCCCGTACAGCAAGACCCTGGTCCTCCAAACGTCTGAGGACACAGATTCAGAAGAACAGACCC TCCCTTTTCCAGTGCCTTCGGAAAGATTGCCGCTCCGTAGAATGAGTCCTTTCTCCTCCACCCTTAATCTACAACCCAGCTTCCCCGGGA GATCCTACTTTGATTTCCGATCCTCACCCCACCAGCTGAGCTTGCATTCCTCTTTACAGTCTCTCAATGCACCGGGATGCAATTTTGAGA CACAATCTGCACCTTACTCTCAATTAGTTGACATTAAAAAGCTGTTGGCAGTGGCCTGGTTTGCTTTCGACCCTGGCTCGGCGCACTCGG ACATCATCCTCTCCAATGACAACCTGACAGTGACCTGTAGTAGCTATGATGACCGGGTGGTGCTAGGGAAGACTGGCTTCTCCAAGGGCA TCCACTACTGGGAGCTCACGGTAGATCGCTATGACAACCACCCTGATCCTGCCTTTGGTGTGGCTCGCATGGACGTGATGAAGGATGTGA TGTTAGGAAAAGACGACAAAGCTTGGGCAATGTATGTGGACAATAACCGGAGCTGGTTCATGCACAACAACTCGCACACCAACAGAACTG AGGGAGGGATCACAAAAGGGGCCACAATTGGGGTCCTCCTCGACTTAAATAGAAAAAACTTGACATTTTTTATCAACGATGAACAACAAG GTCCCATAGCATTTGATAACGTGGAGGGCCTCTTCTTCCCTGCGGTCAGCCTGAACAGGAACGTGCAGGTAAGCACCCTGCCCCTGAGGT TAAACAGCTGTTGCTGGCTACCGGTTCAAAGGCTGCCCAGGGCCGTTCAATCCAACCGAAGAGAGGGAAGCTGACAGCGTAAGGCCCAGA CCAAGATATGAAGGAGAGGAGGAGAGAAAGGGATCAGCACTGGCAACTTATTTTATTCCACGCCAAGCATAAGCACCTTCTTTATACTTG GACGCAGACTAGTTAATAGCACATAGGAAACTAGAATTCTCTTCATGGTTGGCTAATGATCATCTCTGTAGCTCTAGTTTCACCTCTAAA TAGAAACGAAAGCCTTTAACCAACTGGCATGAAAACAAATAGAATGACTTAACAAAGCTCAGGAGTGCCTGAGAAGAGGTGAGGGTGTAA GACTGAAGCATTAGTATCATTATCAGAAAATCTGAGCATCTTCTCTGCACTCTTGTTTGGCTCTGTTAGGTCCTAAGGGGTAGAAAACAT AGTCCCTACCTACAACTTAGTCAATTATTATTTATTATAGAATAAAACAACAGCTTCTCATTTATTCATTCAGCAAATATCTATAGAATG >51100_51100_2_MAN2A1-TRIM9_MAN2A1_chr5_109026253_ENST00000261483_TRIM9_chr14_51492078_ENST00000338969_length(amino acids)=680AA_BP=151 MPRAPCRRSALSLRSSLWGPPFPVAGESRLERGARPRGGRQARAKAKGVWWRRRLGAEQGGDSHPHPRARRSRSPGEGSLRRLLPRVHSA LSPLAEESVLAPSLSRKMKLSRQFTVFGSAIFCVVIFSLYLMLDRGHLDYPRNPRREGSFPQSQLSQALNGLSDRAKEAKEFLVQLRNMV QQIQENSVEFEACLVAQCDALIDALNRRKAQLLARVNKEHEHKLKVVRDQISHCTVKLRQTTGLMEYCLEVIKENDPSGFLQISDALIRR VHLTEDQWGKGTLTPRMTTDFDLSLDNSPLLQSIHQLDFVQVKVPATPILQLEECCTHNNSATLSWKQPPLSTVPADGYILELDDGNGGQ FREVYVGKETMCTVDGLHFNSTYNARVKAFNKTGVSPYSKTLVLQTSEDTDSEEQTLPFPVPSERLPLRRMSPFSSTLNLQPSFPGRSYF DFRSSPHQLSLHSSLQSLNAPGCNFETQSAPYSQLVDIKKLLAVAWFAFDPGSAHSDIILSNDNLTVTCSSYDDRVVLGKTGFSKGIHYW ELTVDRYDNHPDPAFGVARMDVMKDVMLGKDDKAWAMYVDNNRSWFMHNNSHTNRTEGGITKGATIGVLLDLNRKNLTFFINDEQQGPIA -------------------------------------------------------------- >51100_51100_3_MAN2A1-TRIM9_MAN2A1_chr5_109026253_ENST00000261483_TRIM9_chr14_51492078_ENST00000360392_length(transcript)=3954nt_BP=1187nt GGCGGGGCCAGCCGCCCGCTCGGCTCAGGCGCTGCGGGCGCCTATTGACCCAGCGGCTGCTGCGCCGCCGCTGTCTCCTCCTGCTCGTGG CGGGCGGTGCTGGAGCGCCAAGTGGCGCTGGAGAACCGGCGCTTCCTTTCGCCGCTTCCGCCGCCATCTCCGCGTTTGTGGGGCGGGAAA GAGGGAGGGGGCTAGCGGCTGCAGCTGGAGCGGGCTTCTCTCCGGGGACGGTCCTTTCCTCCCTGCTCTCCTTTTCCTTCTTTCCGCGTT GCCGCCGCCCGCCCCCTGCGCCTCCCCGCGGAGCCTGGGTCCGGGAGGGGGAAGGTAGGGGCGGCGGGGGGCGGGAGAGTCTGGCGAGCG GACGCTAGCTCTGAGGAAACTCATCAATCCGTGAGCCCCGGAGTCCGGGGTGCACATCGGCCCAGCCGCAGCGTCGGCGGCGGCGGCGGC AGCAGCACGAAGGGGCTCAGTCGGGGTAGGCGGGGGCGGTGCCGGTGCCGCGGGGGCGGGCCCGACCGTCCCGCCCAGAAGTTGTAGGGC TTGGCTCCTCGCGATCTTGTTCCTTTCCCCTCCGCTTCTCTGACCTAGCTGCGCGGCCCCGGCCCGGGAGCTGCCGAACCCGCGCCTCCC CTGGGTGAGGAGGACACGCCTGCCCTCGTCGAGAAAACTTTTCCTGCCGACTCAGTTGGGGCGGCGGTGGCAGGAAGTGCGGGCAGCGAC CTCTCCTCCGCCTGCCCCGCGCGCCCTGCCGGAGGTCGGCGCTGAGCTTGCGATCAAGTTTGTGGGGGCCCCCCTTCCCAGTTGCCGGCG AGTCTCGCCTCGAGAGGGGCGCCCGACCCCGGGGAGGGCGGCAGGCCAGGGCGAAGGCCAAGGGCGTGTGGTGGCGCCGGAGACTAGGTG CGGAGCAAGGCGGGGACTCGCACCCGCATCCGAGAGCGCGGAGGTCGCGCAGCCCGGGAGAAGGGAGCCTCCGGCGGCTGCTTCCTAGAG TCCACAGTGCGCTGTCTCCTTTGGCTGAGGAGAGTGTCCTGGCCCCGAGTCTATCGAGGAAAATGAAGTTAAGCCGCCAGTTCACCGTGT TCGGCAGTGCGATCTTCTGTGTGGTGATTTTCTCGCTCTACCTGATGCTGGACCGGGGTCACTTAGACTACCCCAGGAACCCGCGCCGCG AGGGCTCCTTCCCTCAGAGCCAGCTCTCCCAGGCGCTGAACGGACTGTCAGACAGGGCCAAAGAAGCCAAGGAGTTTCTGGTACAGCTGC GCAACATGGTCCAGCAGATCCAGGAGAACAGTGTGGAGTTTGAAGCCTGTCTGGTGGCCCAATGTGATGCCCTCATCGATGCCCTCAACA GAAGAAAAGCCCAGCTGCTGGCCCGCGTCAACAAGGAGCATGAGCACAAGCTGAAGGTGGTTCGAGATCAGATCTCTCACTGCACAGTGA AATTGCGCCAGACCACAGGTCTCATGGAGTACTGCTTGGAGGTGATTAAGGAAAATGATCCTAGTGGTTTTTTGCAGATTTCTGACGCCC TCATAAGAAGAGTGCACCTGACTGAGGATCAGTGGGGTAAAGGCACACTCACTCCAAGGATGACCACGGACTTTGACTTGAGTCTGGACA ACAGCCCTCTGCTGCAATCCATCCACCAGCTGGATTTCGTGCAAGTGAAAGCTTCCTCTCCAGTCCCAGCAACCCCTATCCTACAGCTGG AGGAATGTTGTACCCACAACAACAGCGCTACGTTGTCCTGGAAACAGCCACCTCTGTCCACGGTGCCCGCCGATGGATACATTCTGGAGC TGGATGATGGCAACGGTGGTCAATTCCGGGAGGTGTATGTGGGGAAGGAGACAATGTGCACTGTGGATGGTCTTCACTTCAACAGCACAT ACAACGCTCGGGTCAAGGCCTTCAACAAAACAGGAGTCAGCCCGTACAGCAAGACCCTGGTCCTCCAAACGTCTGAGGGTAAGGCCCTTC AGCAGTATCCCTCAGAGCGAGAACTGCGTGGCATCTAAAGTGGCTGGCAAGCCCGGAGGTAACCCCACCACTGCCCACATTCCTGAAGTG TTTCCATGACTTGCTCTGCATTCTGCACAGAGCCGCTGTTCCTCTCCTGCCTTTAGAGAGCCTATGGTATGTGGATGTGATCAAACCAAA GATTCCACATCGGCAGTTCCAATGGCTTGGGCCGGCGGCTTCCTTTGATAACAATCTAAATAAGCTGCAGTTGAAGAAGCTGAAAAATGA AGGCCTGAATGTGCCCCTGGTGTGTAAGACAAATGTATCTAGGCTCTAGAGCAGGCTCCCATTCTCCACCGATACACATCATGTGCCAGT TTTGCCCAGATGATTCTAAATTACTCTGTAGTACTTGCTTGTTCTGAGGGTGGGACCTAGGTTCTTTCCAGTCGTGGATTTGTATGACTG AATGTGTTTCAAATGGGTGGTGGGGTGCTAGAGCTGTTTAGAGAGGGCCTGTTGGCTGCTCCTGGCTTACCCACTTAGACTGCCTCCGTT TCATACCCAAAGCGGAGGCCGTCAGCACCAGGATTGAGACTTCCTGTGGGCACCAAACAGGAAGAGACCAGCAACTTCGCATTACCCGCC ATTTTCATCTTTGCCAGTCCCTTCAGTCTTGGCTAGGCTACCGAGAGCCACCATACAAGGTTCATCTCTAGAGAATTTTTGCTTCTTAGC TATACTTTAAATATTTTGGTCATCAAAGACAAGTAATGTGTCTCAGATGAGAGGCTTGAATTTGATGGCCAGATATAACCTCTGAGGCTT TTAACATTTTCATTTTAAGAGTAAGCAAGACACATGAAATTAAACCTACAAGGAGGTATTTGTGGCTGGTGCCAAAGCATTCTGACACTT TGGGGTGTCATTTTAATCAAAGAAATCACCCCCCCACCTCACCGGGATTCTCCATAACTTCCCTCTGCAGAACTAATTATGTTGATTTTG TTCAAGTTCAAGATGTTAGCTAAAAGAACTATGGTGGTGTTTTTTTTCCCACTTCCCACAAACCTCAACATGTGCCAGTCACCCTAAAAT GCTTCACATGGTTAAAAACAAGCACAATTTTGAATCCTACAGCAAGGATGAAAGGCCCCTAGCCGTGAAAACAGTGCTTTGGGGAGCAGT TGTCAGTCTAAGTGATGCTATTCCAAGAGAAGGATATGCTGAGGGAAAATGCCCACATGACCTGTTCCCATTTGGAGTACAGTGATGTGG TAGCTACAGCTTCCCCCAGAATTATCATTTTAGCACCTTCTCTCAGGGATGACCTATCAGTTTGAGAGCAGTTGCCTCTTTTCTCAAAAT ACCATACTAACTGCTAAAGCCCTCCAAGAGTCTTTCCTAGATGCAACGCAGAAGGCCCTTGCTGGTGATGGCCTCATTTCCCATGTGTGT ACAAGGTGGTTTGATTGAAGAGTGAAGTGCATGCCTGCAGAGCAGAGAGAAATTTGTAGCAATGTTGCTAATATGTGTTATCAGATCTGC GGGAAAACTATTTCTATTCATAATACATCATGGGAATCATACTATTCTGGTAAAAATCAGTTATTAGCATAACAGCCTGAGCACTTATGT CTCTCGTTTCATCCTGCAGGAGGATGTAGCGCCTCAGTTTATTTTAATGTTCATAAGATTATGGTGTCTAATTTAATAAATTACAGGATT GGAACTGCGATCCTTGGTACCACAGTCACAGAACTGGGGGTCATTTTCTAGATGAAACAAACGGAACAAGTTCTCTTCCAACAAAGAAAC TGTACTGTAGAAATTAATTTCCTCCATGAATTTTATATATTGTGTACAAATATAAGGTATGTATCTGAATACAAAGAAAAGCCTATCATC >51100_51100_3_MAN2A1-TRIM9_MAN2A1_chr5_109026253_ENST00000261483_TRIM9_chr14_51492078_ENST00000360392_length(amino acids)=428AA_BP=151 MPRAPCRRSALSLRSSLWGPPFPVAGESRLERGARPRGGRQARAKAKGVWWRRRLGAEQGGDSHPHPRARRSRSPGEGSLRRLLPRVHSA LSPLAEESVLAPSLSRKMKLSRQFTVFGSAIFCVVIFSLYLMLDRGHLDYPRNPRREGSFPQSQLSQALNGLSDRAKEAKEFLVQLRNMV QQIQENSVEFEACLVAQCDALIDALNRRKAQLLARVNKEHEHKLKVVRDQISHCTVKLRQTTGLMEYCLEVIKENDPSGFLQISDALIRR VHLTEDQWGKGTLTPRMTTDFDLSLDNSPLLQSIHQLDFVQVKASSPVPATPILQLEECCTHNNSATLSWKQPPLSTVPADGYILELDDG -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for MAN2A1-TRIM9 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for MAN2A1-TRIM9 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for MAN2A1-TRIM9 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
