
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:MAP2K4-MYH3 (FusionGDB2 ID:51209) |
Fusion Gene Summary for MAP2K4-MYH3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: MAP2K4-MYH3 | Fusion gene ID: 51209 | Hgene | Tgene | Gene symbol | MAP2K4 | MYH3 | Gene ID | 6416 | 4621 |
| Gene name | mitogen-activated protein kinase kinase 4 | myosin heavy chain 3 | |
| Synonyms | JNKK|JNKK1|MAPKK4|MEK4|MKK4|PRKMK4|SAPKK-1|SAPKK1|SEK1|SERK1|SKK1 | CPSFS1A|CPSFS1B|CPSKF1A|CPSKF1B|DA2A|DA2B|DA2B3|DA8|HEMHC|MYHC-EMB|MYHSE1|SMHCE | |
| Cytomap | 17p12 | 17p13.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | dual specificity mitogen-activated protein kinase kinase 4JNK-activated kinase 1JNK-activating kinase 1MAP kinase kinase 4MAPK/ERK kinase 4MAPKK 4MEK 4SAPK/ERK kinase 1c-Jun N-terminal kinase kinase 1stress-activated protein kinase kinase 1 | myosin-3myosin heavy chain, fast skeletal muscle, embryonicmyosin, heavy chain 3, skeletal muscle, embryonicmyosin, heavy polypeptide 3, skeletal muscle, embryonicmyosin, skeletal, heavy chain, embryonic 1 | |
| Modification date | 20200329 | 20200314 | |
| UniProtAcc | P45985 | P11055 | |
| Ensembl transtripts involved in fusion gene | ENST00000353533, ENST00000415385, ENST00000581941, | ENST00000226209, ENST00000583535, | |
| Fusion gene scores | * DoF score | 10 X 4 X 7=280 | 5 X 5 X 3=75 |
| # samples | 13 | 5 | |
| ** MAII score | log2(13/280*10)=-1.10691520391651 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/75*10)=-0.584962500721156 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: MAP2K4 [Title/Abstract] AND MYH3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | MAP2K4(11924318)-MYH3(10539170), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across MAP2K4 (5'-gene) Fusion gene breakpoints across MAP2K4 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
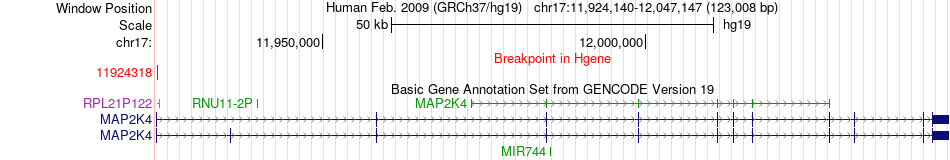 |
 Fusion gene breakpoints across MYH3 (3'-gene) Fusion gene breakpoints across MYH3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
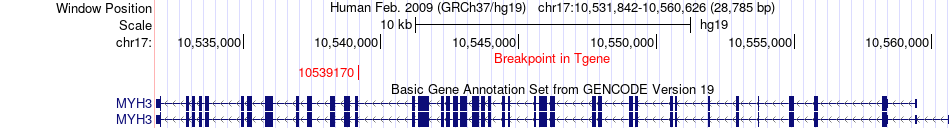 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-61-1914-01A | MAP2K4 | chr17 | 11924318 | + | MYH3 | chr17 | 10539170 | - |
| ChimerDB4 | OV | TCGA-61-1914 | MAP2K4 | chr17 | 11924318 | + | MYH3 | chr17 | 10539170 | - |
Top |
Fusion Gene ORF analysis for MAP2K4-MYH3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000353533 | ENST00000226209 | MAP2K4 | chr17 | 11924318 | + | MYH3 | chr17 | 10539170 | - |
| In-frame | ENST00000353533 | ENST00000583535 | MAP2K4 | chr17 | 11924318 | + | MYH3 | chr17 | 10539170 | - |
| In-frame | ENST00000415385 | ENST00000226209 | MAP2K4 | chr17 | 11924318 | + | MYH3 | chr17 | 10539170 | - |
| In-frame | ENST00000415385 | ENST00000583535 | MAP2K4 | chr17 | 11924318 | + | MYH3 | chr17 | 10539170 | - |
| intron-3CDS | ENST00000581941 | ENST00000226209 | MAP2K4 | chr17 | 11924318 | + | MYH3 | chr17 | 10539170 | - |
| intron-3CDS | ENST00000581941 | ENST00000583535 | MAP2K4 | chr17 | 11924318 | + | MYH3 | chr17 | 10539170 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000353533 | MAP2K4 | chr17 | 11924318 | + | ENST00000226209 | MYH3 | chr17 | 10539170 | - | 2272 | 178 | 63 | 2144 | 693 |
| ENST00000353533 | MAP2K4 | chr17 | 11924318 | + | ENST00000583535 | MYH3 | chr17 | 10539170 | - | 2271 | 178 | 63 | 2144 | 693 |
| ENST00000415385 | MAP2K4 | chr17 | 11924318 | + | ENST00000226209 | MYH3 | chr17 | 10539170 | - | 2262 | 168 | 53 | 2134 | 693 |
| ENST00000415385 | MAP2K4 | chr17 | 11924318 | + | ENST00000583535 | MYH3 | chr17 | 10539170 | - | 2261 | 168 | 53 | 2134 | 693 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000353533 | ENST00000226209 | MAP2K4 | chr17 | 11924318 | + | MYH3 | chr17 | 10539170 | - | 0.004278385 | 0.9957216 |
| ENST00000353533 | ENST00000583535 | MAP2K4 | chr17 | 11924318 | + | MYH3 | chr17 | 10539170 | - | 0.004241232 | 0.9957587 |
| ENST00000415385 | ENST00000226209 | MAP2K4 | chr17 | 11924318 | + | MYH3 | chr17 | 10539170 | - | 0.004244865 | 0.9957552 |
| ENST00000415385 | ENST00000583535 | MAP2K4 | chr17 | 11924318 | + | MYH3 | chr17 | 10539170 | - | 0.004210805 | 0.99578923 |
Top |
Fusion Genomic Features for MAP2K4-MYH3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
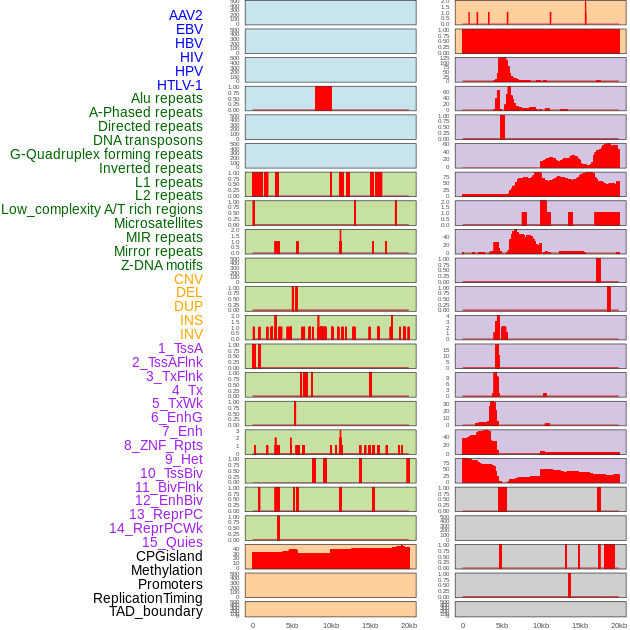 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for MAP2K4-MYH3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:11924318/chr17:10539170) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MAP2K4 | MYH3 |
| FUNCTION: Dual specificity protein kinase which acts as an essential component of the MAP kinase signal transduction pathway. Essential component of the stress-activated protein kinase/c-Jun N-terminal kinase (SAP/JNK) signaling pathway. With MAP2K7/MKK7, is the one of the only known kinase to directly activate the stress-activated protein kinase/c-Jun N-terminal kinases MAPK8/JNK1, MAPK9/JNK2 and MAPK10/JNK3. MAP2K4/MKK4 and MAP2K7/MKK7 both activate the JNKs by phosphorylation, but they differ in their preference for the phosphorylation site in the Thr-Pro-Tyr motif. MAP2K4 shows preference for phosphorylation of the Tyr residue and MAP2K7/MKK7 for the Thr residue. The phosphorylation of the Thr residue by MAP2K7/MKK7 seems to be the prerequisite for JNK activation at least in response to proinflammatory cytokines, while other stimuli activate both MAP2K4/MKK4 and MAP2K7/MKK7 which synergistically phosphorylate JNKs. MAP2K4 is required for maintaining peripheral lymphoid homeostasis. The MKK/JNK signaling pathway is also involved in mitochondrial death signaling pathway, including the release cytochrome c, leading to apoptosis. Whereas MAP2K7/MKK7 exclusively activates JNKs, MAP2K4/MKK4 additionally activates the p38 MAPKs MAPK11, MAPK12, MAPK13 and MAPK14. {ECO:0000269|PubMed:7716521}. | FUNCTION: Muscle contraction. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MAP2K4 | chr17:11924318 | chr17:10539170 | ENST00000353533 | + | 1 | 11 | 5_19 | 38 | 400.0 | Compositional bias | Note=Gly/Ser-rich |
| Hgene | MAP2K4 | chr17:11924318 | chr17:10539170 | ENST00000415385 | + | 1 | 12 | 5_19 | 38 | 411.0 | Compositional bias | Note=Gly/Ser-rich |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MAP2K4 | chr17:11924318 | chr17:10539170 | ENST00000353533 | + | 1 | 11 | 102_367 | 38 | 400.0 | Domain | Protein kinase |
| Hgene | MAP2K4 | chr17:11924318 | chr17:10539170 | ENST00000415385 | + | 1 | 12 | 102_367 | 38 | 411.0 | Domain | Protein kinase |
| Hgene | MAP2K4 | chr17:11924318 | chr17:10539170 | ENST00000353533 | + | 1 | 11 | 108_116 | 38 | 400.0 | Nucleotide binding | ATP |
| Hgene | MAP2K4 | chr17:11924318 | chr17:10539170 | ENST00000415385 | + | 1 | 12 | 108_116 | 38 | 411.0 | Nucleotide binding | ATP |
| Hgene | MAP2K4 | chr17:11924318 | chr17:10539170 | ENST00000353533 | + | 1 | 11 | 364_387 | 38 | 400.0 | Region | Note=DVD domain |
| Hgene | MAP2K4 | chr17:11924318 | chr17:10539170 | ENST00000353533 | + | 1 | 11 | 37_52 | 38 | 400.0 | Region | Note=D domain |
| Hgene | MAP2K4 | chr17:11924318 | chr17:10539170 | ENST00000415385 | + | 1 | 12 | 364_387 | 38 | 411.0 | Region | Note=DVD domain |
| Hgene | MAP2K4 | chr17:11924318 | chr17:10539170 | ENST00000415385 | + | 1 | 12 | 37_52 | 38 | 411.0 | Region | Note=D domain |
| Tgene | MYH3 | chr17:11924318 | chr17:10539170 | ENST00000226209 | 26 | 40 | 840_1933 | 1285 | 1941.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | MYH3 | chr17:11924318 | chr17:10539170 | ENST00000583535 | 27 | 41 | 840_1933 | 1285 | 1941.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | MYH3 | chr17:11924318 | chr17:10539170 | ENST00000226209 | 26 | 40 | 33_82 | 1285 | 1941.0 | Domain | Myosin N-terminal SH3-like | |
| Tgene | MYH3 | chr17:11924318 | chr17:10539170 | ENST00000226209 | 26 | 40 | 782_811 | 1285 | 1941.0 | Domain | IQ | |
| Tgene | MYH3 | chr17:11924318 | chr17:10539170 | ENST00000226209 | 26 | 40 | 86_779 | 1285 | 1941.0 | Domain | Myosin motor | |
| Tgene | MYH3 | chr17:11924318 | chr17:10539170 | ENST00000583535 | 27 | 41 | 33_82 | 1285 | 1941.0 | Domain | Myosin N-terminal SH3-like | |
| Tgene | MYH3 | chr17:11924318 | chr17:10539170 | ENST00000583535 | 27 | 41 | 782_811 | 1285 | 1941.0 | Domain | IQ | |
| Tgene | MYH3 | chr17:11924318 | chr17:10539170 | ENST00000583535 | 27 | 41 | 86_779 | 1285 | 1941.0 | Domain | Myosin motor | |
| Tgene | MYH3 | chr17:11924318 | chr17:10539170 | ENST00000226209 | 26 | 40 | 179_186 | 1285 | 1941.0 | Nucleotide binding | ATP | |
| Tgene | MYH3 | chr17:11924318 | chr17:10539170 | ENST00000583535 | 27 | 41 | 179_186 | 1285 | 1941.0 | Nucleotide binding | ATP | |
| Tgene | MYH3 | chr17:11924318 | chr17:10539170 | ENST00000226209 | 26 | 40 | 656_678 | 1285 | 1941.0 | Region | Note=Actin-binding | |
| Tgene | MYH3 | chr17:11924318 | chr17:10539170 | ENST00000226209 | 26 | 40 | 758_772 | 1285 | 1941.0 | Region | Note=Actin-binding | |
| Tgene | MYH3 | chr17:11924318 | chr17:10539170 | ENST00000583535 | 27 | 41 | 656_678 | 1285 | 1941.0 | Region | Note=Actin-binding | |
| Tgene | MYH3 | chr17:11924318 | chr17:10539170 | ENST00000583535 | 27 | 41 | 758_772 | 1285 | 1941.0 | Region | Note=Actin-binding |
Top |
Fusion Gene Sequence for MAP2K4-MYH3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >51209_51209_1_MAP2K4-MYH3_MAP2K4_chr17_11924318_ENST00000353533_MYH3_chr17_10539170_ENST00000226209_length(transcript)=2272nt_BP=178nt GCGAGAGGCCGAGCTTGCTGCATTGCAGCCGCCGCGGCGCCGCTCGGCTCTTCACTCCCAACAATGGCGGCTCCGAGCCCGAGCGGCGGC GGCGGCTCCGGGGGCGGCAGCGGCAGCGGCACCCCCGGCCCCGTAGGGTCCCCGGCGCCAGGCCACCCGGCCGTCAGCAGCATGCAGGGT GAGCTGAGTCGTCAGCTGGAAGAAAAAGAAAGCATAGTATCCCAACTTTCCAGGAGCAAGCAAGCCTTTACCCAGCAAACAGAAGAGCTC AAGAGGCAGCTGGAGGAAGAGAACAAGGCCAAGAACGCCCTGGCGCACGCCCTGCAGTCCTCCCGCCACGACTGTGACCTGCTGCGGGAA CAGTATGAGGAGGAGCAGGAAGGCAAAGCTGAGCTGCAGAGGGCGCTGTCCAAGGCCAATAGTGAGGTTGCCCAGTGGAGAACCAAATAC GAGACGGACGCCATCCAGCGCACAGAAGAGCTGGAGGAGGCCAAGAAAAAACTTGCTCAGCGCCTTCAAGATTCCGAGGAACAGGTTGAG GCAGTGAATGCTAAATGTGCTTCACTGGAGAAGACCAAGCAGAGGCTGCAAGGAGAGGTGGAGGATCTGATGGTTGATGTTGAAAGAGCC AATTCCTTGGCCGCCGCTCTGGACAAGAAGCAGAGGAACTTTGACAAGGTGTTGGCAGAGTGGAAGACAAAGTGTGAGGAGAGCCAAGCA GAGCTGGAGGCATCCCTGAAGGAGTCCCGCTCCTTGAGCACTGAGCTCTTCAAACTGAAAAATGCCTACGAGGAAGCCTTAGATCAACTT GAAACTGTGAAACGGGAAAATAAGAACTTAGAGCAGGAGATAGCAGATCTCACAGAACAAATTGCTGAAAATGGCAAAACCATCCATGAA CTGGAGAAATCAAGAAAGCAGATTGAGCTGGAAAAGGCTGATATCCAGCTGGCTCTCGAGGAAGCAGAGGCTGCTCTTGAGCATGAAGAA GCCAAGATCCTCCGAATCCAGCTTGAATTGACACAAGTGAAATCAGAAATTGATAGAAAGATCGCCGAGAAGGATGAAGAGATCGAGCAG CTGAAGAGGAACTACCAGAGAACAGTGGAAACCATGCAGAGCGCCCTGGACGCCGAGGTGCGGAGCAGGAATGAAGCCATCCGGCTCAAG AAGAAGATGGAGGGGGACCTGAATGAAATCGAGATCCAGCTGAGCCACGCCAACCGCCAGGCGGCGGAGACCCTCAAACACCTCAGGAGT GTCCAGGGACAGCTGAAGGATACGCAGCTCCACCTGGATGATGCCCTCCGGGGCCAGGAGGACCTGAAGGAGCAGCTGGCGATTGTGGAG CGCAGAGCCAACCTGCTGCAGGCCGAGGTGGAGGAGCTGCGGGCTACTCTGGAGCAGACGGAGAGGGCCCGGAAACTGGCGGAACAGGAG CTCCTGGACTCCAACGAGAGGGTGCAGCTGCTGCATACCCAGAACACCAGCCTCATCCACACCAAGAAGAAGCTGGAGACAGACCTCATG CAGCTCCAGAGTGAGGTAGAAGATGCCAGCAGGGATGCAAGGAACGCTGAGGAGAAGGCCAAGAAGGCCATCACGGACGCTGCCATGATG GCGGAGGAGCTGAAGAAGGAGCAGGACACCAGCGCCCACCTTGAGCGGATGAAGAAGAACCTGGAACAGACGGTGAAGGACCTGCAGCAT CGTCTAGATGAGGCCGAGCAGCTGGCGCTGAAGGGCGGGAAGAAGCAGATCCAGAAACTGGAGACCAGGATCCGAGAGCTGGAGTTTGAA CTTGAGGGAGAGCAGAAGAAGAACACAGAGTCTGTTAAGGGCCTGAGGAAGTATGAGCGGAGGGTCAAGGAGCTGACGTACCAGAGTGAA GAGGACAGGAAGAATGTGCTGAGATTGCAGGATCTGGTGGATAAACTGCAAGTGAAAGTCAAGTCCTACAAGAGGCAGGCGGAGGAGGCT GATGAACAAGCCAATGCTCATCTCACCAAATTCCGAAAGGCTCAGCATGAGCTGGAGGAGGCCGAGGAACGTGCGGATATCGCAGAATCT CAAGTCAACAAGCTCCGCGCTAAGACTCGAGACTTCACCTCCAGCAGGATGGTGGTCCACGAGAGTGAAGAGTGAGCCAGCCCTTCTGGA GCAGGACAGAAGATATGCAAAATGTATATTTTCTTGATTCCTGACCATTGATACTTAATGTCCATGTGACTCTTTTTCACATGCAATAAA >51209_51209_1_MAP2K4-MYH3_MAP2K4_chr17_11924318_ENST00000353533_MYH3_chr17_10539170_ENST00000226209_length(amino acids)=693AA_BP=38 MAAPSPSGGGGSGGGSGSGTPGPVGSPAPGHPAVSSMQGELSRQLEEKESIVSQLSRSKQAFTQQTEELKRQLEEENKAKNALAHALQSS RHDCDLLREQYEEEQEGKAELQRALSKANSEVAQWRTKYETDAIQRTEELEEAKKKLAQRLQDSEEQVEAVNAKCASLEKTKQRLQGEVE DLMVDVERANSLAAALDKKQRNFDKVLAEWKTKCEESQAELEASLKESRSLSTELFKLKNAYEEALDQLETVKRENKNLEQEIADLTEQI AENGKTIHELEKSRKQIELEKADIQLALEEAEAALEHEEAKILRIQLELTQVKSEIDRKIAEKDEEIEQLKRNYQRTVETMQSALDAEVR SRNEAIRLKKKMEGDLNEIEIQLSHANRQAAETLKHLRSVQGQLKDTQLHLDDALRGQEDLKEQLAIVERRANLLQAEVEELRATLEQTE RARKLAEQELLDSNERVQLLHTQNTSLIHTKKKLETDLMQLQSEVEDASRDARNAEEKAKKAITDAAMMAEELKKEQDTSAHLERMKKNL EQTVKDLQHRLDEAEQLALKGGKKQIQKLETRIRELEFELEGEQKKNTESVKGLRKYERRVKELTYQSEEDRKNVLRLQDLVDKLQVKVK -------------------------------------------------------------- >51209_51209_2_MAP2K4-MYH3_MAP2K4_chr17_11924318_ENST00000353533_MYH3_chr17_10539170_ENST00000583535_length(transcript)=2271nt_BP=178nt GCGAGAGGCCGAGCTTGCTGCATTGCAGCCGCCGCGGCGCCGCTCGGCTCTTCACTCCCAACAATGGCGGCTCCGAGCCCGAGCGGCGGC GGCGGCTCCGGGGGCGGCAGCGGCAGCGGCACCCCCGGCCCCGTAGGGTCCCCGGCGCCAGGCCACCCGGCCGTCAGCAGCATGCAGGGT GAGCTGAGTCGTCAGCTGGAAGAAAAAGAAAGCATAGTATCCCAACTTTCCAGGAGCAAGCAAGCCTTTACCCAGCAAACAGAAGAGCTC AAGAGGCAGCTGGAGGAAGAGAACAAGGCCAAGAACGCCCTGGCGCACGCCCTGCAGTCCTCCCGCCACGACTGTGACCTGCTGCGGGAA CAGTATGAGGAGGAGCAGGAAGGCAAAGCTGAGCTGCAGAGGGCGCTGTCCAAGGCCAATAGTGAGGTTGCCCAGTGGAGAACCAAATAC GAGACGGACGCCATCCAGCGCACAGAAGAGCTGGAGGAGGCCAAGAAAAAACTTGCTCAGCGCCTTCAAGATTCCGAGGAACAGGTTGAG GCAGTGAATGCTAAATGTGCTTCACTGGAGAAGACCAAGCAGAGGCTGCAAGGAGAGGTGGAGGATCTGATGGTTGATGTTGAAAGAGCC AATTCCTTGGCCGCCGCTCTGGACAAGAAGCAGAGGAACTTTGACAAGGTGTTGGCAGAGTGGAAGACAAAGTGTGAGGAGAGCCAAGCA GAGCTGGAGGCATCCCTGAAGGAGTCCCGCTCCTTGAGCACTGAGCTCTTCAAACTGAAAAATGCCTACGAGGAAGCCTTAGATCAACTT GAAACTGTGAAACGGGAAAATAAGAACTTAGAGCAGGAGATAGCAGATCTCACAGAACAAATTGCTGAAAATGGCAAAACCATCCATGAA CTGGAGAAATCAAGAAAGCAGATTGAGCTGGAAAAGGCTGATATCCAGCTGGCTCTCGAGGAAGCAGAGGCTGCTCTTGAGCATGAAGAA GCCAAGATCCTCCGAATCCAGCTTGAATTGACACAAGTGAAATCAGAAATTGATAGAAAGATCGCCGAGAAGGATGAAGAGATCGAGCAG CTGAAGAGGAACTACCAGAGAACAGTGGAAACCATGCAGAGCGCCCTGGACGCCGAGGTGCGGAGCAGGAATGAAGCCATCCGGCTCAAG AAGAAGATGGAGGGGGACCTGAATGAAATCGAGATCCAGCTGAGCCACGCCAACCGCCAGGCGGCGGAGACCCTCAAACACCTCAGGAGT GTCCAGGGACAGCTGAAGGATACGCAGCTCCACCTGGATGATGCCCTCCGGGGCCAGGAGGACCTGAAGGAGCAGCTGGCGATTGTGGAG CGCAGAGCCAACCTGCTGCAGGCCGAGGTGGAGGAGCTGCGGGCTACTCTGGAGCAGACGGAGAGGGCCCGGAAACTGGCGGAACAGGAG CTCCTGGACTCCAACGAGAGGGTGCAGCTGCTGCATACCCAGAACACCAGCCTCATCCACACCAAGAAGAAGCTGGAGACAGACCTCATG CAGCTCCAGAGTGAGGTAGAAGATGCCAGCAGGGATGCAAGGAACGCTGAGGAGAAGGCCAAGAAGGCCATCACGGACGCTGCCATGATG GCGGAGGAGCTGAAGAAGGAGCAGGACACCAGCGCCCACCTTGAGCGGATGAAGAAGAACCTGGAACAGACGGTGAAGGACCTGCAGCAT CGTCTAGATGAGGCCGAGCAGCTGGCGCTGAAGGGCGGGAAGAAGCAGATCCAGAAACTGGAGACCAGGATCCGAGAGCTGGAGTTTGAA CTTGAGGGAGAGCAGAAGAAGAACACAGAGTCTGTTAAGGGCCTGAGGAAGTATGAGCGGAGGGTCAAGGAGCTGACGTACCAGAGTGAA GAGGACAGGAAGAATGTGCTGAGATTGCAGGATCTGGTGGATAAACTGCAAGTGAAAGTCAAGTCCTACAAGAGGCAGGCGGAGGAGGCT GATGAACAAGCCAATGCTCATCTCACCAAATTCCGAAAGGCTCAGCATGAGCTGGAGGAGGCCGAGGAACGTGCGGATATCGCAGAATCT CAAGTCAACAAGCTCCGCGCTAAGACTCGAGACTTCACCTCCAGCAGGATGGTGGTCCACGAGAGTGAAGAGTGAGCCAGCCCTTCTGGA GCAGGACAGAAGATATGCAAAATGTATATTTTCTTGATTCCTGACCATTGATACTTAATGTCCATGTGACTCTTTTTCACATGCAATAAA >51209_51209_2_MAP2K4-MYH3_MAP2K4_chr17_11924318_ENST00000353533_MYH3_chr17_10539170_ENST00000583535_length(amino acids)=693AA_BP=38 MAAPSPSGGGGSGGGSGSGTPGPVGSPAPGHPAVSSMQGELSRQLEEKESIVSQLSRSKQAFTQQTEELKRQLEEENKAKNALAHALQSS RHDCDLLREQYEEEQEGKAELQRALSKANSEVAQWRTKYETDAIQRTEELEEAKKKLAQRLQDSEEQVEAVNAKCASLEKTKQRLQGEVE DLMVDVERANSLAAALDKKQRNFDKVLAEWKTKCEESQAELEASLKESRSLSTELFKLKNAYEEALDQLETVKRENKNLEQEIADLTEQI AENGKTIHELEKSRKQIELEKADIQLALEEAEAALEHEEAKILRIQLELTQVKSEIDRKIAEKDEEIEQLKRNYQRTVETMQSALDAEVR SRNEAIRLKKKMEGDLNEIEIQLSHANRQAAETLKHLRSVQGQLKDTQLHLDDALRGQEDLKEQLAIVERRANLLQAEVEELRATLEQTE RARKLAEQELLDSNERVQLLHTQNTSLIHTKKKLETDLMQLQSEVEDASRDARNAEEKAKKAITDAAMMAEELKKEQDTSAHLERMKKNL EQTVKDLQHRLDEAEQLALKGGKKQIQKLETRIRELEFELEGEQKKNTESVKGLRKYERRVKELTYQSEEDRKNVLRLQDLVDKLQVKVK -------------------------------------------------------------- >51209_51209_3_MAP2K4-MYH3_MAP2K4_chr17_11924318_ENST00000415385_MYH3_chr17_10539170_ENST00000226209_length(transcript)=2262nt_BP=168nt GAGCTTGCTGCATTGCAGCCGCCGCGGCGCCGCTCGGCTCTTCACTCCCAACAATGGCGGCTCCGAGCCCGAGCGGCGGCGGCGGCTCCG GGGGCGGCAGCGGCAGCGGCACCCCCGGCCCCGTAGGGTCCCCGGCGCCAGGCCACCCGGCCGTCAGCAGCATGCAGGGTGAGCTGAGTC GTCAGCTGGAAGAAAAAGAAAGCATAGTATCCCAACTTTCCAGGAGCAAGCAAGCCTTTACCCAGCAAACAGAAGAGCTCAAGAGGCAGC TGGAGGAAGAGAACAAGGCCAAGAACGCCCTGGCGCACGCCCTGCAGTCCTCCCGCCACGACTGTGACCTGCTGCGGGAACAGTATGAGG AGGAGCAGGAAGGCAAAGCTGAGCTGCAGAGGGCGCTGTCCAAGGCCAATAGTGAGGTTGCCCAGTGGAGAACCAAATACGAGACGGACG CCATCCAGCGCACAGAAGAGCTGGAGGAGGCCAAGAAAAAACTTGCTCAGCGCCTTCAAGATTCCGAGGAACAGGTTGAGGCAGTGAATG CTAAATGTGCTTCACTGGAGAAGACCAAGCAGAGGCTGCAAGGAGAGGTGGAGGATCTGATGGTTGATGTTGAAAGAGCCAATTCCTTGG CCGCCGCTCTGGACAAGAAGCAGAGGAACTTTGACAAGGTGTTGGCAGAGTGGAAGACAAAGTGTGAGGAGAGCCAAGCAGAGCTGGAGG CATCCCTGAAGGAGTCCCGCTCCTTGAGCACTGAGCTCTTCAAACTGAAAAATGCCTACGAGGAAGCCTTAGATCAACTTGAAACTGTGA AACGGGAAAATAAGAACTTAGAGCAGGAGATAGCAGATCTCACAGAACAAATTGCTGAAAATGGCAAAACCATCCATGAACTGGAGAAAT CAAGAAAGCAGATTGAGCTGGAAAAGGCTGATATCCAGCTGGCTCTCGAGGAAGCAGAGGCTGCTCTTGAGCATGAAGAAGCCAAGATCC TCCGAATCCAGCTTGAATTGACACAAGTGAAATCAGAAATTGATAGAAAGATCGCCGAGAAGGATGAAGAGATCGAGCAGCTGAAGAGGA ACTACCAGAGAACAGTGGAAACCATGCAGAGCGCCCTGGACGCCGAGGTGCGGAGCAGGAATGAAGCCATCCGGCTCAAGAAGAAGATGG AGGGGGACCTGAATGAAATCGAGATCCAGCTGAGCCACGCCAACCGCCAGGCGGCGGAGACCCTCAAACACCTCAGGAGTGTCCAGGGAC AGCTGAAGGATACGCAGCTCCACCTGGATGATGCCCTCCGGGGCCAGGAGGACCTGAAGGAGCAGCTGGCGATTGTGGAGCGCAGAGCCA ACCTGCTGCAGGCCGAGGTGGAGGAGCTGCGGGCTACTCTGGAGCAGACGGAGAGGGCCCGGAAACTGGCGGAACAGGAGCTCCTGGACT CCAACGAGAGGGTGCAGCTGCTGCATACCCAGAACACCAGCCTCATCCACACCAAGAAGAAGCTGGAGACAGACCTCATGCAGCTCCAGA GTGAGGTAGAAGATGCCAGCAGGGATGCAAGGAACGCTGAGGAGAAGGCCAAGAAGGCCATCACGGACGCTGCCATGATGGCGGAGGAGC TGAAGAAGGAGCAGGACACCAGCGCCCACCTTGAGCGGATGAAGAAGAACCTGGAACAGACGGTGAAGGACCTGCAGCATCGTCTAGATG AGGCCGAGCAGCTGGCGCTGAAGGGCGGGAAGAAGCAGATCCAGAAACTGGAGACCAGGATCCGAGAGCTGGAGTTTGAACTTGAGGGAG AGCAGAAGAAGAACACAGAGTCTGTTAAGGGCCTGAGGAAGTATGAGCGGAGGGTCAAGGAGCTGACGTACCAGAGTGAAGAGGACAGGA AGAATGTGCTGAGATTGCAGGATCTGGTGGATAAACTGCAAGTGAAAGTCAAGTCCTACAAGAGGCAGGCGGAGGAGGCTGATGAACAAG CCAATGCTCATCTCACCAAATTCCGAAAGGCTCAGCATGAGCTGGAGGAGGCCGAGGAACGTGCGGATATCGCAGAATCTCAAGTCAACA AGCTCCGCGCTAAGACTCGAGACTTCACCTCCAGCAGGATGGTGGTCCACGAGAGTGAAGAGTGAGCCAGCCCTTCTGGAGCAGGACAGA AGATATGCAAAATGTATATTTTCTTGATTCCTGACCATTGATACTTAATGTCCATGTGACTCTTTTTCACATGCAATAAACTTTGCTTTG >51209_51209_3_MAP2K4-MYH3_MAP2K4_chr17_11924318_ENST00000415385_MYH3_chr17_10539170_ENST00000226209_length(amino acids)=693AA_BP=38 MAAPSPSGGGGSGGGSGSGTPGPVGSPAPGHPAVSSMQGELSRQLEEKESIVSQLSRSKQAFTQQTEELKRQLEEENKAKNALAHALQSS RHDCDLLREQYEEEQEGKAELQRALSKANSEVAQWRTKYETDAIQRTEELEEAKKKLAQRLQDSEEQVEAVNAKCASLEKTKQRLQGEVE DLMVDVERANSLAAALDKKQRNFDKVLAEWKTKCEESQAELEASLKESRSLSTELFKLKNAYEEALDQLETVKRENKNLEQEIADLTEQI AENGKTIHELEKSRKQIELEKADIQLALEEAEAALEHEEAKILRIQLELTQVKSEIDRKIAEKDEEIEQLKRNYQRTVETMQSALDAEVR SRNEAIRLKKKMEGDLNEIEIQLSHANRQAAETLKHLRSVQGQLKDTQLHLDDALRGQEDLKEQLAIVERRANLLQAEVEELRATLEQTE RARKLAEQELLDSNERVQLLHTQNTSLIHTKKKLETDLMQLQSEVEDASRDARNAEEKAKKAITDAAMMAEELKKEQDTSAHLERMKKNL EQTVKDLQHRLDEAEQLALKGGKKQIQKLETRIRELEFELEGEQKKNTESVKGLRKYERRVKELTYQSEEDRKNVLRLQDLVDKLQVKVK -------------------------------------------------------------- >51209_51209_4_MAP2K4-MYH3_MAP2K4_chr17_11924318_ENST00000415385_MYH3_chr17_10539170_ENST00000583535_length(transcript)=2261nt_BP=168nt GAGCTTGCTGCATTGCAGCCGCCGCGGCGCCGCTCGGCTCTTCACTCCCAACAATGGCGGCTCCGAGCCCGAGCGGCGGCGGCGGCTCCG GGGGCGGCAGCGGCAGCGGCACCCCCGGCCCCGTAGGGTCCCCGGCGCCAGGCCACCCGGCCGTCAGCAGCATGCAGGGTGAGCTGAGTC GTCAGCTGGAAGAAAAAGAAAGCATAGTATCCCAACTTTCCAGGAGCAAGCAAGCCTTTACCCAGCAAACAGAAGAGCTCAAGAGGCAGC TGGAGGAAGAGAACAAGGCCAAGAACGCCCTGGCGCACGCCCTGCAGTCCTCCCGCCACGACTGTGACCTGCTGCGGGAACAGTATGAGG AGGAGCAGGAAGGCAAAGCTGAGCTGCAGAGGGCGCTGTCCAAGGCCAATAGTGAGGTTGCCCAGTGGAGAACCAAATACGAGACGGACG CCATCCAGCGCACAGAAGAGCTGGAGGAGGCCAAGAAAAAACTTGCTCAGCGCCTTCAAGATTCCGAGGAACAGGTTGAGGCAGTGAATG CTAAATGTGCTTCACTGGAGAAGACCAAGCAGAGGCTGCAAGGAGAGGTGGAGGATCTGATGGTTGATGTTGAAAGAGCCAATTCCTTGG CCGCCGCTCTGGACAAGAAGCAGAGGAACTTTGACAAGGTGTTGGCAGAGTGGAAGACAAAGTGTGAGGAGAGCCAAGCAGAGCTGGAGG CATCCCTGAAGGAGTCCCGCTCCTTGAGCACTGAGCTCTTCAAACTGAAAAATGCCTACGAGGAAGCCTTAGATCAACTTGAAACTGTGA AACGGGAAAATAAGAACTTAGAGCAGGAGATAGCAGATCTCACAGAACAAATTGCTGAAAATGGCAAAACCATCCATGAACTGGAGAAAT CAAGAAAGCAGATTGAGCTGGAAAAGGCTGATATCCAGCTGGCTCTCGAGGAAGCAGAGGCTGCTCTTGAGCATGAAGAAGCCAAGATCC TCCGAATCCAGCTTGAATTGACACAAGTGAAATCAGAAATTGATAGAAAGATCGCCGAGAAGGATGAAGAGATCGAGCAGCTGAAGAGGA ACTACCAGAGAACAGTGGAAACCATGCAGAGCGCCCTGGACGCCGAGGTGCGGAGCAGGAATGAAGCCATCCGGCTCAAGAAGAAGATGG AGGGGGACCTGAATGAAATCGAGATCCAGCTGAGCCACGCCAACCGCCAGGCGGCGGAGACCCTCAAACACCTCAGGAGTGTCCAGGGAC AGCTGAAGGATACGCAGCTCCACCTGGATGATGCCCTCCGGGGCCAGGAGGACCTGAAGGAGCAGCTGGCGATTGTGGAGCGCAGAGCCA ACCTGCTGCAGGCCGAGGTGGAGGAGCTGCGGGCTACTCTGGAGCAGACGGAGAGGGCCCGGAAACTGGCGGAACAGGAGCTCCTGGACT CCAACGAGAGGGTGCAGCTGCTGCATACCCAGAACACCAGCCTCATCCACACCAAGAAGAAGCTGGAGACAGACCTCATGCAGCTCCAGA GTGAGGTAGAAGATGCCAGCAGGGATGCAAGGAACGCTGAGGAGAAGGCCAAGAAGGCCATCACGGACGCTGCCATGATGGCGGAGGAGC TGAAGAAGGAGCAGGACACCAGCGCCCACCTTGAGCGGATGAAGAAGAACCTGGAACAGACGGTGAAGGACCTGCAGCATCGTCTAGATG AGGCCGAGCAGCTGGCGCTGAAGGGCGGGAAGAAGCAGATCCAGAAACTGGAGACCAGGATCCGAGAGCTGGAGTTTGAACTTGAGGGAG AGCAGAAGAAGAACACAGAGTCTGTTAAGGGCCTGAGGAAGTATGAGCGGAGGGTCAAGGAGCTGACGTACCAGAGTGAAGAGGACAGGA AGAATGTGCTGAGATTGCAGGATCTGGTGGATAAACTGCAAGTGAAAGTCAAGTCCTACAAGAGGCAGGCGGAGGAGGCTGATGAACAAG CCAATGCTCATCTCACCAAATTCCGAAAGGCTCAGCATGAGCTGGAGGAGGCCGAGGAACGTGCGGATATCGCAGAATCTCAAGTCAACA AGCTCCGCGCTAAGACTCGAGACTTCACCTCCAGCAGGATGGTGGTCCACGAGAGTGAAGAGTGAGCCAGCCCTTCTGGAGCAGGACAGA AGATATGCAAAATGTATATTTTCTTGATTCCTGACCATTGATACTTAATGTCCATGTGACTCTTTTTCACATGCAATAAACTTTGCTTTG >51209_51209_4_MAP2K4-MYH3_MAP2K4_chr17_11924318_ENST00000415385_MYH3_chr17_10539170_ENST00000583535_length(amino acids)=693AA_BP=38 MAAPSPSGGGGSGGGSGSGTPGPVGSPAPGHPAVSSMQGELSRQLEEKESIVSQLSRSKQAFTQQTEELKRQLEEENKAKNALAHALQSS RHDCDLLREQYEEEQEGKAELQRALSKANSEVAQWRTKYETDAIQRTEELEEAKKKLAQRLQDSEEQVEAVNAKCASLEKTKQRLQGEVE DLMVDVERANSLAAALDKKQRNFDKVLAEWKTKCEESQAELEASLKESRSLSTELFKLKNAYEEALDQLETVKRENKNLEQEIADLTEQI AENGKTIHELEKSRKQIELEKADIQLALEEAEAALEHEEAKILRIQLELTQVKSEIDRKIAEKDEEIEQLKRNYQRTVETMQSALDAEVR SRNEAIRLKKKMEGDLNEIEIQLSHANRQAAETLKHLRSVQGQLKDTQLHLDDALRGQEDLKEQLAIVERRANLLQAEVEELRATLEQTE RARKLAEQELLDSNERVQLLHTQNTSLIHTKKKLETDLMQLQSEVEDASRDARNAEEKAKKAITDAAMMAEELKKEQDTSAHLERMKKNL EQTVKDLQHRLDEAEQLALKGGKKQIQKLETRIRELEFELEGEQKKNTESVKGLRKYERRVKELTYQSEEDRKNVLRLQDLVDKLQVKVK -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for MAP2K4-MYH3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for MAP2K4-MYH3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for MAP2K4-MYH3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
