
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:MAT2B-FBXL17 (FusionGDB2 ID:51874) |
Fusion Gene Summary for MAT2B-FBXL17 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: MAT2B-FBXL17 | Fusion gene ID: 51874 | Hgene | Tgene | Gene symbol | MAT2B | FBXL17 | Gene ID | 27430 | 64839 |
| Gene name | methionine adenosyltransferase 2B | F-box and leucine rich repeat protein 17 | |
| Synonyms | MAT-II|MATIIbeta|Nbla02999|SDR23E1|TGR | FBXO13|Fbl17|Fbx13 | |
| Cytomap | 5q34 | 5q21.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | methionine adenosyltransferase 2 subunit betaMAT II betabeta regulatory subunit of methionine adenosyltransferasedTDP-4-keto-6-deoxy-D-glucose 4-reductasemethionine adenosyltransferase II, betaputative dTDP-4-keto-6-deoxy-D-glucose 4-reductaseputati | F-box/LRR-repeat protein 17F-box only protein 13 | |
| Modification date | 20200315 | 20200313 | |
| UniProtAcc | Q9NZL9 | Q9UF56 | |
| Ensembl transtripts involved in fusion gene | ENST00000280969, ENST00000321757, ENST00000518095, ENST00000521838, | ENST00000359660, ENST00000542267, ENST00000496714, | |
| Fusion gene scores | * DoF score | 5 X 3 X 5=75 | 14 X 13 X 5=910 |
| # samples | 6 | 13 | |
| ** MAII score | log2(6/75*10)=-0.321928094887362 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(13/910*10)=-2.8073549220576 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: MAT2B [Title/Abstract] AND FBXL17 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | MAT2B(162939202)-FBXL17(107216880), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | MAT2B-FBXL17 seems lost the major protein functional domain in Hgene partner, which is a cell metabolism gene due to the frame-shifted ORF. MAT2B-FBXL17 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | MAT2B | GO:0006556 | S-adenosylmethionine biosynthetic process | 10644686|25075345 |
| Tgene | FBXL17 | GO:0000209 | protein polyubiquitination | 27234298 |
| Tgene | FBXL17 | GO:0006515 | protein quality control for misfolded or incompletely synthesized proteins | 30190310 |
| Tgene | FBXL17 | GO:0016567 | protein ubiquitination | 24035498|30190310 |
| Tgene | FBXL17 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 24035498|27234298 |
| Tgene | FBXL17 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 30190310 |
 Fusion gene breakpoints across MAT2B (5'-gene) Fusion gene breakpoints across MAT2B (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
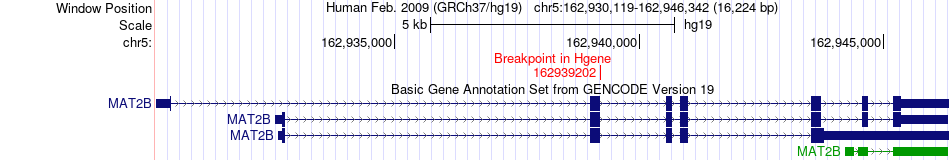 |
 Fusion gene breakpoints across FBXL17 (3'-gene) Fusion gene breakpoints across FBXL17 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
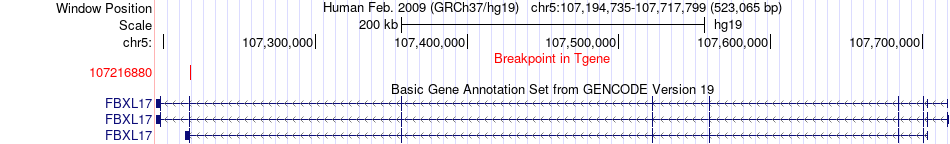 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | KIRC | TCGA-BP-4986-01A | MAT2B | chr5 | 162939202 | + | FBXL17 | chr5 | 107216880 | - |
Top |
Fusion Gene ORF analysis for MAT2B-FBXL17 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| Frame-shift | ENST00000280969 | ENST00000359660 | MAT2B | chr5 | 162939202 | + | FBXL17 | chr5 | 107216880 | - |
| Frame-shift | ENST00000280969 | ENST00000542267 | MAT2B | chr5 | 162939202 | + | FBXL17 | chr5 | 107216880 | - |
| Frame-shift | ENST00000321757 | ENST00000359660 | MAT2B | chr5 | 162939202 | + | FBXL17 | chr5 | 107216880 | - |
| Frame-shift | ENST00000321757 | ENST00000542267 | MAT2B | chr5 | 162939202 | + | FBXL17 | chr5 | 107216880 | - |
| Frame-shift | ENST00000518095 | ENST00000359660 | MAT2B | chr5 | 162939202 | + | FBXL17 | chr5 | 107216880 | - |
| Frame-shift | ENST00000518095 | ENST00000542267 | MAT2B | chr5 | 162939202 | + | FBXL17 | chr5 | 107216880 | - |
| In-frame | ENST00000280969 | ENST00000496714 | MAT2B | chr5 | 162939202 | + | FBXL17 | chr5 | 107216880 | - |
| In-frame | ENST00000321757 | ENST00000496714 | MAT2B | chr5 | 162939202 | + | FBXL17 | chr5 | 107216880 | - |
| In-frame | ENST00000518095 | ENST00000496714 | MAT2B | chr5 | 162939202 | + | FBXL17 | chr5 | 107216880 | - |
| intron-3CDS | ENST00000521838 | ENST00000359660 | MAT2B | chr5 | 162939202 | + | FBXL17 | chr5 | 107216880 | - |
| intron-3CDS | ENST00000521838 | ENST00000496714 | MAT2B | chr5 | 162939202 | + | FBXL17 | chr5 | 107216880 | - |
| intron-3CDS | ENST00000521838 | ENST00000542267 | MAT2B | chr5 | 162939202 | + | FBXL17 | chr5 | 107216880 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000280969 | MAT2B | chr5 | 162939202 | + | ENST00000496714 | FBXL17 | chr5 | 107216880 | - | 3449 | 506 | 1553 | 1936 | 127 |
| ENST00000321757 | MAT2B | chr5 | 162939202 | + | ENST00000496714 | FBXL17 | chr5 | 107216880 | - | 3340 | 397 | 429 | 40 | 129 |
| ENST00000518095 | MAT2B | chr5 | 162939202 | + | ENST00000496714 | FBXL17 | chr5 | 107216880 | - | 3274 | 331 | 1378 | 1761 | 127 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000280969 | ENST00000496714 | MAT2B | chr5 | 162939202 | + | FBXL17 | chr5 | 107216880 | - | 0.026344782 | 0.97365516 |
| ENST00000321757 | ENST00000496714 | MAT2B | chr5 | 162939202 | + | FBXL17 | chr5 | 107216880 | - | 0.026706867 | 0.9732931 |
| ENST00000518095 | ENST00000496714 | MAT2B | chr5 | 162939202 | + | FBXL17 | chr5 | 107216880 | - | 0.027326424 | 0.97267354 |
Top |
Fusion Genomic Features for MAT2B-FBXL17 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
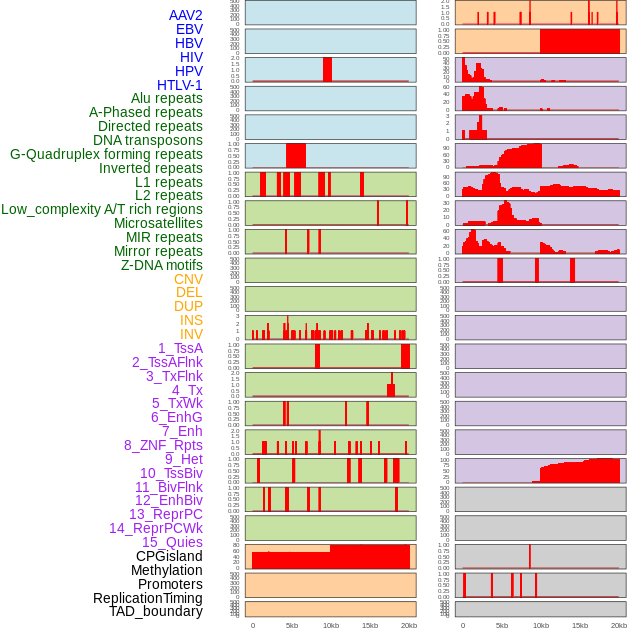 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for MAT2B-FBXL17 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr5:162939202/chr5:107216880) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MAT2B | FBXL17 |
| FUNCTION: Regulatory subunit of S-adenosylmethionine synthetase 2, an enzyme that catalyzes the formation of S-adenosylmethionine from methionine and ATP. Regulates MAT2A catalytic activity by changing its kinetic properties, increasing its affinity for L-methionine (PubMed:10644686, PubMed:23189196, PubMed:25075345). Can bind NADP (in vitro) (PubMed:23189196, PubMed:23425511). {ECO:0000269|PubMed:10644686, ECO:0000269|PubMed:23189196, ECO:0000269|PubMed:23425511, ECO:0000269|PubMed:25075345}. | FUNCTION: Substrate-recognition component of the SCF(FBXL17) E3 ubiquitin ligase complex, a key component of a quality control pathway required to ensure functional dimerization of BTB domain-containing proteins (dimerization quality control, DQC) (PubMed:30190310). FBXL17 specifically recognizes and binds a conserved degron of non-consecutive residues present at the interface of BTB dimers of aberrant composition: aberrant BTB dimer are then ubiquitinated by the SCF(FBXL17) complex and degraded by the proteaseome (PubMed:30190310). The ability of the SCF(FBXL17) complex to eliminate compromised BTB dimers is required for the differentiation and survival of neural crest and neuronal cells (By similarity). The SCF(FBXL17) complex mediates ubiquitination and degradation of BACH1 (PubMed:24035498, PubMed:30190310). The SCF(FBXL17) complex is also involved in the regulation of the hedgehog/smoothened (Hh) signaling pathway by mediating the ubiquitination and degradation of SUFU, allowing the release of GLI1 from SUFU for proper Hh signal transduction (PubMed:27234298). The SCF(FBXL17) complex mediates ubiquitination and degradation of PRMT1 (By similarity). {ECO:0000250|UniProtKB:B1H1X1, ECO:0000250|UniProtKB:Q9QZN1, ECO:0000269|PubMed:24035498, ECO:0000269|PubMed:27234298, ECO:0000269|PubMed:30190310}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MAT2B | chr5:162939202 | chr5:107216880 | ENST00000280969 | + | 2 | 7 | 37_40 | 75 | 324.0 | Nucleotide binding | NADP |
| Hgene | MAT2B | chr5:162939202 | chr5:107216880 | ENST00000280969 | + | 2 | 7 | 71_72 | 75 | 324.0 | Nucleotide binding | NADP |
| Hgene | MAT2B | chr5:162939202 | chr5:107216880 | ENST00000321757 | + | 2 | 7 | 37_40 | 86 | 335.0 | Nucleotide binding | NADP |
| Hgene | MAT2B | chr5:162939202 | chr5:107216880 | ENST00000321757 | + | 2 | 7 | 71_72 | 86 | 335.0 | Nucleotide binding | NADP |
| Hgene | MAT2B | chr5:162939202 | chr5:107216880 | ENST00000518095 | + | 2 | 5 | 37_40 | 86 | 260.0 | Nucleotide binding | NADP |
| Hgene | MAT2B | chr5:162939202 | chr5:107216880 | ENST00000518095 | + | 2 | 5 | 71_72 | 86 | 260.0 | Nucleotide binding | NADP |
| Tgene | FBXL17 | chr5:162939202 | chr5:107216880 | ENST00000359660 | 6 | 9 | 216_236 | 209 | 304.0 | Compositional bias | Note=Gly-rich | |
| Tgene | FBXL17 | chr5:162939202 | chr5:107216880 | ENST00000496714 | 5 | 7 | 216_236 | 209 | 315.0 | Compositional bias | Note=Gly-rich | |
| Tgene | FBXL17 | chr5:162939202 | chr5:107216880 | ENST00000359660 | 6 | 9 | 318_365 | 209 | 304.0 | Domain | F-box | |
| Tgene | FBXL17 | chr5:162939202 | chr5:107216880 | ENST00000496714 | 5 | 7 | 318_365 | 209 | 315.0 | Domain | F-box |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | FBXL17 | chr5:162939202 | chr5:107216880 | ENST00000359660 | 6 | 9 | 25_28 | 209 | 304.0 | Compositional bias | Note=Poly-Arg | |
| Tgene | FBXL17 | chr5:162939202 | chr5:107216880 | ENST00000359660 | 6 | 9 | 29_86 | 209 | 304.0 | Compositional bias | Note=Pro-rich | |
| Tgene | FBXL17 | chr5:162939202 | chr5:107216880 | ENST00000359660 | 6 | 9 | 90_151 | 209 | 304.0 | Compositional bias | Note=Ala-rich | |
| Tgene | FBXL17 | chr5:162939202 | chr5:107216880 | ENST00000496714 | 5 | 7 | 25_28 | 209 | 315.0 | Compositional bias | Note=Poly-Arg | |
| Tgene | FBXL17 | chr5:162939202 | chr5:107216880 | ENST00000496714 | 5 | 7 | 29_86 | 209 | 315.0 | Compositional bias | Note=Pro-rich | |
| Tgene | FBXL17 | chr5:162939202 | chr5:107216880 | ENST00000496714 | 5 | 7 | 90_151 | 209 | 315.0 | Compositional bias | Note=Ala-rich | |
| Tgene | FBXL17 | chr5:162939202 | chr5:107216880 | ENST00000542267 | 6 | 9 | 216_236 | 607 | 702.0 | Compositional bias | Note=Gly-rich | |
| Tgene | FBXL17 | chr5:162939202 | chr5:107216880 | ENST00000542267 | 6 | 9 | 25_28 | 607 | 702.0 | Compositional bias | Note=Poly-Arg | |
| Tgene | FBXL17 | chr5:162939202 | chr5:107216880 | ENST00000542267 | 6 | 9 | 29_86 | 607 | 702.0 | Compositional bias | Note=Pro-rich | |
| Tgene | FBXL17 | chr5:162939202 | chr5:107216880 | ENST00000542267 | 6 | 9 | 90_151 | 607 | 702.0 | Compositional bias | Note=Ala-rich | |
| Tgene | FBXL17 | chr5:162939202 | chr5:107216880 | ENST00000542267 | 6 | 9 | 318_365 | 607 | 702.0 | Domain | F-box |
Top |
Fusion Gene Sequence for MAT2B-FBXL17 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >51874_51874_1_MAT2B-FBXL17_MAT2B_chr5_162939202_ENST00000280969_FBXL17_chr5_107216880_ENST00000496714_length(transcript)=3449nt_BP=506nt GCAAATAGACCCAGGCTTGACTGGCAATTAAACATGAAACTTCTCATTGGGTATTTTCGAGACTACTACGGGGAATCAGCTACCAGCTTA CTGCCATGTGGAGAACTGCACGAGATTCCGGGATTGGAATCAAAATGCTAATTTAAAAGGTCAAGTGAAGCTGCTCCTCACGTTTTGGCG TGCCTGCGCTCTCTGCAGGCAGAAGCGAACAAAGACCCAGCAAGAGAAGGCAGAGGCTAAGACCCATCCCGTATCTGCTCTCCTGAAATA ATTCTGGAGTCATGCCTGAAATGCCAGAGGACATGGAGCAGGAGGAAGTTAACATCCCTAATAGGAGGGTTCTGGTTACTGGTGCCACTG GGCTTCTTGGCAGAGCTGTACACAAAGAATTTCAGCAGAATAATTGGCATGCAGTTGGCTGTGGTTTCAGAAGAGCAAGACCAAAATTTG AACAGGTTAATCTGTTGGATTCTAATGCAGTTCATCACATCATTCATGATTTTCAGCACTGATAGCCATTGGGCGATACAGCATGACAAT AGAGACTGTGGATGTCGGATGGTGTAAAGAAATCACAGACCAAGGAGCCACCCTGATTGCACAGAGCAGCAAGTCTCTGAGATATTTGGG GCTGATGAGATGTGATAAAGTAAGAGTTGACTATCAAGTTGTTTGCTTTCTACATATCAGTATAGTTAATAGCTTGATGTCATATCCTCT CTCCTTCTCTATCTCCACCCCCGTGTATTATATATTATATATACATTTTATATGTATATATGCAATTATAGCTATGCATTGTTTGCCTGC TTTTGTAAACTAATCTGAGATTATCTTTGTATTTCCCCACTTAATAATGAATTTTGTGCTTAAGTAGAAAGAAAAAGTAATCATATACAT AGTCAACTAGAGTTTGGCATCCTATTCATAAGTCCATGGACCAAAATGTAAAAGATTACATGACAAACTGCAATTTGCAGGGTCTAACCG TAAAATTGTATTCTGGGTTTCAAGTTTCCAGCAACAGTAAAGGTGGAAAGAACAGTGCCAAATTGGTTTTTCACAAGTGTGTAATTTTGG TACTGAACATCTATGTATTCTGGCCATTCGGGACCAACAAAGGGCCAGTAAGGGGGTATGTGGGAATTTGCATCTCCCCATACATCAGAT GCCACTCCCAGCACATGATAAGAAGTGGTTCCAGAGGTCCTGGTGGAAGGCCTGTAGAACCAGCCAGGGATGTGAGCATGAGTTTAGGAA ATGGTGTAGCATTTAACGTGGGACCCTTTGAAGACTGAGAAAACAGATAATAAAAATCTATACTGACACCTGTAATCCCAGCACATTGGG AGGCCAAGGCAGAAGGATCACTTGAGCCCAGGAATTTGAGACCAGCCTGGACAACATGGGAAAACCCTGTCTCTATCCCCCTCTCCCCCA ACAAAAAAAAATTAGCCAGGCATAATGGCACACACCTGTAGTCCCAGCTACTCGGGAAGCTAAGGTGGGAGGATCACCTGAGCTCAAGAG GCCGAGGTTGCAGTGAGCCGTGATTGTGCCGCAACCACCTCAGCCTGGATGACGGAGCACGACTGTGTCCCAAACAAACAAAAAAAACCT ATACTGGACACCAGGCACTCCACAGGTGTTTTCTTCTTTGTTTATTCCATGTTTTACCTATAACAAGTCTAAGAAATAGTTATTTGCATT GTGAAGGGGAGAAAATGGAGGCTCAGAAAGACCAGTCAGTGGCCCCAGGATACTCTGGCAAGACTTCATGCCGGGTCTTTGGAGTCTGTA TCCAGTGCTGCTGGCACAACCCAGGGGGCAGTCTGTGGCCACGGGGACTGCTTCCCTTGGAAAAGGTCCAGTTCAGATACTTGCTCAGAA AGTGCCGCCATCTTGTACTAATTTACACATGTGTGCTACTTTTGTAAATGGTGATTAGGAACAAATCTAGCAAAATAATGTGAAACTAAA TAGACACGCCATTCAAAGGGCATGCATCTTCTAAATGTAGCAAATGTGGGGGCAAATGAAGTATGGCCCAAAGCTCTTGTGTCCTTTGTG ATTTTTTGTTAATTGGCTAATTCTTTTCTCCAAACACTTTAGAATCCCAACTTATACGAGACCTTTGCCAGCTAGTTATGTTTAGAGAAA TGTTATTATATATTCTCAGCCCAGCCCAGAAGCGACTTACTTTGTAACTAAGCAACAGCACTGAGATCCGGGGCTTGCGAGTCTCCCACA AATCTTTCCACTAACCAACCTAATGCCGCTGCTAACCCTGCCACTTTAAAATGACTTCTTTTAAGACAAGTTAAAGGGTCCACCATAGGC ATAGTCCTCTAGAAACAGGAATCAATGACAGCAGAACTCCTCAGCTTCCAGCAGAAGGAACTGCCACATGAATGCTAACTAAAGCTACCA AAATGTACACATTCATACATATGTGTCCGCAGCCATTCTCAGAAATTGTCATATTTCTAACCTGTAATCTTTAGATGGTTAAGATCAGCT CCATCTTCGGAGATACACTGTAGAAGTTGTATGTTACAGTTACTACAGGAGAATGTTATTCCAGACATATTTGCTGACCAGCATTTTGCT GATTTTTAGTTAAGAAATGGGTTATTTTCTACCCAAATGCAAATTAGTCCAAAATTAAATAGAGACAGTATTCAGATTAGCTGTATCCCT GAGGGTTTTAGCCCTTTTCCTGGGCTGCCTCTGAATCCTTTCTCTAACCAGAGAGCCACTCTGAGAACCCATTGGCTCCAGCACGACGCC GTCTGGTCAGAGCCTGCTGCAGTGCAGCCCAGGGAGATGCTGGACCCAGCCTTGCTGTCTGGCTTCCTCAGACTTAAAGCAGCTCAGAGC CAAACACGAGAGGCTCTGTGAGGATGGAAGACAGTTCCAAACCACACACAAAGCACAAGTAAAGCCTTGCACAGAAAAGCCTGCTTGTTG GTTTAGTTGGATGGAAATCTAAAGTAGCTATTCAGTTTTCAGATTTTGTATCATGCCATCCATCTCCAGATTTTAAGTTACCATCAAGAT TCAAAGGAAGATATGAGTAACAAAGAAAAGAAATTAGTAGCAACGGTATGGTCTGCGGAGAGCCAGAGTTCAGGTTCAGATCTGGGCTTT GCACTTACTAGTTGTGTCACCGCGGGCAGTTACTAACCTCTCAGTGCCTGTTCCCTCGTTTGCAAAATGAGGATGATACTAGTACCTATC TTGTAGAATTGTGAGCATGTGAATGAGTTAACATACATAAAGCACTTAGAATAATGTCTATTATATATAAAATGCTCTGTTACTGATTAG TCTTATCATCCATATCATTATATTAACCTGTGTTGTGATACTGGAATTTTCCAATGAGAAAAAAAAATTATATTGCCTACTATCCCATTT >51874_51874_1_MAT2B-FBXL17_MAT2B_chr5_162939202_ENST00000280969_FBXL17_chr5_107216880_ENST00000496714_length(amino acids)=127AA_BP= MCRNHLSLDDGARLCPKQTKKTYTGHQALHRCFLLCLFHVLPITSLRNSYLHCEGEKMEAQKDQSVAPGYSGKTSCRVFGVCIQCCWHNP -------------------------------------------------------------- >51874_51874_2_MAT2B-FBXL17_MAT2B_chr5_162939202_ENST00000321757_FBXL17_chr5_107216880_ENST00000496714_length(transcript)=3340nt_BP=397nt GCAGCGGAAGCCGGAAGCGGCGAGCGGGGTCGTTCTGGGCCTAGGGGAGGCGGGCCGAGGGCGTCTGAGCTGAGGCCCGCGTCGATCCTG GGTTGGAGGAGGTGGCGGCCGCTGAGGCTGCGGCGTGAAGACGGCGGGCATGGTGGGGCGGGAGAAAGAGCTCTCTATACACTTTGTTCC CGGGAGCTGTCGGCTGGTGGAGGAGGAAGTTAACATCCCTAATAGGAGGGTTCTGGTTACTGGTGCCACTGGGCTTCTTGGCAGAGCTGT ACACAAAGAATTTCAGCAGAATAATTGGCATGCAGTTGGCTGTGGTTTCAGAAGAGCAAGACCAAAATTTGAACAGGTTAATCTGTTGGA TTCTAATGCAGTTCATCACATCATTCATGATTTTCAGCACTGATAGCCATTGGGCGATACAGCATGACAATAGAGACTGTGGATGTCGGA TGGTGTAAAGAAATCACAGACCAAGGAGCCACCCTGATTGCACAGAGCAGCAAGTCTCTGAGATATTTGGGGCTGATGAGATGTGATAAA GTAAGAGTTGACTATCAAGTTGTTTGCTTTCTACATATCAGTATAGTTAATAGCTTGATGTCATATCCTCTCTCCTTCTCTATCTCCACC CCCGTGTATTATATATTATATATACATTTTATATGTATATATGCAATTATAGCTATGCATTGTTTGCCTGCTTTTGTAAACTAATCTGAG ATTATCTTTGTATTTCCCCACTTAATAATGAATTTTGTGCTTAAGTAGAAAGAAAAAGTAATCATATACATAGTCAACTAGAGTTTGGCA TCCTATTCATAAGTCCATGGACCAAAATGTAAAAGATTACATGACAAACTGCAATTTGCAGGGTCTAACCGTAAAATTGTATTCTGGGTT TCAAGTTTCCAGCAACAGTAAAGGTGGAAAGAACAGTGCCAAATTGGTTTTTCACAAGTGTGTAATTTTGGTACTGAACATCTATGTATT CTGGCCATTCGGGACCAACAAAGGGCCAGTAAGGGGGTATGTGGGAATTTGCATCTCCCCATACATCAGATGCCACTCCCAGCACATGAT AAGAAGTGGTTCCAGAGGTCCTGGTGGAAGGCCTGTAGAACCAGCCAGGGATGTGAGCATGAGTTTAGGAAATGGTGTAGCATTTAACGT GGGACCCTTTGAAGACTGAGAAAACAGATAATAAAAATCTATACTGACACCTGTAATCCCAGCACATTGGGAGGCCAAGGCAGAAGGATC ACTTGAGCCCAGGAATTTGAGACCAGCCTGGACAACATGGGAAAACCCTGTCTCTATCCCCCTCTCCCCCAACAAAAAAAAATTAGCCAG GCATAATGGCACACACCTGTAGTCCCAGCTACTCGGGAAGCTAAGGTGGGAGGATCACCTGAGCTCAAGAGGCCGAGGTTGCAGTGAGCC GTGATTGTGCCGCAACCACCTCAGCCTGGATGACGGAGCACGACTGTGTCCCAAACAAACAAAAAAAACCTATACTGGACACCAGGCACT CCACAGGTGTTTTCTTCTTTGTTTATTCCATGTTTTACCTATAACAAGTCTAAGAAATAGTTATTTGCATTGTGAAGGGGAGAAAATGGA GGCTCAGAAAGACCAGTCAGTGGCCCCAGGATACTCTGGCAAGACTTCATGCCGGGTCTTTGGAGTCTGTATCCAGTGCTGCTGGCACAA CCCAGGGGGCAGTCTGTGGCCACGGGGACTGCTTCCCTTGGAAAAGGTCCAGTTCAGATACTTGCTCAGAAAGTGCCGCCATCTTGTACT AATTTACACATGTGTGCTACTTTTGTAAATGGTGATTAGGAACAAATCTAGCAAAATAATGTGAAACTAAATAGACACGCCATTCAAAGG GCATGCATCTTCTAAATGTAGCAAATGTGGGGGCAAATGAAGTATGGCCCAAAGCTCTTGTGTCCTTTGTGATTTTTTGTTAATTGGCTA ATTCTTTTCTCCAAACACTTTAGAATCCCAACTTATACGAGACCTTTGCCAGCTAGTTATGTTTAGAGAAATGTTATTATATATTCTCAG CCCAGCCCAGAAGCGACTTACTTTGTAACTAAGCAACAGCACTGAGATCCGGGGCTTGCGAGTCTCCCACAAATCTTTCCACTAACCAAC CTAATGCCGCTGCTAACCCTGCCACTTTAAAATGACTTCTTTTAAGACAAGTTAAAGGGTCCACCATAGGCATAGTCCTCTAGAAACAGG AATCAATGACAGCAGAACTCCTCAGCTTCCAGCAGAAGGAACTGCCACATGAATGCTAACTAAAGCTACCAAAATGTACACATTCATACA TATGTGTCCGCAGCCATTCTCAGAAATTGTCATATTTCTAACCTGTAATCTTTAGATGGTTAAGATCAGCTCCATCTTCGGAGATACACT GTAGAAGTTGTATGTTACAGTTACTACAGGAGAATGTTATTCCAGACATATTTGCTGACCAGCATTTTGCTGATTTTTAGTTAAGAAATG GGTTATTTTCTACCCAAATGCAAATTAGTCCAAAATTAAATAGAGACAGTATTCAGATTAGCTGTATCCCTGAGGGTTTTAGCCCTTTTC CTGGGCTGCCTCTGAATCCTTTCTCTAACCAGAGAGCCACTCTGAGAACCCATTGGCTCCAGCACGACGCCGTCTGGTCAGAGCCTGCTG CAGTGCAGCCCAGGGAGATGCTGGACCCAGCCTTGCTGTCTGGCTTCCTCAGACTTAAAGCAGCTCAGAGCCAAACACGAGAGGCTCTGT GAGGATGGAAGACAGTTCCAAACCACACACAAAGCACAAGTAAAGCCTTGCACAGAAAAGCCTGCTTGTTGGTTTAGTTGGATGGAAATC TAAAGTAGCTATTCAGTTTTCAGATTTTGTATCATGCCATCCATCTCCAGATTTTAAGTTACCATCAAGATTCAAAGGAAGATATGAGTA ACAAAGAAAAGAAATTAGTAGCAACGGTATGGTCTGCGGAGAGCCAGAGTTCAGGTTCAGATCTGGGCTTTGCACTTACTAGTTGTGTCA CCGCGGGCAGTTACTAACCTCTCAGTGCCTGTTCCCTCGTTTGCAAAATGAGGATGATACTAGTACCTATCTTGTAGAATTGTGAGCATG TGAATGAGTTAACATACATAAAGCACTTAGAATAATGTCTATTATATATAAAATGCTCTGTTACTGATTAGTCTTATCATCCATATCATT ATATTAACCTGTGTTGTGATACTGGAATTTTCCAATGAGAAAAAAAAATTATATTGCCTACTATCCCATTTTGAAAGACAATTAAAAAGA >51874_51874_2_MAT2B-FBXL17_MAT2B_chr5_162939202_ENST00000321757_FBXL17_chr5_107216880_ENST00000496714_length(amino acids)=129AA_BP= MSCCIAQWLSVLKIMNDVMNCIRIQQINLFKFWSCSSETTANCMPIILLKFFVYSSAKKPSGTSNQNPPIRDVNFLLHQPTAPGNKVYRE -------------------------------------------------------------- >51874_51874_3_MAT2B-FBXL17_MAT2B_chr5_162939202_ENST00000518095_FBXL17_chr5_107216880_ENST00000496714_length(transcript)=3274nt_BP=331nt GAGCTGAGGCCCGCGTCGATCCTGGGTTGGAGGAGGTGGCGGCCGCTGAGGCTGCGGCGTGAAGACGGCGGGCATGGTGGGGCGGGAGAA AGAGCTCTCTATACACTTTGTTCCCGGGAGCTGTCGGCTGGTGGAGGAGGAAGTTAACATCCCTAATAGGAGGGTTCTGGTTACTGGTGC CACTGGGCTTCTTGGCAGAGCTGTACACAAAGAATTTCAGCAGAATAATTGGCATGCAGTTGGCTGTGGTTTCAGAAGAGCAAGACCAAA ATTTGAACAGGTTAATCTGTTGGATTCTAATGCAGTTCATCACATCATTCATGATTTTCAGCACTGATAGCCATTGGGCGATACAGCATG ACAATAGAGACTGTGGATGTCGGATGGTGTAAAGAAATCACAGACCAAGGAGCCACCCTGATTGCACAGAGCAGCAAGTCTCTGAGATAT TTGGGGCTGATGAGATGTGATAAAGTAAGAGTTGACTATCAAGTTGTTTGCTTTCTACATATCAGTATAGTTAATAGCTTGATGTCATAT CCTCTCTCCTTCTCTATCTCCACCCCCGTGTATTATATATTATATATACATTTTATATGTATATATGCAATTATAGCTATGCATTGTTTG CCTGCTTTTGTAAACTAATCTGAGATTATCTTTGTATTTCCCCACTTAATAATGAATTTTGTGCTTAAGTAGAAAGAAAAAGTAATCATA TACATAGTCAACTAGAGTTTGGCATCCTATTCATAAGTCCATGGACCAAAATGTAAAAGATTACATGACAAACTGCAATTTGCAGGGTCT AACCGTAAAATTGTATTCTGGGTTTCAAGTTTCCAGCAACAGTAAAGGTGGAAAGAACAGTGCCAAATTGGTTTTTCACAAGTGTGTAAT TTTGGTACTGAACATCTATGTATTCTGGCCATTCGGGACCAACAAAGGGCCAGTAAGGGGGTATGTGGGAATTTGCATCTCCCCATACAT CAGATGCCACTCCCAGCACATGATAAGAAGTGGTTCCAGAGGTCCTGGTGGAAGGCCTGTAGAACCAGCCAGGGATGTGAGCATGAGTTT AGGAAATGGTGTAGCATTTAACGTGGGACCCTTTGAAGACTGAGAAAACAGATAATAAAAATCTATACTGACACCTGTAATCCCAGCACA TTGGGAGGCCAAGGCAGAAGGATCACTTGAGCCCAGGAATTTGAGACCAGCCTGGACAACATGGGAAAACCCTGTCTCTATCCCCCTCTC CCCCAACAAAAAAAAATTAGCCAGGCATAATGGCACACACCTGTAGTCCCAGCTACTCGGGAAGCTAAGGTGGGAGGATCACCTGAGCTC AAGAGGCCGAGGTTGCAGTGAGCCGTGATTGTGCCGCAACCACCTCAGCCTGGATGACGGAGCACGACTGTGTCCCAAACAAACAAAAAA AACCTATACTGGACACCAGGCACTCCACAGGTGTTTTCTTCTTTGTTTATTCCATGTTTTACCTATAACAAGTCTAAGAAATAGTTATTT GCATTGTGAAGGGGAGAAAATGGAGGCTCAGAAAGACCAGTCAGTGGCCCCAGGATACTCTGGCAAGACTTCATGCCGGGTCTTTGGAGT CTGTATCCAGTGCTGCTGGCACAACCCAGGGGGCAGTCTGTGGCCACGGGGACTGCTTCCCTTGGAAAAGGTCCAGTTCAGATACTTGCT CAGAAAGTGCCGCCATCTTGTACTAATTTACACATGTGTGCTACTTTTGTAAATGGTGATTAGGAACAAATCTAGCAAAATAATGTGAAA CTAAATAGACACGCCATTCAAAGGGCATGCATCTTCTAAATGTAGCAAATGTGGGGGCAAATGAAGTATGGCCCAAAGCTCTTGTGTCCT TTGTGATTTTTTGTTAATTGGCTAATTCTTTTCTCCAAACACTTTAGAATCCCAACTTATACGAGACCTTTGCCAGCTAGTTATGTTTAG AGAAATGTTATTATATATTCTCAGCCCAGCCCAGAAGCGACTTACTTTGTAACTAAGCAACAGCACTGAGATCCGGGGCTTGCGAGTCTC CCACAAATCTTTCCACTAACCAACCTAATGCCGCTGCTAACCCTGCCACTTTAAAATGACTTCTTTTAAGACAAGTTAAAGGGTCCACCA TAGGCATAGTCCTCTAGAAACAGGAATCAATGACAGCAGAACTCCTCAGCTTCCAGCAGAAGGAACTGCCACATGAATGCTAACTAAAGC TACCAAAATGTACACATTCATACATATGTGTCCGCAGCCATTCTCAGAAATTGTCATATTTCTAACCTGTAATCTTTAGATGGTTAAGAT CAGCTCCATCTTCGGAGATACACTGTAGAAGTTGTATGTTACAGTTACTACAGGAGAATGTTATTCCAGACATATTTGCTGACCAGCATT TTGCTGATTTTTAGTTAAGAAATGGGTTATTTTCTACCCAAATGCAAATTAGTCCAAAATTAAATAGAGACAGTATTCAGATTAGCTGTA TCCCTGAGGGTTTTAGCCCTTTTCCTGGGCTGCCTCTGAATCCTTTCTCTAACCAGAGAGCCACTCTGAGAACCCATTGGCTCCAGCACG ACGCCGTCTGGTCAGAGCCTGCTGCAGTGCAGCCCAGGGAGATGCTGGACCCAGCCTTGCTGTCTGGCTTCCTCAGACTTAAAGCAGCTC AGAGCCAAACACGAGAGGCTCTGTGAGGATGGAAGACAGTTCCAAACCACACACAAAGCACAAGTAAAGCCTTGCACAGAAAAGCCTGCT TGTTGGTTTAGTTGGATGGAAATCTAAAGTAGCTATTCAGTTTTCAGATTTTGTATCATGCCATCCATCTCCAGATTTTAAGTTACCATC AAGATTCAAAGGAAGATATGAGTAACAAAGAAAAGAAATTAGTAGCAACGGTATGGTCTGCGGAGAGCCAGAGTTCAGGTTCAGATCTGG GCTTTGCACTTACTAGTTGTGTCACCGCGGGCAGTTACTAACCTCTCAGTGCCTGTTCCCTCGTTTGCAAAATGAGGATGATACTAGTAC CTATCTTGTAGAATTGTGAGCATGTGAATGAGTTAACATACATAAAGCACTTAGAATAATGTCTATTATATATAAAATGCTCTGTTACTG ATTAGTCTTATCATCCATATCATTATATTAACCTGTGTTGTGATACTGGAATTTTCCAATGAGAAAAAAAAATTATATTGCCTACTATCC >51874_51874_3_MAT2B-FBXL17_MAT2B_chr5_162939202_ENST00000518095_FBXL17_chr5_107216880_ENST00000496714_length(amino acids)=127AA_BP= MCRNHLSLDDGARLCPKQTKKTYTGHQALHRCFLLCLFHVLPITSLRNSYLHCEGEKMEAQKDQSVAPGYSGKTSCRVFGVCIQCCWHNP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for MAT2B-FBXL17 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for MAT2B-FBXL17 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for MAT2B-FBXL17 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
