
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:MAVS-STK24 (FusionGDB2 ID:51934) |
Fusion Gene Summary for MAVS-STK24 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: MAVS-STK24 | Fusion gene ID: 51934 | Hgene | Tgene | Gene symbol | MAVS | STK24 | Gene ID | 57506 | 8428 |
| Gene name | mitochondrial antiviral signaling protein | serine/threonine kinase 24 | |
| Synonyms | CARDIF|IPS-1|IPS1|VISA | HEL-S-95|MST3|MST3B|STE20|STK3 | |
| Cytomap | 20p13 | 13q32.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | mitochondrial antiviral-signaling proteinCARD adapter inducing interferon betaCARD adaptor inducing IFN-betaIFN-B promoter stimulator 1interferon beta promoter stimulator protein 1putative NF-kappa-B-activating protein 031Nvirus-induced signaling ad | serine/threonine-protein kinase 24STE20-like kinase 3STE20-like kinase MST3epididymis secretory protein Li 95mammalian STE20-like protein kinase 3serine/threonine kinase 24 (STE20 homolog, yeast)sterile 20-like kinase 3 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q7Z434 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000358134, ENST00000416600, ENST00000428216, | ENST00000397517, ENST00000539966, ENST00000376547, ENST00000481288, | |
| Fusion gene scores | * DoF score | 4 X 4 X 2=32 | 15 X 8 X 8=960 |
| # samples | 4 | 18 | |
| ** MAII score | log2(4/32*10)=0.321928094887362 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(18/960*10)=-2.41503749927884 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: MAVS [Title/Abstract] AND STK24 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | MAVS(3845435)-STK24(99171727), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | MAVS | GO:0001934 | positive regulation of protein phosphorylation | 16127453 |
| Hgene | MAVS | GO:0002230 | positive regulation of defense response to virus by host | 16127453 |
| Hgene | MAVS | GO:0032727 | positive regulation of interferon-alpha production | 16127453 |
| Hgene | MAVS | GO:0032728 | positive regulation of interferon-beta production | 16127453 |
| Hgene | MAVS | GO:0032757 | positive regulation of interleukin-8 production | 16127453 |
| Hgene | MAVS | GO:0032760 | positive regulation of tumor necrosis factor production | 16127453 |
| Hgene | MAVS | GO:0042307 | positive regulation of protein import into nucleus | 18818105 |
| Hgene | MAVS | GO:0045071 | negative regulation of viral genome replication | 20451243 |
| Hgene | MAVS | GO:0045944 | positive regulation of transcription by RNA polymerase II | 16127453|18818105 |
| Hgene | MAVS | GO:0051091 | positive regulation of DNA-binding transcription factor activity | 16127453|18818105 |
| Hgene | MAVS | GO:0051607 | defense response to virus | 20451243 |
| Hgene | MAVS | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway | 16127453|20451243 |
| Hgene | MAVS | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production | 16127453 |
| Hgene | MAVS | GO:0071660 | positive regulation of IP-10 production | 16127453 |
| Tgene | STK24 | GO:0006468 | protein phosphorylation | 19604147 |
| Tgene | STK24 | GO:0042542 | response to hydrogen peroxide | 22291017 |
| Tgene | STK24 | GO:0046777 | protein autophosphorylation | 17046825|17657516 |
 Fusion gene breakpoints across MAVS (5'-gene) Fusion gene breakpoints across MAVS (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
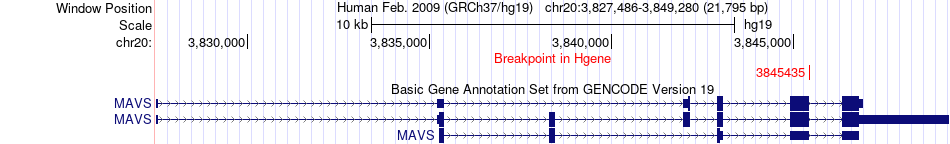 |
 Fusion gene breakpoints across STK24 (3'-gene) Fusion gene breakpoints across STK24 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
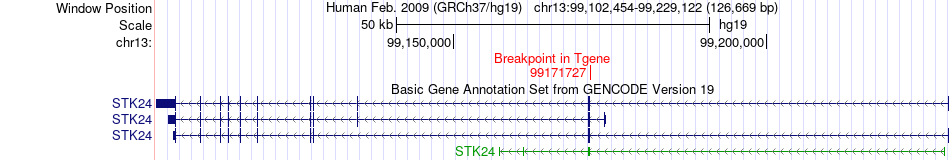 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-29-1770-01A | MAVS | chr20 | 3845435 | + | STK24 | chr13 | 99171727 | - |
Top |
Fusion Gene ORF analysis for MAVS-STK24 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000358134 | ENST00000397517 | MAVS | chr20 | 3845435 | + | STK24 | chr13 | 99171727 | - |
| 3UTR-3CDS | ENST00000358134 | ENST00000539966 | MAVS | chr20 | 3845435 | + | STK24 | chr13 | 99171727 | - |
| 3UTR-5UTR | ENST00000358134 | ENST00000376547 | MAVS | chr20 | 3845435 | + | STK24 | chr13 | 99171727 | - |
| 3UTR-5UTR | ENST00000358134 | ENST00000481288 | MAVS | chr20 | 3845435 | + | STK24 | chr13 | 99171727 | - |
| 5CDS-5UTR | ENST00000416600 | ENST00000376547 | MAVS | chr20 | 3845435 | + | STK24 | chr13 | 99171727 | - |
| 5CDS-5UTR | ENST00000416600 | ENST00000481288 | MAVS | chr20 | 3845435 | + | STK24 | chr13 | 99171727 | - |
| 5CDS-5UTR | ENST00000428216 | ENST00000376547 | MAVS | chr20 | 3845435 | + | STK24 | chr13 | 99171727 | - |
| 5CDS-5UTR | ENST00000428216 | ENST00000481288 | MAVS | chr20 | 3845435 | + | STK24 | chr13 | 99171727 | - |
| In-frame | ENST00000416600 | ENST00000397517 | MAVS | chr20 | 3845435 | + | STK24 | chr13 | 99171727 | - |
| In-frame | ENST00000416600 | ENST00000539966 | MAVS | chr20 | 3845435 | + | STK24 | chr13 | 99171727 | - |
| In-frame | ENST00000428216 | ENST00000397517 | MAVS | chr20 | 3845435 | + | STK24 | chr13 | 99171727 | - |
| In-frame | ENST00000428216 | ENST00000539966 | MAVS | chr20 | 3845435 | + | STK24 | chr13 | 99171727 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000416600 | MAVS | chr20 | 3845435 | + | ENST00000397517 | STK24 | chr13 | 99171727 | - | 5340 | 1114 | 166 | 2367 | 733 |
| ENST00000416600 | MAVS | chr20 | 3845435 | + | ENST00000539966 | STK24 | chr13 | 99171727 | - | 2569 | 1114 | 166 | 2310 | 714 |
| ENST00000428216 | MAVS | chr20 | 3845435 | + | ENST00000397517 | STK24 | chr13 | 99171727 | - | 5512 | 1286 | 128 | 2539 | 803 |
| ENST00000428216 | MAVS | chr20 | 3845435 | + | ENST00000539966 | STK24 | chr13 | 99171727 | - | 2741 | 1286 | 128 | 2482 | 784 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000416600 | ENST00000397517 | MAVS | chr20 | 3845435 | + | STK24 | chr13 | 99171727 | - | 0.001667766 | 0.9983322 |
| ENST00000416600 | ENST00000539966 | MAVS | chr20 | 3845435 | + | STK24 | chr13 | 99171727 | - | 0.014837329 | 0.9851627 |
| ENST00000428216 | ENST00000397517 | MAVS | chr20 | 3845435 | + | STK24 | chr13 | 99171727 | - | 0.001468472 | 0.99853146 |
| ENST00000428216 | ENST00000539966 | MAVS | chr20 | 3845435 | + | STK24 | chr13 | 99171727 | - | 0.008875679 | 0.99112433 |
Top |
Fusion Genomic Features for MAVS-STK24 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
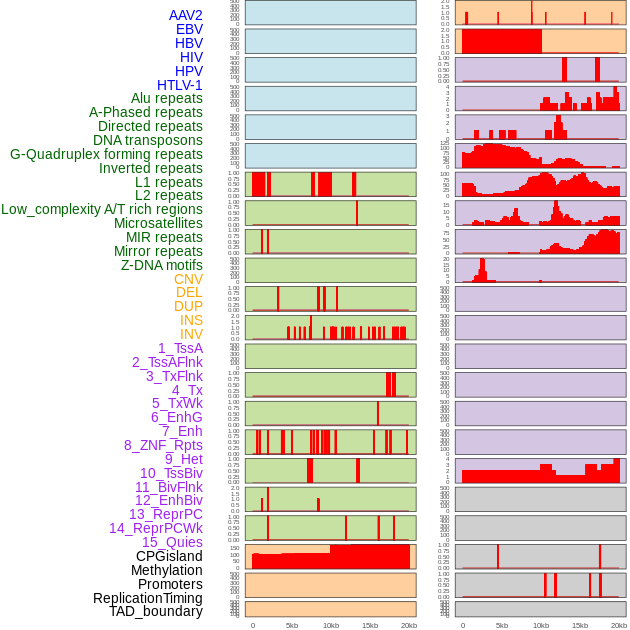 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for MAVS-STK24 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr20:3845435/chr13:99171727) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MAVS | . |
| FUNCTION: Required for innate immune defense against viruses (PubMed:16125763, PubMed:16127453, PubMed:16153868, PubMed:16177806, PubMed:19631370, PubMed:20451243, PubMed:23087404, PubMed:20127681, PubMed:21170385). Acts downstream of DHX33, DDX58/RIG-I and IFIH1/MDA5, which detect intracellular dsRNA produced during viral replication, to coordinate pathways leading to the activation of NF-kappa-B, IRF3 and IRF7, and to the subsequent induction of antiviral cytokines such as IFNB and RANTES (CCL5) (PubMed:16125763, PubMed:16127453, PubMed:16153868, PubMed:16177806, PubMed:19631370, PubMed:20451243, PubMed:23087404, PubMed:25636800, PubMed:20127681, PubMed:21170385, PubMed:20628368). Peroxisomal and mitochondrial MAVS act sequentially to create an antiviral cellular state (PubMed:20451243). Upon viral infection, peroxisomal MAVS induces the rapid interferon-independent expression of defense factors that provide short-term protection, whereas mitochondrial MAVS activates an interferon-dependent signaling pathway with delayed kinetics, which amplifies and stabilizes the antiviral response (PubMed:20451243). May activate the same pathways following detection of extracellular dsRNA by TLR3 (PubMed:16153868). May protect cells from apoptosis (PubMed:16125763). {ECO:0000269|PubMed:16125763, ECO:0000269|PubMed:16127453, ECO:0000269|PubMed:16153868, ECO:0000269|PubMed:16177806, ECO:0000269|PubMed:19631370, ECO:0000269|PubMed:20127681, ECO:0000269|PubMed:20451243, ECO:0000269|PubMed:20628368, ECO:0000269|PubMed:21170385, ECO:0000269|PubMed:23087404, ECO:0000269|PubMed:25636800}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MAVS | chr20:3845435 | chr13:99171727 | ENST00000358134 | + | 4 | 5 | 103_153 | 328 | 360.3333333333333 | Compositional bias | Note=Pro-rich |
| Hgene | MAVS | chr20:3845435 | chr13:99171727 | ENST00000416600 | + | 5 | 6 | 103_153 | 245 | 400.0 | Compositional bias | Note=Pro-rich |
| Hgene | MAVS | chr20:3845435 | chr13:99171727 | ENST00000428216 | + | 6 | 7 | 103_153 | 386 | 541.0 | Compositional bias | Note=Pro-rich |
| Tgene | STK24 | chr20:3845435 | chr13:99171727 | ENST00000376547 | 0 | 11 | 36_286 | 26 | 444.0 | Domain | Protein kinase | |
| Tgene | STK24 | chr20:3845435 | chr13:99171727 | ENST00000397517 | 0 | 11 | 36_286 | 14 | 432.0 | Domain | Protein kinase | |
| Tgene | STK24 | chr20:3845435 | chr13:99171727 | ENST00000376547 | 0 | 11 | 278_292 | 26 | 444.0 | Motif | Note=Bipartite nuclear localization signal | |
| Tgene | STK24 | chr20:3845435 | chr13:99171727 | ENST00000376547 | 0 | 11 | 335_386 | 26 | 444.0 | Motif | Note=Nuclear export signal (NES) | |
| Tgene | STK24 | chr20:3845435 | chr13:99171727 | ENST00000397517 | 0 | 11 | 278_292 | 14 | 432.0 | Motif | Note=Bipartite nuclear localization signal | |
| Tgene | STK24 | chr20:3845435 | chr13:99171727 | ENST00000397517 | 0 | 11 | 335_386 | 14 | 432.0 | Motif | Note=Nuclear export signal (NES) | |
| Tgene | STK24 | chr20:3845435 | chr13:99171727 | ENST00000376547 | 0 | 11 | 112_114 | 26 | 444.0 | Nucleotide binding | Note=ATP | |
| Tgene | STK24 | chr20:3845435 | chr13:99171727 | ENST00000376547 | 0 | 11 | 42_50 | 26 | 444.0 | Nucleotide binding | Note=ATP | |
| Tgene | STK24 | chr20:3845435 | chr13:99171727 | ENST00000397517 | 0 | 11 | 112_114 | 14 | 432.0 | Nucleotide binding | Note=ATP | |
| Tgene | STK24 | chr20:3845435 | chr13:99171727 | ENST00000397517 | 0 | 11 | 42_50 | 14 | 432.0 | Nucleotide binding | Note=ATP |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MAVS | chr20:3845435 | chr13:99171727 | ENST00000358134 | + | 4 | 5 | 439_442 | 328 | 360.3333333333333 | Motif | pLxIS motif |
| Hgene | MAVS | chr20:3845435 | chr13:99171727 | ENST00000416600 | + | 5 | 6 | 439_442 | 245 | 400.0 | Motif | pLxIS motif |
| Hgene | MAVS | chr20:3845435 | chr13:99171727 | ENST00000428216 | + | 6 | 7 | 439_442 | 386 | 541.0 | Motif | pLxIS motif |
| Hgene | MAVS | chr20:3845435 | chr13:99171727 | ENST00000358134 | + | 4 | 5 | 1_513 | 328 | 360.3333333333333 | Topological domain | Cytoplasmic |
| Hgene | MAVS | chr20:3845435 | chr13:99171727 | ENST00000358134 | + | 4 | 5 | 535_540 | 328 | 360.3333333333333 | Topological domain | Mitochondrial intermembrane |
| Hgene | MAVS | chr20:3845435 | chr13:99171727 | ENST00000416600 | + | 5 | 6 | 1_513 | 245 | 400.0 | Topological domain | Cytoplasmic |
| Hgene | MAVS | chr20:3845435 | chr13:99171727 | ENST00000416600 | + | 5 | 6 | 535_540 | 245 | 400.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | MAVS | chr20:3845435 | chr13:99171727 | ENST00000428216 | + | 6 | 7 | 1_513 | 386 | 541.0 | Topological domain | Cytoplasmic |
| Hgene | MAVS | chr20:3845435 | chr13:99171727 | ENST00000428216 | + | 6 | 7 | 535_540 | 386 | 541.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | MAVS | chr20:3845435 | chr13:99171727 | ENST00000358134 | + | 4 | 5 | 514_534 | 328 | 360.3333333333333 | Transmembrane | Helical |
| Hgene | MAVS | chr20:3845435 | chr13:99171727 | ENST00000416600 | + | 5 | 6 | 514_534 | 245 | 400.0 | Transmembrane | Helical |
| Hgene | MAVS | chr20:3845435 | chr13:99171727 | ENST00000428216 | + | 6 | 7 | 514_534 | 386 | 541.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for MAVS-STK24 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >51934_51934_1_MAVS-STK24_MAVS_chr20_3845435_ENST00000416600_STK24_chr13_99171727_ENST00000397517_length(transcript)=5340nt_BP=1114nt GCTGCGGGAAGGGTCCTGGGCCCCGGGCGGCGGTCGCCAGGTCTCAGGGCCGGGGGTACCCGAGTCTCGTTTCCTCTCAGTCCATCCACC CTTCATGGGGCCAGAGCCCTCTCTCCAGAATCTGAGCAGCAATGCCGTTTGCTGAAGACAAGACCTATAAGTATATCTGCCGCAATTTCA GCAATTTTTGCAATGTGGATGTTGTAGAGATTCTGCCTTACCTGCCCTGCCTCACAGCAAGAGACCAGGGACCTCGGACCGTCCCCCAGA CCCACTGGAGCCACCGTCACTTCCTGCTGAGAGGCCAGGGCCCCCCACACCTGCTGCGGCCCACAGCATCCCCTACAACAGCTGCAGAGA GAAGGAGCCAAGTTACCCCATGCCTGTCCAGGAGACCCAGGCGCCAGAGTCCCCAGGAGAGAATTCAGAGCAAGCCCTGCAGACGCTCAG CCCCAGAGCCATCCCAAGGAATCCAGATGGTGGCCCCCTGGAGTCCTCCTCTGACCTGGCAGCCCTCAGCCCTCTGACCTCCAGCGGGCA TCAGGAGCAGGACACAGAACTGGGCAGTACCCACACAGCAGGTGCGACCTCCAGCCTCACACCATCCCGTGGGCCTGTGTCTCCATCTGT CTCCTTCCAGCCCCTGGCCCGTTCCACCCCCAGGGCAAGCCGCTTGCCTGGACCCACAGGGTCAGTTGTATCTACTGGCACCTCCTTCTC CTCCTCATCCCCTGGCTTGGCCTCTGCAGGGGCTGCAGAGGGTAAACAGGGTGCAGAGAGTGACCAGGCCGAGCCTATCATCTGCTCCAG TGGGGCAGAGGCACCTGCCAACTCTCTGCCCTCCAAAGTGCCTACCACCTTGATGCCTGTGAACACAGTGGCCCTGAAAGTGCCTGCCAA CCCAGCATCTGTCAGCACAGTGCCCTCCAAGTTGCCAACTAGCTCAAAGCCCCCTGGTGCAGTGCCTTCTAATGCGCTCACCAATCCAGC ACCATCCAAATTGCCCATCAACTCAACCCGTGCTGGCATGGTGCCATCCAAAGTGCCTACTAGCATGGTGCTCACCAAGGTGTCTGCCAG CACAGTCCCCACTGACGGGAGCAGCAGAAATGAGAACCTAAAGGCAGACCCAGAAGAGCTTTTTACAAAACTAGAGAAAATTGGGAAGGG CTCCTTTGGAGAGGTGTTCAAAGGCATTGACAATCGGACTCAGAAAGTGGTTGCCATAAAGATCATTGATCTGGAAGAAGCTGAAGATGA GATAGAGGACATTCAACAAGAAATCACAGTGCTGAGTCAGTGTGACAGTCCATATGTAACCAAATATTATGGATCCTATCTGAAGGATAC AAAATTATGGATAATAATGGAATATCTTGGTGGAGGCTCCGCACTAGATCTATTAGAACCTGGCCCATTAGATGAAACCCAGATCGCTAC TATATTAAGAGAAATACTGAAAGGACTCGATTATCTCCATTCGGAGAAGAAAATCCACAGAGACATTAAAGCGGCCAACGTCCTGCTGTC TGAGCATGGCGAGGTGAAGCTGGCGGACTTTGGCGTGGCTGGCCAGCTGACAGACACCCAGATCAAAAGGAACACCTTCGTGGGCACCCC ATTCTGGATGGCACCCGAGGTCATCAAACAGTCGGCCTATGACTCGAAGGCAGACATCTGGTCCCTGGGCATAACAGCTATTGAACTTGC AAGAGGGGAACCACCTCATTCCGAGCTGCACCCCATGAAAGTTTTATTCCTCATTCCAAAGAACAACCCACCGACGTTGGAAGGAAACTA CAGTAAACCCCTCAAGGAGTTTGTGGAGGCCTGTTTGAATAAGGAGCCGAGCTTTAGACCCACTGCTAAGGAGTTATTGAAGCACAAGTT TATACTACGCAATGCAAAGAAAACTTCCTACTTGACCGAGCTCATCGACAGGTACAAGAGATGGAAGGCCGAGCAGAGCCATGACGACTC GAGCTCCGAGGATTCCGACGCGGAAACAGATGGCCAAGCCTCGGGGGGCAGTGATTCTGGGGACTGGATCTTCACAATCCGAGAAAAAGA TCCCAAGAATCTCGAGAATGGAGCTCTTCAGCCATCGGACTTGGACAGAAATAAGATGAAAGACATCCCGAAGAGGCCTTTCTCTCAGTG TTTATCTACAATTATTTCTCCTCTGTTTGCAGAGTTGAAGGAGAAGAGCCAGGCGTGCGGAGGGAACTTGGGGTCCATTGAAGAGCTGCG AGGGGCCATCTACCTAGCGGAGGAGGCGTGCCCTGGCATCTCCGACACCATGGTGGCCCAGCTCGTGCAGCGGCTCCAGAGATACTCTCT AAGTGGTGGAGGAACTTCATCCCACTGAAATTCCTTTGGCATTTGGGGTTTTGTTTTTCCTTTTTTCCTTCTTCATCCTCCTCCTTTTTT AAAAGTCAACGAGAGCCTTCGCTGACTCCACCGAAGAGGTGCGCCACTGGGAGCCACCCCAGCGCCAGGCGCCCGTCCAGGGACACACAC AGTCTTCACTGTGCTGCAGCCAGATGAAGTCTCTCAGATGGGTGGGGAGGGTCAGCTCCTTCCAGCGATCATTTTATTTTATTTTATTAC TTTTGTTTTTAATTTTAACCATAGTGCACATATTCCAGGAAAGTGTCTTTAAAAACAAAAACAAACCCTGAAATGTATATTTGGGATTAT GATAAGGCAACTAAAGACATGAAACCTCAGGTATCCTGCTTTAAGTTGATAACTCCCTCTGGGAGCTGGAGAATCGCTCTGGTGGATGGG TGTACAGATTTGTATATAATGTCATTTTTACGGAAACCCTTTCGGCGTGCATAAGGAATCACTGTGTACAAACTGGCCAAGTGCTTCTGT AGATAACGTCAGTGGAGTAAATATTCGACAGGCCATAACTTGAGTCTATTGCCTTGCCTTTATTACATGTACATTTTGAATTCTGTGACC AGTGATTTGGGTTTTATTTTGTATTTGCAGGGTTTGTCATTAATAATTAATGCCCCTCTCTTACAGAACACTCCTATTTGTACCTCAACA AATGCAAATTTTCCCCGTTTGCCCTACGCCCCTTTTGGTACACCTAGAGGTTGATTTCCTTTTTCATCGATGGTACTATTTCTTAGTGTT TTAAATTGGAACATATCTTGCCTCATGAAGCTTTAAATTATAATTTTCAGTTTCTCCCCATGAAGCGCTCTCGTCTGACATTTGTTTGGA ATCGTGCCACTGCTGGTCTGCGCCAGATGTACCGTCCTTTCCAATACGATTTTCTGTTGCACCTTGTAGTGGATTCTGCATATCATCTTT CCCACCTAAAAATGTCTGAATGCTTACACAAATAAATTTTATAACACGCTTATTTTGCATACTCCTTGAAATGTGACTCTTCAGAGGACA GGGCACCTGCTGTGTATGTGTGGCCGTGCGTGTGTACTCGTGGCTGTGTGTGTGTGATGAGACACTTTGGAAGACTCCAGGGAGAAGTCC CCAGGCCTGGAGCTGCCGAGTGCCCAGGTCAGCGCCCTGGACTGCTTGCGCACTTGCTCACCGAGATGATGCAGTTGGAGGTTGCTGATC TGTGCGATTGCTGTAGCGGTTGCCGGGGACCTTAAGAGTTATTTTGCTTCTCTGGAAGGGGCCTATGCTTGCTAGGCAGGCAGCCAGTGT GTCTGTTTTTCTTGGTTTGCTGTGGGACCTTGCTTGGCGAGGGGGAAAATCTCTGGGTTTCTGGAGTGGGAGGGTTCGTGCAGCAGCTGT TGACTGGTACATGAAGCATTCTTTTATGTTTGTTGAAGCTGATGATTGACATCTCCCGTGGGTGTGCCAGTTCTTGTGGAGTTAAGACAG GATTTTTGGAAGCAAGGAAGTTAGTGGGTGAGCTTGGGGATGTAGCTCAGCTATCTGCTGGTCTAGTGGCCTCTAAGCTATAGGGAGGGG ACAGAGCCCTGAGCTACAGATGCTTGAGTGGGTTATTGTGTCGGTTTGCTAGTGCAGTCTGGTTTTTAAGCTCTAAAATTGAGGTATTTT ATTAGAAGTGGATTTGGGTTGAACTCTTAATTTGTATAAGGGATATATTTTGGTTGGGGAAATAGAACTGAGTTGCTAATTCTTATTGTA CTCATTACTCCATACAAGAATGTTATGTTGAATAATAAAATTGGAGAAGATTTCATTTTGTGTTTCCAGGGAGTATTCTGTGTGGGGAAC TGTTTCCTTACGTGAGGCCGGCGGCATAAGTCAAAGATGAGTTTTGTCCTTGCGAATCACACAGATTGAGTCTGTGTTCCCCAGGGTGTG CCGTTACCTGATTTTTAAGTGAGCCAGGGCGGACAGCAGCTTTTCTGATTTACAGAGTTCTTCAGATTTACAAATGGACAATGACATCAC AGTTTTTAGCACTGAAGCCAGTCTCATGCTAGTAACAGTGGGTGAGCCGCTCGAGGGACTGGGTTCTAATGAATACTGGTATGAACGGGG AGTCTCTGCAGTCGCCAGACAAATCATACTCAGCCCCTTCCCCCGTAGAGCAACAAGTGGTTCTTTTAGAGTTGACTGGCAGCATTTCCT GTCGGGGGAGGTGGGGTTTGATGGAGTTAGAAAGCTCGCCTCTGTGTACATTCTCTCCTGGGCTGTTACTTTCTGTAGACGCACAAAATC AGCCCCAATGTTTTTAAGGGCATCTTAGCCAAGGAAGCTGGCTTTTGTGTCGCCACTTCCAGGCCTGCATTAAGAGAGAGCCCAGGCACC AGGGCTACCACTGGAACCTGCCTCAGCGTCAACTGCTGCTGGTCTGTAGCCAGGCCCAGCCTTTGAGACGGGTTTACTGTCACCAGTAGC CTCTCAGTGCCAGCCCTGAGCTGCTCCTGGCTCAGCTGCCCAGAGCCTGCAGCCTGGGGAGGTACTCAGCCTCTGGGAGACGAGGGCCGT GGACTGGGTGGCTGGTAGCTCCTGCGTTTTTGAGCTGTGTCCTGGCTGGCTGCTGCCAATGAGGTGGACACCAGTGTGGTTTGGGGTGCA CTGGCCACTTCTTGCTGGGTTCTGATTTTCTTGGAAGTGCATCTGCCTTCCTTATCCAATAGTTTTATCCCTGCATTGCTCTTGTGAAGT GGCTGGTTTGGTTCTGTATGTAGCATTTTGTACCTTTCCTCTGGCAAAACACTGTCAGTTTATAAACATTTTTTATATTTCCCTCCTTTA AAAACAGCTTGTGTATTTCTGCTATAAAATGTGTCAGCAAAGGCAGAGTGACCTAATAGGGCATGTTCTTAAGCACAGGGACTGTATCAT >51934_51934_1_MAVS-STK24_MAVS_chr20_3845435_ENST00000416600_STK24_chr13_99171727_ENST00000397517_length(amino acids)=733AA_BP=316 MPQFQQFLQCGCCRDSALPALPHSKRPGTSDRPPDPLEPPSLPAERPGPPTPAAAHSIPYNSCREKEPSYPMPVQETQAPESPGENSEQA LQTLSPRAIPRNPDGGPLESSSDLAALSPLTSSGHQEQDTELGSTHTAGATSSLTPSRGPVSPSVSFQPLARSTPRASRLPGPTGSVVST GTSFSSSSPGLASAGAAEGKQGAESDQAEPIICSSGAEAPANSLPSKVPTTLMPVNTVALKVPANPASVSTVPSKLPTSSKPPGAVPSNA LTNPAPSKLPINSTRAGMVPSKVPTSMVLTKVSASTVPTDGSSRNENLKADPEELFTKLEKIGKGSFGEVFKGIDNRTQKVVAIKIIDLE EAEDEIEDIQQEITVLSQCDSPYVTKYYGSYLKDTKLWIIMEYLGGGSALDLLEPGPLDETQIATILREILKGLDYLHSEKKIHRDIKAA NVLLSEHGEVKLADFGVAGQLTDTQIKRNTFVGTPFWMAPEVIKQSAYDSKADIWSLGITAIELARGEPPHSELHPMKVLFLIPKNNPPT LEGNYSKPLKEFVEACLNKEPSFRPTAKELLKHKFILRNAKKTSYLTELIDRYKRWKAEQSHDDSSSEDSDAETDGQASGGSDSGDWIFT IREKDPKNLENGALQPSDLDRNKMKDIPKRPFSQCLSTIISPLFAELKEKSQACGGNLGSIEELRGAIYLAEEACPGISDTMVAQLVQRL -------------------------------------------------------------- >51934_51934_2_MAVS-STK24_MAVS_chr20_3845435_ENST00000416600_STK24_chr13_99171727_ENST00000539966_length(transcript)=2569nt_BP=1114nt GCTGCGGGAAGGGTCCTGGGCCCCGGGCGGCGGTCGCCAGGTCTCAGGGCCGGGGGTACCCGAGTCTCGTTTCCTCTCAGTCCATCCACC CTTCATGGGGCCAGAGCCCTCTCTCCAGAATCTGAGCAGCAATGCCGTTTGCTGAAGACAAGACCTATAAGTATATCTGCCGCAATTTCA GCAATTTTTGCAATGTGGATGTTGTAGAGATTCTGCCTTACCTGCCCTGCCTCACAGCAAGAGACCAGGGACCTCGGACCGTCCCCCAGA CCCACTGGAGCCACCGTCACTTCCTGCTGAGAGGCCAGGGCCCCCCACACCTGCTGCGGCCCACAGCATCCCCTACAACAGCTGCAGAGA GAAGGAGCCAAGTTACCCCATGCCTGTCCAGGAGACCCAGGCGCCAGAGTCCCCAGGAGAGAATTCAGAGCAAGCCCTGCAGACGCTCAG CCCCAGAGCCATCCCAAGGAATCCAGATGGTGGCCCCCTGGAGTCCTCCTCTGACCTGGCAGCCCTCAGCCCTCTGACCTCCAGCGGGCA TCAGGAGCAGGACACAGAACTGGGCAGTACCCACACAGCAGGTGCGACCTCCAGCCTCACACCATCCCGTGGGCCTGTGTCTCCATCTGT CTCCTTCCAGCCCCTGGCCCGTTCCACCCCCAGGGCAAGCCGCTTGCCTGGACCCACAGGGTCAGTTGTATCTACTGGCACCTCCTTCTC CTCCTCATCCCCTGGCTTGGCCTCTGCAGGGGCTGCAGAGGGTAAACAGGGTGCAGAGAGTGACCAGGCCGAGCCTATCATCTGCTCCAG TGGGGCAGAGGCACCTGCCAACTCTCTGCCCTCCAAAGTGCCTACCACCTTGATGCCTGTGAACACAGTGGCCCTGAAAGTGCCTGCCAA CCCAGCATCTGTCAGCACAGTGCCCTCCAAGTTGCCAACTAGCTCAAAGCCCCCTGGTGCAGTGCCTTCTAATGCGCTCACCAATCCAGC ACCATCCAAATTGCCCATCAACTCAACCCGTGCTGGCATGGTGCCATCCAAAGTGCCTACTAGCATGGTGCTCACCAAGGTGTCTGCCAG CACAGTCCCCACTGACGGGAGCAGCAGAAATGAGAACCTAAAGGCAGACCCAGAAGAGCTTTTTACAAAACTAGAGAAAATTGGGAAGGG CTCCTTTGGAGAGGTGTTCAAAGGCATTGACAATCGGACTCAGAAAGTGGTTGCCATAAAGATCATTGATCTGGAAGAAGCTGAAGATGA GATAGAGGACATTCAACAAGAAATCACAGTGCTGAGTCAGTGTGACAGTCCATATGTAACCAAATATTATGGATCCTATCTGAAGTTAGA ACCTGGCCCATTAGATGAAACCCAGATCGCTACTATATTAAGAGAAATACTGAAAGGACTCGATTATCTCCATTCGGAGAAGAAAATCCA CAGAGACATTAAAGCGGCCAACGTCCTGCTGTCTGAGCATGGCGAGGTGAAGCTGGCGGACTTTGGCGTGGCTGGCCAGCTGACAGACAC CCAGATCAAAAGGAACACCTTCGTGGGCACCCCATTCTGGATGGCACCCGAGGTCATCAAACAGTCGGCCTATGACTCGAAGGCAGACAT CTGGTCCCTGGGCATAACAGCTATTGAACTTGCAAGAGGGGAACCACCTCATTCCGAGCTGCACCCCATGAAAGTTTTATTCCTCATTCC AAAGAACAACCCACCGACGTTGGAAGGAAACTACAGTAAACCCCTCAAGGAGTTTGTGGAGGCCTGTTTGAATAAGGAGCCGAGCTTTAG ACCCACTGCTAAGGAGTTATTGAAGCACAAGTTTATACTACGCAATGCAAAGAAAACTTCCTACTTGACCGAGCTCATCGACAGGTACAA GAGATGGAAGGCCGAGCAGAGCCATGACGACTCGAGCTCCGAGGATTCCGACGCGGAAACAGATGGCCAAGCCTCGGGGGGCAGTGATTC TGGGGACTGGATCTTCACAATCCGAGAAAAAGATCCCAAGAATCTCGAGAATGGAGCTCTTCAGCCATCGGACTTGGACAGAAATAAGAT GAAAGACATCCCGAAGAGGCCTTTCTCTCAGTGTTTATCTACAATTATTTCTCCTCTGTTTGCAGAGTTGAAGGAGAAGAGCCAGGCGTG CGGAGGGAACTTGGGGTCCATTGAAGAGCTGCGAGGGGCCATCTACCTAGCGGAGGAGGCGTGCCCTGGCATCTCCGACACCATGGTGGC CCAGCTCGTGCAGCGGCTCCAGAGATACTCTCTAAGTGGTGGAGGAACTTCATCCCACTGAAATTCCTTTGGCATTTGGGGTTTTGTTTT TCCTTTTTTCCTTCTTCATCCTCCTCCTTTTTTAAAAGTCAACGAGAGCCTTCGCTGACTCCACCGAAGAGGTGCGCCACTGGGAGCCAC CCCAGCGCCAGGCGCCCGTCCAGGGACACACACAGTCTTCACTGTGCTGCAGCCAGATGAAGTCTCTCAGATGGGTGGGGAGGGTCAGCT >51934_51934_2_MAVS-STK24_MAVS_chr20_3845435_ENST00000416600_STK24_chr13_99171727_ENST00000539966_length(amino acids)=714AA_BP=316 MPQFQQFLQCGCCRDSALPALPHSKRPGTSDRPPDPLEPPSLPAERPGPPTPAAAHSIPYNSCREKEPSYPMPVQETQAPESPGENSEQA LQTLSPRAIPRNPDGGPLESSSDLAALSPLTSSGHQEQDTELGSTHTAGATSSLTPSRGPVSPSVSFQPLARSTPRASRLPGPTGSVVST GTSFSSSSPGLASAGAAEGKQGAESDQAEPIICSSGAEAPANSLPSKVPTTLMPVNTVALKVPANPASVSTVPSKLPTSSKPPGAVPSNA LTNPAPSKLPINSTRAGMVPSKVPTSMVLTKVSASTVPTDGSSRNENLKADPEELFTKLEKIGKGSFGEVFKGIDNRTQKVVAIKIIDLE EAEDEIEDIQQEITVLSQCDSPYVTKYYGSYLKLEPGPLDETQIATILREILKGLDYLHSEKKIHRDIKAANVLLSEHGEVKLADFGVAG QLTDTQIKRNTFVGTPFWMAPEVIKQSAYDSKADIWSLGITAIELARGEPPHSELHPMKVLFLIPKNNPPTLEGNYSKPLKEFVEACLNK EPSFRPTAKELLKHKFILRNAKKTSYLTELIDRYKRWKAEQSHDDSSSEDSDAETDGQASGGSDSGDWIFTIREKDPKNLENGALQPSDL -------------------------------------------------------------- >51934_51934_3_MAVS-STK24_MAVS_chr20_3845435_ENST00000428216_STK24_chr13_99171727_ENST00000397517_length(transcript)=5512nt_BP=1286nt GCGGGAAGGGTCCTGGGCCCCGGGCGGCGGTCGCCAGGTCTCAGGGCCGGGGGTACCCGAGTCTCGTTTCCTCTCAGTCCATCCACCCTT CATGGGGCCAGAGCCCTCTCTCCAGAATCTGAGCAGCAATGCCGTTTGCTGAAGACAAGACCTATAAGTATATCTGCCGCAATTTCAGCA ATTTTTGCAATGTGGATGTTGTAGAGATTCTGCCTTACCTGCCCTGCCTCACAGCAAGAGACCAGGATCGACTGCGGGCCACCTGCACAC TCTCAGGGAACCGGGACACCCTCTGGCATCTCTTCAATACCCTTCAGCGGCGGCCCGGCTGGGTGGAGTACTTCATTGCGGCACTGAGGG GCTGTGAGCTAGTTGATCTCGCGGACGAAGTGGCCTCTGTCTACCAGAGCTACCAGCCTCGGACCTCGGACCGTCCCCCAGACCCACTGG AGCCACCGTCACTTCCTGCTGAGAGGCCAGGGCCCCCCACACCTGCTGCGGCCCACAGCATCCCCTACAACAGCTGCAGAGAGAAGGAGC CAAGTTACCCCATGCCTGTCCAGGAGACCCAGGCGCCAGAGTCCCCAGGAGAGAATTCAGAGCAAGCCCTGCAGACGCTCAGCCCCAGAG CCATCCCAAGGAATCCAGATGGTGGCCCCCTGGAGTCCTCCTCTGACCTGGCAGCCCTCAGCCCTCTGACCTCCAGCGGGCATCAGGAGC AGGACACAGAACTGGGCAGTACCCACACAGCAGGTGCGACCTCCAGCCTCACACCATCCCGTGGGCCTGTGTCTCCATCTGTCTCCTTCC AGCCCCTGGCCCGTTCCACCCCCAGGGCAAGCCGCTTGCCTGGACCCACAGGGTCAGTTGTATCTACTGGCACCTCCTTCTCCTCCTCAT CCCCTGGCTTGGCCTCTGCAGGGGCTGCAGAGGGTAAACAGGGTGCAGAGAGTGACCAGGCCGAGCCTATCATCTGCTCCAGTGGGGCAG AGGCACCTGCCAACTCTCTGCCCTCCAAAGTGCCTACCACCTTGATGCCTGTGAACACAGTGGCCCTGAAAGTGCCTGCCAACCCAGCAT CTGTCAGCACAGTGCCCTCCAAGTTGCCAACTAGCTCAAAGCCCCCTGGTGCAGTGCCTTCTAATGCGCTCACCAATCCAGCACCATCCA AATTGCCCATCAACTCAACCCGTGCTGGCATGGTGCCATCCAAAGTGCCTACTAGCATGGTGCTCACCAAGGTGTCTGCCAGCACAGTCC CCACTGACGGGAGCAGCAGAAATGAGAACCTAAAGGCAGACCCAGAAGAGCTTTTTACAAAACTAGAGAAAATTGGGAAGGGCTCCTTTG GAGAGGTGTTCAAAGGCATTGACAATCGGACTCAGAAAGTGGTTGCCATAAAGATCATTGATCTGGAAGAAGCTGAAGATGAGATAGAGG ACATTCAACAAGAAATCACAGTGCTGAGTCAGTGTGACAGTCCATATGTAACCAAATATTATGGATCCTATCTGAAGGATACAAAATTAT GGATAATAATGGAATATCTTGGTGGAGGCTCCGCACTAGATCTATTAGAACCTGGCCCATTAGATGAAACCCAGATCGCTACTATATTAA GAGAAATACTGAAAGGACTCGATTATCTCCATTCGGAGAAGAAAATCCACAGAGACATTAAAGCGGCCAACGTCCTGCTGTCTGAGCATG GCGAGGTGAAGCTGGCGGACTTTGGCGTGGCTGGCCAGCTGACAGACACCCAGATCAAAAGGAACACCTTCGTGGGCACCCCATTCTGGA TGGCACCCGAGGTCATCAAACAGTCGGCCTATGACTCGAAGGCAGACATCTGGTCCCTGGGCATAACAGCTATTGAACTTGCAAGAGGGG AACCACCTCATTCCGAGCTGCACCCCATGAAAGTTTTATTCCTCATTCCAAAGAACAACCCACCGACGTTGGAAGGAAACTACAGTAAAC CCCTCAAGGAGTTTGTGGAGGCCTGTTTGAATAAGGAGCCGAGCTTTAGACCCACTGCTAAGGAGTTATTGAAGCACAAGTTTATACTAC GCAATGCAAAGAAAACTTCCTACTTGACCGAGCTCATCGACAGGTACAAGAGATGGAAGGCCGAGCAGAGCCATGACGACTCGAGCTCCG AGGATTCCGACGCGGAAACAGATGGCCAAGCCTCGGGGGGCAGTGATTCTGGGGACTGGATCTTCACAATCCGAGAAAAAGATCCCAAGA ATCTCGAGAATGGAGCTCTTCAGCCATCGGACTTGGACAGAAATAAGATGAAAGACATCCCGAAGAGGCCTTTCTCTCAGTGTTTATCTA CAATTATTTCTCCTCTGTTTGCAGAGTTGAAGGAGAAGAGCCAGGCGTGCGGAGGGAACTTGGGGTCCATTGAAGAGCTGCGAGGGGCCA TCTACCTAGCGGAGGAGGCGTGCCCTGGCATCTCCGACACCATGGTGGCCCAGCTCGTGCAGCGGCTCCAGAGATACTCTCTAAGTGGTG GAGGAACTTCATCCCACTGAAATTCCTTTGGCATTTGGGGTTTTGTTTTTCCTTTTTTCCTTCTTCATCCTCCTCCTTTTTTAAAAGTCA ACGAGAGCCTTCGCTGACTCCACCGAAGAGGTGCGCCACTGGGAGCCACCCCAGCGCCAGGCGCCCGTCCAGGGACACACACAGTCTTCA CTGTGCTGCAGCCAGATGAAGTCTCTCAGATGGGTGGGGAGGGTCAGCTCCTTCCAGCGATCATTTTATTTTATTTTATTACTTTTGTTT TTAATTTTAACCATAGTGCACATATTCCAGGAAAGTGTCTTTAAAAACAAAAACAAACCCTGAAATGTATATTTGGGATTATGATAAGGC AACTAAAGACATGAAACCTCAGGTATCCTGCTTTAAGTTGATAACTCCCTCTGGGAGCTGGAGAATCGCTCTGGTGGATGGGTGTACAGA TTTGTATATAATGTCATTTTTACGGAAACCCTTTCGGCGTGCATAAGGAATCACTGTGTACAAACTGGCCAAGTGCTTCTGTAGATAACG TCAGTGGAGTAAATATTCGACAGGCCATAACTTGAGTCTATTGCCTTGCCTTTATTACATGTACATTTTGAATTCTGTGACCAGTGATTT GGGTTTTATTTTGTATTTGCAGGGTTTGTCATTAATAATTAATGCCCCTCTCTTACAGAACACTCCTATTTGTACCTCAACAAATGCAAA TTTTCCCCGTTTGCCCTACGCCCCTTTTGGTACACCTAGAGGTTGATTTCCTTTTTCATCGATGGTACTATTTCTTAGTGTTTTAAATTG GAACATATCTTGCCTCATGAAGCTTTAAATTATAATTTTCAGTTTCTCCCCATGAAGCGCTCTCGTCTGACATTTGTTTGGAATCGTGCC ACTGCTGGTCTGCGCCAGATGTACCGTCCTTTCCAATACGATTTTCTGTTGCACCTTGTAGTGGATTCTGCATATCATCTTTCCCACCTA AAAATGTCTGAATGCTTACACAAATAAATTTTATAACACGCTTATTTTGCATACTCCTTGAAATGTGACTCTTCAGAGGACAGGGCACCT GCTGTGTATGTGTGGCCGTGCGTGTGTACTCGTGGCTGTGTGTGTGTGATGAGACACTTTGGAAGACTCCAGGGAGAAGTCCCCAGGCCT GGAGCTGCCGAGTGCCCAGGTCAGCGCCCTGGACTGCTTGCGCACTTGCTCACCGAGATGATGCAGTTGGAGGTTGCTGATCTGTGCGAT TGCTGTAGCGGTTGCCGGGGACCTTAAGAGTTATTTTGCTTCTCTGGAAGGGGCCTATGCTTGCTAGGCAGGCAGCCAGTGTGTCTGTTT TTCTTGGTTTGCTGTGGGACCTTGCTTGGCGAGGGGGAAAATCTCTGGGTTTCTGGAGTGGGAGGGTTCGTGCAGCAGCTGTTGACTGGT ACATGAAGCATTCTTTTATGTTTGTTGAAGCTGATGATTGACATCTCCCGTGGGTGTGCCAGTTCTTGTGGAGTTAAGACAGGATTTTTG GAAGCAAGGAAGTTAGTGGGTGAGCTTGGGGATGTAGCTCAGCTATCTGCTGGTCTAGTGGCCTCTAAGCTATAGGGAGGGGACAGAGCC CTGAGCTACAGATGCTTGAGTGGGTTATTGTGTCGGTTTGCTAGTGCAGTCTGGTTTTTAAGCTCTAAAATTGAGGTATTTTATTAGAAG TGGATTTGGGTTGAACTCTTAATTTGTATAAGGGATATATTTTGGTTGGGGAAATAGAACTGAGTTGCTAATTCTTATTGTACTCATTAC TCCATACAAGAATGTTATGTTGAATAATAAAATTGGAGAAGATTTCATTTTGTGTTTCCAGGGAGTATTCTGTGTGGGGAACTGTTTCCT TACGTGAGGCCGGCGGCATAAGTCAAAGATGAGTTTTGTCCTTGCGAATCACACAGATTGAGTCTGTGTTCCCCAGGGTGTGCCGTTACC TGATTTTTAAGTGAGCCAGGGCGGACAGCAGCTTTTCTGATTTACAGAGTTCTTCAGATTTACAAATGGACAATGACATCACAGTTTTTA GCACTGAAGCCAGTCTCATGCTAGTAACAGTGGGTGAGCCGCTCGAGGGACTGGGTTCTAATGAATACTGGTATGAACGGGGAGTCTCTG CAGTCGCCAGACAAATCATACTCAGCCCCTTCCCCCGTAGAGCAACAAGTGGTTCTTTTAGAGTTGACTGGCAGCATTTCCTGTCGGGGG AGGTGGGGTTTGATGGAGTTAGAAAGCTCGCCTCTGTGTACATTCTCTCCTGGGCTGTTACTTTCTGTAGACGCACAAAATCAGCCCCAA TGTTTTTAAGGGCATCTTAGCCAAGGAAGCTGGCTTTTGTGTCGCCACTTCCAGGCCTGCATTAAGAGAGAGCCCAGGCACCAGGGCTAC CACTGGAACCTGCCTCAGCGTCAACTGCTGCTGGTCTGTAGCCAGGCCCAGCCTTTGAGACGGGTTTACTGTCACCAGTAGCCTCTCAGT GCCAGCCCTGAGCTGCTCCTGGCTCAGCTGCCCAGAGCCTGCAGCCTGGGGAGGTACTCAGCCTCTGGGAGACGAGGGCCGTGGACTGGG TGGCTGGTAGCTCCTGCGTTTTTGAGCTGTGTCCTGGCTGGCTGCTGCCAATGAGGTGGACACCAGTGTGGTTTGGGGTGCACTGGCCAC TTCTTGCTGGGTTCTGATTTTCTTGGAAGTGCATCTGCCTTCCTTATCCAATAGTTTTATCCCTGCATTGCTCTTGTGAAGTGGCTGGTT TGGTTCTGTATGTAGCATTTTGTACCTTTCCTCTGGCAAAACACTGTCAGTTTATAAACATTTTTTATATTTCCCTCCTTTAAAAACAGC TTGTGTATTTCTGCTATAAAATGTGTCAGCAAAGGCAGAGTGACCTAATAGGGCATGTTCTTAAGCACAGGGACTGTATCATGCAGGGGC >51934_51934_3_MAVS-STK24_MAVS_chr20_3845435_ENST00000428216_STK24_chr13_99171727_ENST00000397517_length(amino acids)=803AA_BP=386 MPFAEDKTYKYICRNFSNFCNVDVVEILPYLPCLTARDQDRLRATCTLSGNRDTLWHLFNTLQRRPGWVEYFIAALRGCELVDLADEVAS VYQSYQPRTSDRPPDPLEPPSLPAERPGPPTPAAAHSIPYNSCREKEPSYPMPVQETQAPESPGENSEQALQTLSPRAIPRNPDGGPLES SSDLAALSPLTSSGHQEQDTELGSTHTAGATSSLTPSRGPVSPSVSFQPLARSTPRASRLPGPTGSVVSTGTSFSSSSPGLASAGAAEGK QGAESDQAEPIICSSGAEAPANSLPSKVPTTLMPVNTVALKVPANPASVSTVPSKLPTSSKPPGAVPSNALTNPAPSKLPINSTRAGMVP SKVPTSMVLTKVSASTVPTDGSSRNENLKADPEELFTKLEKIGKGSFGEVFKGIDNRTQKVVAIKIIDLEEAEDEIEDIQQEITVLSQCD SPYVTKYYGSYLKDTKLWIIMEYLGGGSALDLLEPGPLDETQIATILREILKGLDYLHSEKKIHRDIKAANVLLSEHGEVKLADFGVAGQ LTDTQIKRNTFVGTPFWMAPEVIKQSAYDSKADIWSLGITAIELARGEPPHSELHPMKVLFLIPKNNPPTLEGNYSKPLKEFVEACLNKE PSFRPTAKELLKHKFILRNAKKTSYLTELIDRYKRWKAEQSHDDSSSEDSDAETDGQASGGSDSGDWIFTIREKDPKNLENGALQPSDLD -------------------------------------------------------------- >51934_51934_4_MAVS-STK24_MAVS_chr20_3845435_ENST00000428216_STK24_chr13_99171727_ENST00000539966_length(transcript)=2741nt_BP=1286nt GCGGGAAGGGTCCTGGGCCCCGGGCGGCGGTCGCCAGGTCTCAGGGCCGGGGGTACCCGAGTCTCGTTTCCTCTCAGTCCATCCACCCTT CATGGGGCCAGAGCCCTCTCTCCAGAATCTGAGCAGCAATGCCGTTTGCTGAAGACAAGACCTATAAGTATATCTGCCGCAATTTCAGCA ATTTTTGCAATGTGGATGTTGTAGAGATTCTGCCTTACCTGCCCTGCCTCACAGCAAGAGACCAGGATCGACTGCGGGCCACCTGCACAC TCTCAGGGAACCGGGACACCCTCTGGCATCTCTTCAATACCCTTCAGCGGCGGCCCGGCTGGGTGGAGTACTTCATTGCGGCACTGAGGG GCTGTGAGCTAGTTGATCTCGCGGACGAAGTGGCCTCTGTCTACCAGAGCTACCAGCCTCGGACCTCGGACCGTCCCCCAGACCCACTGG AGCCACCGTCACTTCCTGCTGAGAGGCCAGGGCCCCCCACACCTGCTGCGGCCCACAGCATCCCCTACAACAGCTGCAGAGAGAAGGAGC CAAGTTACCCCATGCCTGTCCAGGAGACCCAGGCGCCAGAGTCCCCAGGAGAGAATTCAGAGCAAGCCCTGCAGACGCTCAGCCCCAGAG CCATCCCAAGGAATCCAGATGGTGGCCCCCTGGAGTCCTCCTCTGACCTGGCAGCCCTCAGCCCTCTGACCTCCAGCGGGCATCAGGAGC AGGACACAGAACTGGGCAGTACCCACACAGCAGGTGCGACCTCCAGCCTCACACCATCCCGTGGGCCTGTGTCTCCATCTGTCTCCTTCC AGCCCCTGGCCCGTTCCACCCCCAGGGCAAGCCGCTTGCCTGGACCCACAGGGTCAGTTGTATCTACTGGCACCTCCTTCTCCTCCTCAT CCCCTGGCTTGGCCTCTGCAGGGGCTGCAGAGGGTAAACAGGGTGCAGAGAGTGACCAGGCCGAGCCTATCATCTGCTCCAGTGGGGCAG AGGCACCTGCCAACTCTCTGCCCTCCAAAGTGCCTACCACCTTGATGCCTGTGAACACAGTGGCCCTGAAAGTGCCTGCCAACCCAGCAT CTGTCAGCACAGTGCCCTCCAAGTTGCCAACTAGCTCAAAGCCCCCTGGTGCAGTGCCTTCTAATGCGCTCACCAATCCAGCACCATCCA AATTGCCCATCAACTCAACCCGTGCTGGCATGGTGCCATCCAAAGTGCCTACTAGCATGGTGCTCACCAAGGTGTCTGCCAGCACAGTCC CCACTGACGGGAGCAGCAGAAATGAGAACCTAAAGGCAGACCCAGAAGAGCTTTTTACAAAACTAGAGAAAATTGGGAAGGGCTCCTTTG GAGAGGTGTTCAAAGGCATTGACAATCGGACTCAGAAAGTGGTTGCCATAAAGATCATTGATCTGGAAGAAGCTGAAGATGAGATAGAGG ACATTCAACAAGAAATCACAGTGCTGAGTCAGTGTGACAGTCCATATGTAACCAAATATTATGGATCCTATCTGAAGTTAGAACCTGGCC CATTAGATGAAACCCAGATCGCTACTATATTAAGAGAAATACTGAAAGGACTCGATTATCTCCATTCGGAGAAGAAAATCCACAGAGACA TTAAAGCGGCCAACGTCCTGCTGTCTGAGCATGGCGAGGTGAAGCTGGCGGACTTTGGCGTGGCTGGCCAGCTGACAGACACCCAGATCA AAAGGAACACCTTCGTGGGCACCCCATTCTGGATGGCACCCGAGGTCATCAAACAGTCGGCCTATGACTCGAAGGCAGACATCTGGTCCC TGGGCATAACAGCTATTGAACTTGCAAGAGGGGAACCACCTCATTCCGAGCTGCACCCCATGAAAGTTTTATTCCTCATTCCAAAGAACA ACCCACCGACGTTGGAAGGAAACTACAGTAAACCCCTCAAGGAGTTTGTGGAGGCCTGTTTGAATAAGGAGCCGAGCTTTAGACCCACTG CTAAGGAGTTATTGAAGCACAAGTTTATACTACGCAATGCAAAGAAAACTTCCTACTTGACCGAGCTCATCGACAGGTACAAGAGATGGA AGGCCGAGCAGAGCCATGACGACTCGAGCTCCGAGGATTCCGACGCGGAAACAGATGGCCAAGCCTCGGGGGGCAGTGATTCTGGGGACT GGATCTTCACAATCCGAGAAAAAGATCCCAAGAATCTCGAGAATGGAGCTCTTCAGCCATCGGACTTGGACAGAAATAAGATGAAAGACA TCCCGAAGAGGCCTTTCTCTCAGTGTTTATCTACAATTATTTCTCCTCTGTTTGCAGAGTTGAAGGAGAAGAGCCAGGCGTGCGGAGGGA ACTTGGGGTCCATTGAAGAGCTGCGAGGGGCCATCTACCTAGCGGAGGAGGCGTGCCCTGGCATCTCCGACACCATGGTGGCCCAGCTCG TGCAGCGGCTCCAGAGATACTCTCTAAGTGGTGGAGGAACTTCATCCCACTGAAATTCCTTTGGCATTTGGGGTTTTGTTTTTCCTTTTT TCCTTCTTCATCCTCCTCCTTTTTTAAAAGTCAACGAGAGCCTTCGCTGACTCCACCGAAGAGGTGCGCCACTGGGAGCCACCCCAGCGC CAGGCGCCCGTCCAGGGACACACACAGTCTTCACTGTGCTGCAGCCAGATGAAGTCTCTCAGATGGGTGGGGAGGGTCAGCTCCTTCCAG >51934_51934_4_MAVS-STK24_MAVS_chr20_3845435_ENST00000428216_STK24_chr13_99171727_ENST00000539966_length(amino acids)=784AA_BP=386 MPFAEDKTYKYICRNFSNFCNVDVVEILPYLPCLTARDQDRLRATCTLSGNRDTLWHLFNTLQRRPGWVEYFIAALRGCELVDLADEVAS VYQSYQPRTSDRPPDPLEPPSLPAERPGPPTPAAAHSIPYNSCREKEPSYPMPVQETQAPESPGENSEQALQTLSPRAIPRNPDGGPLES SSDLAALSPLTSSGHQEQDTELGSTHTAGATSSLTPSRGPVSPSVSFQPLARSTPRASRLPGPTGSVVSTGTSFSSSSPGLASAGAAEGK QGAESDQAEPIICSSGAEAPANSLPSKVPTTLMPVNTVALKVPANPASVSTVPSKLPTSSKPPGAVPSNALTNPAPSKLPINSTRAGMVP SKVPTSMVLTKVSASTVPTDGSSRNENLKADPEELFTKLEKIGKGSFGEVFKGIDNRTQKVVAIKIIDLEEAEDEIEDIQQEITVLSQCD SPYVTKYYGSYLKLEPGPLDETQIATILREILKGLDYLHSEKKIHRDIKAANVLLSEHGEVKLADFGVAGQLTDTQIKRNTFVGTPFWMA PEVIKQSAYDSKADIWSLGITAIELARGEPPHSELHPMKVLFLIPKNNPPTLEGNYSKPLKEFVEACLNKEPSFRPTAKELLKHKFILRN AKKTSYLTELIDRYKRWKAEQSHDDSSSEDSDAETDGQASGGSDSGDWIFTIREKDPKNLENGALQPSDLDRNKMKDIPKRPFSQCLSTI -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for MAVS-STK24 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Hgene | MAVS | chr20:3845435 | chr13:99171727 | ENST00000358134 | + | 4 | 5 | 143_147 | 328.3333333333333 | 360.3333333333333 | TRAF2 |
| Hgene | MAVS | chr20:3845435 | chr13:99171727 | ENST00000416600 | + | 5 | 6 | 143_147 | 245.0 | 400.0 | TRAF2 |
| Hgene | MAVS | chr20:3845435 | chr13:99171727 | ENST00000428216 | + | 6 | 7 | 143_147 | 386.0 | 541.0 | TRAF2 |
| Hgene | MAVS | chr20:3845435 | chr13:99171727 | ENST00000358134 | + | 4 | 5 | 153_158 | 328.3333333333333 | 360.3333333333333 | TRAF6 |
| Hgene | MAVS | chr20:3845435 | chr13:99171727 | ENST00000416600 | + | 5 | 6 | 153_158 | 245.0 | 400.0 | TRAF6 |
| Hgene | MAVS | chr20:3845435 | chr13:99171727 | ENST00000428216 | + | 6 | 7 | 153_158 | 386.0 | 541.0 | TRAF6 |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | MAVS | chr20:3845435 | chr13:99171727 | ENST00000358134 | + | 4 | 5 | 455_460 | 328.3333333333333 | 360.3333333333333 | TRAF6 |
| Hgene | MAVS | chr20:3845435 | chr13:99171727 | ENST00000416600 | + | 5 | 6 | 455_460 | 245.0 | 400.0 | TRAF6 |
| Hgene | MAVS | chr20:3845435 | chr13:99171727 | ENST00000428216 | + | 6 | 7 | 455_460 | 386.0 | 541.0 | TRAF6 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for MAVS-STK24 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for MAVS-STK24 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
