
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:MCF2L2-DMRT1 (FusionGDB2 ID:52171) |
Fusion Gene Summary for MCF2L2-DMRT1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: MCF2L2-DMRT1 | Fusion gene ID: 52171 | Hgene | Tgene | Gene symbol | MCF2L2 | DMRT1 | Gene ID | 23101 | 1761 |
| Gene name | MCF.2 cell line derived transforming sequence-like 2 | doublesex and mab-3 related transcription factor 1 | |
| Synonyms | ARHGEF22 | CT154|DMT1 | |
| Cytomap | 3q27.1 | 9p24.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | probable guanine nucleotide exchange factor MCF2L2MCF2-transforming sequence-like protein 2dbs-related Rho family guanine nucleotide exchange factor | doublesex- and mab-3-related transcription factor 1DM domain expressed in testis 1DM domain expressed in testis protein 1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q86YR7 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000328913, ENST00000414362, ENST00000447025, ENST00000473233, ENST00000468976, | ENST00000382276, ENST00000569227, | |
| Fusion gene scores | * DoF score | 5 X 6 X 3=90 | 3 X 5 X 3=45 |
| # samples | 4 | 5 | |
| ** MAII score | log2(4/90*10)=-1.16992500144231 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/45*10)=0.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: MCF2L2 [Title/Abstract] AND DMRT1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | MCF2L2(182994660)-DMRT1(967985), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | MCF2L2-DMRT1 seems lost the major protein functional domain in Tgene partner, which is a transcription factor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across MCF2L2 (5'-gene) Fusion gene breakpoints across MCF2L2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
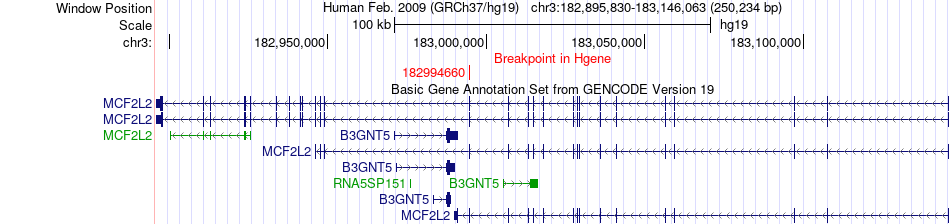 |
 Fusion gene breakpoints across DMRT1 (3'-gene) Fusion gene breakpoints across DMRT1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
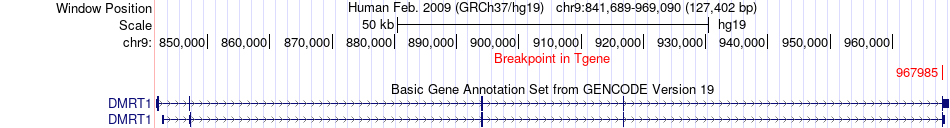 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SKCM | TCGA-EE-A29H-06A | MCF2L2 | chr3 | 182994660 | - | DMRT1 | chr9 | 967985 | + |
Top |
Fusion Gene ORF analysis for MCF2L2-DMRT1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| Frame-shift | ENST00000328913 | ENST00000382276 | MCF2L2 | chr3 | 182994660 | - | DMRT1 | chr9 | 967985 | + |
| Frame-shift | ENST00000414362 | ENST00000382276 | MCF2L2 | chr3 | 182994660 | - | DMRT1 | chr9 | 967985 | + |
| Frame-shift | ENST00000447025 | ENST00000382276 | MCF2L2 | chr3 | 182994660 | - | DMRT1 | chr9 | 967985 | + |
| Frame-shift | ENST00000473233 | ENST00000382276 | MCF2L2 | chr3 | 182994660 | - | DMRT1 | chr9 | 967985 | + |
| In-frame | ENST00000328913 | ENST00000569227 | MCF2L2 | chr3 | 182994660 | - | DMRT1 | chr9 | 967985 | + |
| In-frame | ENST00000414362 | ENST00000569227 | MCF2L2 | chr3 | 182994660 | - | DMRT1 | chr9 | 967985 | + |
| In-frame | ENST00000447025 | ENST00000569227 | MCF2L2 | chr3 | 182994660 | - | DMRT1 | chr9 | 967985 | + |
| In-frame | ENST00000473233 | ENST00000569227 | MCF2L2 | chr3 | 182994660 | - | DMRT1 | chr9 | 967985 | + |
| intron-3CDS | ENST00000468976 | ENST00000382276 | MCF2L2 | chr3 | 182994660 | - | DMRT1 | chr9 | 967985 | + |
| intron-3CDS | ENST00000468976 | ENST00000569227 | MCF2L2 | chr3 | 182994660 | - | DMRT1 | chr9 | 967985 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000328913 | MCF2L2 | chr3 | 182994660 | - | ENST00000569227 | DMRT1 | chr9 | 967985 | + | 2691 | 2160 | 70 | 2223 | 717 |
| ENST00000473233 | MCF2L2 | chr3 | 182994660 | - | ENST00000569227 | DMRT1 | chr9 | 967985 | + | 2393 | 1862 | 0 | 1925 | 641 |
| ENST00000447025 | MCF2L2 | chr3 | 182994660 | - | ENST00000569227 | DMRT1 | chr9 | 967985 | + | 2501 | 1970 | 18 | 2033 | 671 |
| ENST00000414362 | MCF2L2 | chr3 | 182994660 | - | ENST00000569227 | DMRT1 | chr9 | 967985 | + | 2517 | 1986 | 34 | 2049 | 671 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000328913 | ENST00000569227 | MCF2L2 | chr3 | 182994660 | - | DMRT1 | chr9 | 967985 | + | 0.006610779 | 0.99338925 |
| ENST00000473233 | ENST00000569227 | MCF2L2 | chr3 | 182994660 | - | DMRT1 | chr9 | 967985 | + | 0.006363613 | 0.99363637 |
| ENST00000447025 | ENST00000569227 | MCF2L2 | chr3 | 182994660 | - | DMRT1 | chr9 | 967985 | + | 0.006383888 | 0.9936161 |
| ENST00000414362 | ENST00000569227 | MCF2L2 | chr3 | 182994660 | - | DMRT1 | chr9 | 967985 | + | 0.006478539 | 0.9935215 |
Top |
Fusion Genomic Features for MCF2L2-DMRT1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
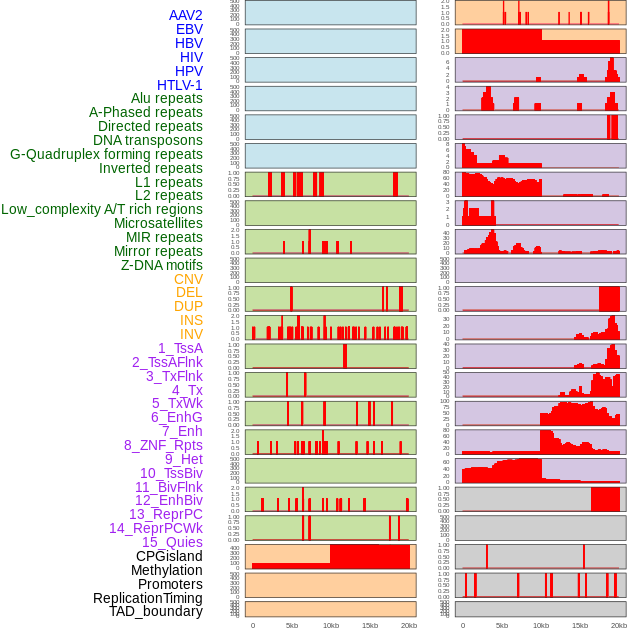 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
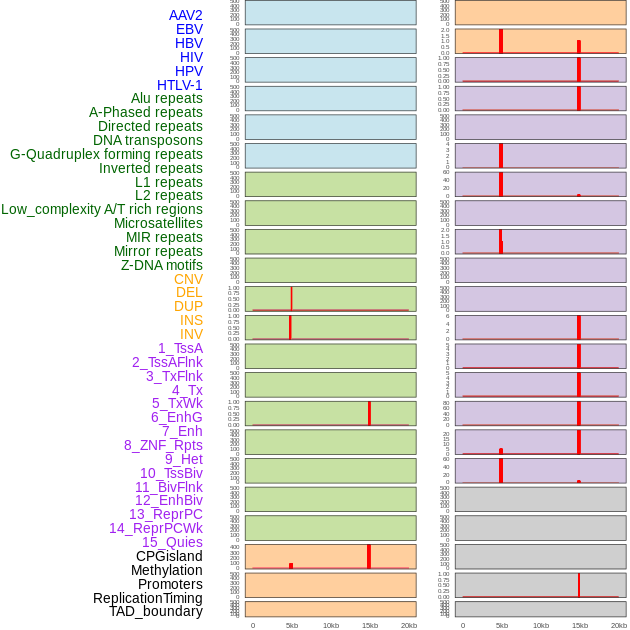 |
Top |
Fusion Protein Features for MCF2L2-DMRT1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:182994660/chr9:967985) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MCF2L2 | . |
| FUNCTION: Probably functions as a guanine nucleotide exchange factor. {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MCF2L2 | chr3:182994660 | chr9:967985 | ENST00000328913 | - | 15 | 30 | 11_193 | 620 | 1115.0 | Domain | CRAL-TRIO |
| Hgene | MCF2L2 | chr3:182994660 | chr9:967985 | ENST00000414362 | - | 15 | 16 | 11_193 | 620 | 629.0 | Domain | CRAL-TRIO |
| Hgene | MCF2L2 | chr3:182994660 | chr9:967985 | ENST00000447025 | - | 15 | 18 | 11_193 | 620 | 712.0 | Domain | CRAL-TRIO |
| Hgene | MCF2L2 | chr3:182994660 | chr9:967985 | ENST00000473233 | - | 15 | 29 | 11_193 | 620 | 1075.0 | Domain | CRAL-TRIO |
| Hgene | MCF2L2 | chr3:182994660 | chr9:967985 | ENST00000328913 | - | 15 | 30 | 323_428 | 620 | 1115.0 | Repeat | Note=Spectrin |
| Hgene | MCF2L2 | chr3:182994660 | chr9:967985 | ENST00000414362 | - | 15 | 16 | 323_428 | 620 | 629.0 | Repeat | Note=Spectrin |
| Hgene | MCF2L2 | chr3:182994660 | chr9:967985 | ENST00000447025 | - | 15 | 18 | 323_428 | 620 | 712.0 | Repeat | Note=Spectrin |
| Hgene | MCF2L2 | chr3:182994660 | chr9:967985 | ENST00000473233 | - | 15 | 29 | 323_428 | 620 | 1075.0 | Repeat | Note=Spectrin |
| Tgene | DMRT1 | chr3:182994660 | chr9:967985 | ENST00000382276 | 3 | 5 | 322_347 | 322 | 374.0 | Compositional bias | Note=Pro/Ser-rich |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MCF2L2 | chr3:182994660 | chr9:967985 | ENST00000328913 | - | 15 | 30 | 619_822 | 620 | 1115.0 | Domain | DH |
| Hgene | MCF2L2 | chr3:182994660 | chr9:967985 | ENST00000328913 | - | 15 | 30 | 834_954 | 620 | 1115.0 | Domain | PH |
| Hgene | MCF2L2 | chr3:182994660 | chr9:967985 | ENST00000414362 | - | 15 | 16 | 619_822 | 620 | 629.0 | Domain | DH |
| Hgene | MCF2L2 | chr3:182994660 | chr9:967985 | ENST00000414362 | - | 15 | 16 | 834_954 | 620 | 629.0 | Domain | PH |
| Hgene | MCF2L2 | chr3:182994660 | chr9:967985 | ENST00000447025 | - | 15 | 18 | 619_822 | 620 | 712.0 | Domain | DH |
| Hgene | MCF2L2 | chr3:182994660 | chr9:967985 | ENST00000447025 | - | 15 | 18 | 834_954 | 620 | 712.0 | Domain | PH |
| Hgene | MCF2L2 | chr3:182994660 | chr9:967985 | ENST00000473233 | - | 15 | 29 | 619_822 | 620 | 1075.0 | Domain | DH |
| Hgene | MCF2L2 | chr3:182994660 | chr9:967985 | ENST00000473233 | - | 15 | 29 | 834_954 | 620 | 1075.0 | Domain | PH |
| Tgene | DMRT1 | chr3:182994660 | chr9:967985 | ENST00000382276 | 3 | 5 | 72_118 | 322 | 374.0 | DNA binding | DM |
Top |
Fusion Gene Sequence for MCF2L2-DMRT1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >52171_52171_1_MCF2L2-DMRT1_MCF2L2_chr3_182994660_ENST00000328913_DMRT1_chr9_967985_ENST00000569227_length(transcript)=2691nt_BP=2160nt CGGGGCGCGGTTCCGGCGAGATTGGCGACTTGCTTCCTTTTGCAGAACGGCCGCTCCTCTCTCGGTCCAGCTGCGACGCTGGGAAAGCGC CAGACTTGGTCTGTCACGGGAGCCCCCGCGGGGCGTGGCGAGGGAGAGCTGGCCTAGGCTGCGAGGGACTCTCGTTTCTTGCCTGGAGAG CCAGGAAGAGGGCGAGGGCAGAGCATCCTTGGGCGGAGATGCCTTTAAAAAATCATCCACCGCAGCGGTAGAAACAGTTTTGTTTGGCTT TATTTATACGGAATGGTTTTTCAGTGAAATGCTGTCTTGCTTAAAAGAAGAGATGCCTCCCCAGGAGCTCACCCGGCGACTGGCCACAGT GATCACTCATGTCGATGAAATTATGCAGCAGGAAGTCAGACCCCTGATGGCGGTGGAGATAATAGAACAACTTCACAGACAATTTGCCAT TCTTTCAGGAGGCCGAGGGGAGGATGGCGCCCCCATCATCACGTTCCCAGAGTTTTCGGGGTTCAAACACATCCCAGATGAAGACTTCCT GAATGTCATGACCTACCTGACTAGCATCCCCAGTGTGGAGGCTGCCAGCATTGGATTCATTGTTGTTATCGACAGACGAAGAGACAAGTG GAGCTCCGTAAAGGCATCCTTGACACGAATAGCTGTGGCATTTCCAGGAAACTTACAGCTCATATTCATCCTTCGTCCATCTCGCTTTAT CCAGAGGACATTCACTGACATTGGCATTAAATACTATCGAAATGAGTTTAAAACGAAAGTGCCGATCATCATGGTAAACTCTGTCTCTGA CCTTCACGGCTACATCGACAAAAGCCAACTGACCCGGGAATTAGGGGGGACTTTGGAATATCGCCACGGTCAGTGGGTAAATCACCGCAC TGCCATCGAAAACTTTGCCTTGACCTTGAAGACCACTGCCCAGATGCTGCAGACGTTTGGGTCCTGCCTGGCCACAGCAGAGCTGCCCAG AAGCATGCTATCCACGGAAGACCTTCTCATGTCCCACACAAGGCAGCGGGACAAGCTGCAGGATGAGCTGAAATTACTTGGAAAGCAGGG GACCACATTGCTGTCATGCATCCAAGAACCAGCAACCAAATGTCCCAACAGCAAACTCAATCTCAACCAACTTGAGAATGTAACTACCAT GGAAAGGTTATTAGTTCAACTGGATGAAACAGAAAAAGCCTTTAGTCACTTTTGGTCTGAGCATCATCTGAAGCTTAACCAGTGCCTACA ACTACAGCATTTTGAGCACGATTTTTGTAAGGCTAAGCTTGCCCTGGATAATTTGCTGGAAGAGCAAGCAGAGTTTACAGGCATTGGAGA CAGCGTGATGCACGTGGAGCAGATTCTTAAGGAACACAAAAAACTGGAGGAAAAAAGCCAGGAGCCCCTGGAAAAGGCCCAGCTGCTGGC ACTGGTTGGGGACCAGCTCATCCAAAGCCACCATTATGCAGCAGATGCCATCAGGCCCCGGTGTGTGGAGCTCAGGCACCTCTGTGACGA TTTCATCAATGGAAACAAGAAAAAATGGGACATTTTAGGAAAGTCCTTAGAGTTTCATAGACAGCTGGACAAGGTCAGCCAATGGTGTGA GGCAGGAATCTACCTCTTGGCTTCCCAAGCTGTAGACAAGTGCCAGTCTCGAGAAGGGGTTGATATCGCCTTGAACGACATTGCGACATT CCTGGGCACAGTCAAGGAGTACCCGTTGCTCAGCCCCAAGGAGTTTTACAACGAGTTTGAGTTGCTGCTCACCCTCGATGCAAAGGCCAA AGCCCAGAAAGTTTTGCAGAGGCTGGATGATGTCCAGGAAATATTTCACAAGAGGCAAGTGAGTCTGATGAAACTGGCAGCCAAACAGAC TCGTCCAGTGCAACCTGTGGCCCCACATCCTGAGTCTTCACCAAAATGGGTGTCATCAAAAACCAGCCAGCCCTCCACCTCGGTCCCTCT AGCTCGTCCTCTGAGAACGTCTGAGGAACCTTATACGGAGACAGAGTTGAACTCCCGGGGAAAGGAAGATGATGAGACTAAATTTGAAGT CAAGAGTGAAGAAATCTTTGAAAGCCATCATGAAAGGGGGAACCCTGAGCTGGAGCAGCAGGCCAGGCTCGGAGACCTTTCCCCCCGCAG TATTCTCGCCGCCCAGCAGTCAAGATTCTGGCTTGGTTTCCCTCTCGAGCAGCTCTCCTATTAGTAACAAGAGCACAAAGGCAGTGCTTG AATGTGAGCCTGCGTCGGAGCCCAGCAGCTTCACAGTCACTCCCGTCATCGAGGAGGACGAGTGAGCAGTGCCTGCTGCCGATGGCGGTT CACTTGGAGTAACAGGCTTATTCCACTTTCCATGGGGTTTGTTAATATTTTGCATTGACTCATACTATCTTAACTGTTGAGAACGTATTT GGTTTATATTCCTTAGAGTTTAGTCCAGAGGCTGTAACACATTTGTAATACTTTAGGGTCCGTGACTACCATCTGCATGGTTTAAGTGCT TTACTCACGGAGTTTAAATAATAGTGTTCATTTTTTTAATGACACTGGTTTCATGTAGTTTTCAAGAAATAAAAGAATTCATTCAAGTGA >52171_52171_1_MCF2L2-DMRT1_MCF2L2_chr3_182994660_ENST00000328913_DMRT1_chr9_967985_ENST00000569227_length(amino acids)=717AA_BP=693 MRRWESARLGLSREPPRGVARESWPRLRGTLVSCLESQEEGEGRASLGGDAFKKSSTAAVETVLFGFIYTEWFFSEMLSCLKEEMPPQEL TRRLATVITHVDEIMQQEVRPLMAVEIIEQLHRQFAILSGGRGEDGAPIITFPEFSGFKHIPDEDFLNVMTYLTSIPSVEAASIGFIVVI DRRRDKWSSVKASLTRIAVAFPGNLQLIFILRPSRFIQRTFTDIGIKYYRNEFKTKVPIIMVNSVSDLHGYIDKSQLTRELGGTLEYRHG QWVNHRTAIENFALTLKTTAQMLQTFGSCLATAELPRSMLSTEDLLMSHTRQRDKLQDELKLLGKQGTTLLSCIQEPATKCPNSKLNLNQ LENVTTMERLLVQLDETEKAFSHFWSEHHLKLNQCLQLQHFEHDFCKAKLALDNLLEEQAEFTGIGDSVMHVEQILKEHKKLEEKSQEPL EKAQLLALVGDQLIQSHHYAADAIRPRCVELRHLCDDFINGNKKKWDILGKSLEFHRQLDKVSQWCEAGIYLLASQAVDKCQSREGVDIA LNDIATFLGTVKEYPLLSPKEFYNEFELLLTLDAKAKAQKVLQRLDDVQEIFHKRQVSLMKLAAKQTRPVQPVAPHPESSPKWVSSKTSQ -------------------------------------------------------------- >52171_52171_2_MCF2L2-DMRT1_MCF2L2_chr3_182994660_ENST00000414362_DMRT1_chr9_967985_ENST00000569227_length(transcript)=2517nt_BP=1986nt GGAGAGCCAGGAAGAGGGCGAGGGCAGAGCATCCTTGGGCGGAGATGCCTTTAAAAAATCATCCACCGCAGCGGTAGAAACAGTTTTGTT TGGCTTTATTTATACGGAATGGTTTTTCAGTGAAATGCTGTCTTGCTTAAAAGAAGAGATGCCTCCCCAGGAGCTCACCCGGCGACTGGC CACAGTGATCACTCATGTCGATGAAATTATGCAGCAGGAAGTCAGACCCCTGATGGCGGTGGAGATAATAGAACAACTTCACAGACAATT TGCCATTCTTTCAGGAGGCCGAGGGGAGGATGGCGCCCCCATCATCACGTTCCCAGAGTTTTCGGGGTTCAAACACATCCCAGATGAAGA CTTCCTGAATGTCATGACCTACCTGACTAGCATCCCCAGTGTGGAGGCTGCCAGCATTGGATTCATTGTTGTTATCGACAGACGAAGAGA CAAGTGGAGCTCCGTAAAGGCATCCTTGACACGAATAGCTGTGGCATTTCCAGGAAACTTACAGCTCATATTCATCCTTCGTCCATCTCG CTTTATCCAGAGGACATTCACTGACATTGGCATTAAATACTATCGAAATGAGTTTAAAACGAAAGTGCCGATCATCATGGTAAACTCTGT CTCTGACCTTCACGGCTACATCGACAAAAGCCAACTGACCCGGGAATTAGGGGGGACTTTGGAATATCGCCACGGTCAGTGGGTAAATCA CCGCACTGCCATCGAAAACTTTGCCTTGACCTTGAAGACCACTGCCCAGATGCTGCAGACGTTTGGGTCCTGCCTGGCCACAGCAGAGCT GCCCAGAAGCATGCTATCCACGGAAGACCTTCTCATGTCCCACACAAGGCAGCGGGACAAGCTGCAGGATGAGCTGAAATTACTTGGAAA GCAGGGGACCACATTGCTGTCATGCATCCAAGAACCAGCAACCAAATGTCCCAACAGCAAACTCAATCTCAACCAACTTGAGAATGTAAC TACCATGGAAAGGTTATTAGTTCAACTGGATGAAACAGAAAAAGCCTTTAGTCACTTTTGGTCTGAGCATCATCTGAAGCTTAACCAGTG CCTACAACTACAGCATTTTGAGCACGATTTTTGTAAGGCTAAGCTTGCCCTGGATAATTTGCTGGAAGAGCAAGCAGAGTTTACAGGCAT TGGAGACAGCGTGATGCACGTGGAGCAGATTCTTAAGGAACACAAAAAACTGGAGGAAAAAAGCCAGGAGCCCCTGGAAAAGGCCCAGCT GCTGGCACTGGTTGGGGACCAGCTCATCCAAAGCCACCATTATGCAGCAGATGCCATCAGGCCCCGGTGTGTGGAGCTCAGGCACCTCTG TGACGATTTCATCAATGGAAACAAGAAAAAATGGGACATTTTAGGAAAGTCCTTAGAGTTTCATAGACAGCTGGACAAGGTCAGCCAATG GTGTGAGGCAGGAATCTACCTCTTGGCTTCCCAAGCTGTAGACAAGTGCCAGTCTCGAGAAGGGGTTGATATCGCCTTGAACGACATTGC GACATTCCTGGGCACAGTCAAGGAGTACCCGTTGCTCAGCCCCAAGGAGTTTTACAACGAGTTTGAGTTGCTGCTCACCCTCGATGCAAA GGCCAAAGCCCAGAAAGTTTTGCAGAGGCTGGATGATGTCCAGGAAATATTTCACAAGAGGCAAGTGAGTCTGATGAAACTGGCAGCCAA ACAGACTCGTCCAGTGCAACCTGTGGCCCCACATCCTGAGTCTTCACCAAAATGGGTGTCATCAAAAACCAGCCAGCCCTCCACCTCGGT CCCTCTAGCTCGTCCTCTGAGAACGTCTGAGGAACCTTATACGGAGACAGAGTTGAACTCCCGGGGAAAGGAAGATGATGAGACTAAATT TGAAGTCAAGAGTGAAGAAATCTTTGAAAGCCATCATGAAAGGGGGAACCCTGAGCTGGAGCAGCAGGCCAGGCTCGGAGACCTTTCCCC CCGCAGTATTCTCGCCGCCCAGCAGTCAAGATTCTGGCTTGGTTTCCCTCTCGAGCAGCTCTCCTATTAGTAACAAGAGCACAAAGGCAG TGCTTGAATGTGAGCCTGCGTCGGAGCCCAGCAGCTTCACAGTCACTCCCGTCATCGAGGAGGACGAGTGAGCAGTGCCTGCTGCCGATG GCGGTTCACTTGGAGTAACAGGCTTATTCCACTTTCCATGGGGTTTGTTAATATTTTGCATTGACTCATACTATCTTAACTGTTGAGAAC GTATTTGGTTTATATTCCTTAGAGTTTAGTCCAGAGGCTGTAACACATTTGTAATACTTTAGGGTCCGTGACTACCATCTGCATGGTTTA AGTGCTTTACTCACGGAGTTTAAATAATAGTGTTCATTTTTTTAATGACACTGGTTTCATGTAGTTTTCAAGAAATAAAAGAATTCATTC >52171_52171_2_MCF2L2-DMRT1_MCF2L2_chr3_182994660_ENST00000414362_DMRT1_chr9_967985_ENST00000569227_length(amino acids)=671AA_BP=647 MGGDAFKKSSTAAVETVLFGFIYTEWFFSEMLSCLKEEMPPQELTRRLATVITHVDEIMQQEVRPLMAVEIIEQLHRQFAILSGGRGEDG APIITFPEFSGFKHIPDEDFLNVMTYLTSIPSVEAASIGFIVVIDRRRDKWSSVKASLTRIAVAFPGNLQLIFILRPSRFIQRTFTDIGI KYYRNEFKTKVPIIMVNSVSDLHGYIDKSQLTRELGGTLEYRHGQWVNHRTAIENFALTLKTTAQMLQTFGSCLATAELPRSMLSTEDLL MSHTRQRDKLQDELKLLGKQGTTLLSCIQEPATKCPNSKLNLNQLENVTTMERLLVQLDETEKAFSHFWSEHHLKLNQCLQLQHFEHDFC KAKLALDNLLEEQAEFTGIGDSVMHVEQILKEHKKLEEKSQEPLEKAQLLALVGDQLIQSHHYAADAIRPRCVELRHLCDDFINGNKKKW DILGKSLEFHRQLDKVSQWCEAGIYLLASQAVDKCQSREGVDIALNDIATFLGTVKEYPLLSPKEFYNEFELLLTLDAKAKAQKVLQRLD DVQEIFHKRQVSLMKLAAKQTRPVQPVAPHPESSPKWVSSKTSQPSTSVPLARPLRTSEEPYTETELNSRGKEDDETKFEVKSEEIFESH -------------------------------------------------------------- >52171_52171_3_MCF2L2-DMRT1_MCF2L2_chr3_182994660_ENST00000447025_DMRT1_chr9_967985_ENST00000569227_length(transcript)=2501nt_BP=1970nt GGCGAGGGCAGAGCATCCTTGGGCGGAGATGCCTTTAAAAAATCATCCACCGCAGCGGTAGAAACAGTTTTGTTTGGCTTTATTTATACG GAATGGTTTTTCAGTGAAATGCTGTCTTGCTTAAAAGAAGAGATGCCTCCCCAGGAGCTCACCCGGCGACTGGCCACAGTGATCACTCAT GTCGATGAAATTATGCAGCAGGAAGTCAGACCCCTGATGGCGGTGGAGATAATAGAACAACTTCACAGACAATTTGCCATTCTTTCAGGA GGCCGAGGGGAGGATGGCGCCCCCATCATCACGTTCCCAGAGTTTTCGGGGTTCAAACACATCCCAGATGAAGACTTCCTGAATGTCATG ACCTACCTGACTAGCATCCCCAGTGTGGAGGCTGCCAGCATTGGATTCATTGTTGTTATCGACAGACGAAGAGACAAGTGGAGCTCCGTA AAGGCATCCTTGACACGAATAGCTGTGGCATTTCCAGGAAACTTACAGCTCATATTCATCCTTCGTCCATCTCGCTTTATCCAGAGGACA TTCACTGACATTGGCATTAAATACTATCGAAATGAGTTTAAAACGAAAGTGCCGATCATCATGGTAAACTCTGTCTCTGACCTTCACGGC TACATCGACAAAAGCCAACTGACCCGGGAATTAGGGGGGACTTTGGAATATCGCCACGGTCAGTGGGTAAATCACCGCACTGCCATCGAA AACTTTGCCTTGACCTTGAAGACCACTGCCCAGATGCTGCAGACGTTTGGGTCCTGCCTGGCCACAGCAGAGCTGCCCAGAAGCATGCTA TCCACGGAAGACCTTCTCATGTCCCACACAAGGCAGCGGGACAAGCTGCAGGATGAGCTGAAATTACTTGGAAAGCAGGGGACCACATTG CTGTCATGCATCCAAGAACCAGCAACCAAATGTCCCAACAGCAAACTCAATCTCAACCAACTTGAGAATGTAACTACCATGGAAAGGTTA TTAGTTCAACTGGATGAAACAGAAAAAGCCTTTAGTCACTTTTGGTCTGAGCATCATCTGAAGCTTAACCAGTGCCTACAACTACAGCAT TTTGAGCACGATTTTTGTAAGGCTAAGCTTGCCCTGGATAATTTGCTGGAAGAGCAAGCAGAGTTTACAGGCATTGGAGACAGCGTGATG CACGTGGAGCAGATTCTTAAGGAACACAAAAAACTGGAGGAAAAAAGCCAGGAGCCCCTGGAAAAGGCCCAGCTGCTGGCACTGGTTGGG GACCAGCTCATCCAAAGCCACCATTATGCAGCAGATGCCATCAGGCCCCGGTGTGTGGAGCTCAGGCACCTCTGTGACGATTTCATCAAT GGAAACAAGAAAAAATGGGACATTTTAGGAAAGTCCTTAGAGTTTCATAGACAGCTGGACAAGGTCAGCCAATGGTGTGAGGCAGGAATC TACCTCTTGGCTTCCCAAGCTGTAGACAAGTGCCAGTCTCGAGAAGGGGTTGATATCGCCTTGAACGACATTGCGACATTCCTGGGCACA GTCAAGGAGTACCCGTTGCTCAGCCCCAAGGAGTTTTACAACGAGTTTGAGTTGCTGCTCACCCTCGATGCAAAGGCCAAAGCCCAGAAA GTTTTGCAGAGGCTGGATGATGTCCAGGAAATATTTCACAAGAGGCAAGTGAGTCTGATGAAACTGGCAGCCAAACAGACTCGTCCAGTG CAACCTGTGGCCCCACATCCTGAGTCTTCACCAAAATGGGTGTCATCAAAAACCAGCCAGCCCTCCACCTCGGTCCCTCTAGCTCGTCCT CTGAGAACGTCTGAGGAACCTTATACGGAGACAGAGTTGAACTCCCGGGGAAAGGAAGATGATGAGACTAAATTTGAAGTCAAGAGTGAA GAAATCTTTGAAAGCCATCATGAAAGGGGGAACCCTGAGCTGGAGCAGCAGGCCAGGCTCGGAGACCTTTCCCCCCGCAGTATTCTCGCC GCCCAGCAGTCAAGATTCTGGCTTGGTTTCCCTCTCGAGCAGCTCTCCTATTAGTAACAAGAGCACAAAGGCAGTGCTTGAATGTGAGCC TGCGTCGGAGCCCAGCAGCTTCACAGTCACTCCCGTCATCGAGGAGGACGAGTGAGCAGTGCCTGCTGCCGATGGCGGTTCACTTGGAGT AACAGGCTTATTCCACTTTCCATGGGGTTTGTTAATATTTTGCATTGACTCATACTATCTTAACTGTTGAGAACGTATTTGGTTTATATT CCTTAGAGTTTAGTCCAGAGGCTGTAACACATTTGTAATACTTTAGGGTCCGTGACTACCATCTGCATGGTTTAAGTGCTTTACTCACGG AGTTTAAATAATAGTGTTCATTTTTTTAATGACACTGGTTTCATGTAGTTTTCAAGAAATAAAAGAATTCATTCAAGTGAAGCCATTTGT >52171_52171_3_MCF2L2-DMRT1_MCF2L2_chr3_182994660_ENST00000447025_DMRT1_chr9_967985_ENST00000569227_length(amino acids)=671AA_BP=647 MGGDAFKKSSTAAVETVLFGFIYTEWFFSEMLSCLKEEMPPQELTRRLATVITHVDEIMQQEVRPLMAVEIIEQLHRQFAILSGGRGEDG APIITFPEFSGFKHIPDEDFLNVMTYLTSIPSVEAASIGFIVVIDRRRDKWSSVKASLTRIAVAFPGNLQLIFILRPSRFIQRTFTDIGI KYYRNEFKTKVPIIMVNSVSDLHGYIDKSQLTRELGGTLEYRHGQWVNHRTAIENFALTLKTTAQMLQTFGSCLATAELPRSMLSTEDLL MSHTRQRDKLQDELKLLGKQGTTLLSCIQEPATKCPNSKLNLNQLENVTTMERLLVQLDETEKAFSHFWSEHHLKLNQCLQLQHFEHDFC KAKLALDNLLEEQAEFTGIGDSVMHVEQILKEHKKLEEKSQEPLEKAQLLALVGDQLIQSHHYAADAIRPRCVELRHLCDDFINGNKKKW DILGKSLEFHRQLDKVSQWCEAGIYLLASQAVDKCQSREGVDIALNDIATFLGTVKEYPLLSPKEFYNEFELLLTLDAKAKAQKVLQRLD DVQEIFHKRQVSLMKLAAKQTRPVQPVAPHPESSPKWVSSKTSQPSTSVPLARPLRTSEEPYTETELNSRGKEDDETKFEVKSEEIFESH -------------------------------------------------------------- >52171_52171_4_MCF2L2-DMRT1_MCF2L2_chr3_182994660_ENST00000473233_DMRT1_chr9_967985_ENST00000569227_length(transcript)=2393nt_BP=1862nt ATGCTGTCTTGCTTAAAAGAAGAGATGCCTCCCCAGGAGCTCACCCGGCGACTGGCCACAGTGATCACTCATGTCGATGAAATTATGCAG CAGGAAGTCAGACCCCTGATGGCGGTGGAGATAATAGAACAACTTCACAGACAATTTGCCATTCTTTCAGGAGGCCGAGGGGAGGATGGC GCCCCCATCATCACGTTCCCAGAGTTTTCGGGGTTCAAACACATCCCAGATGAAGACTTCCTGAATGTCATGACCTACCTGACTAGCATC CCCAGTGTGGAGGCTGCCAGCATTGGATTCATTGTTGTTATCGACAGACGAAGAGACAAGTGGAGCTCCGTAAAGGCATCCTTGACACGA ATAGCTGTGGCATTTCCAGGAAACTTACAGCTCATATTCATCCTTCGTCCATCTCGCTTTATCCAGAGGACATTCACTGACATTGGCATT AAATACTATCGAAATGAGTTTAAAACGAAAGTGCCGATCATCATGGTAAACTCTGTCTCTGACCTTCACGGCTACATCGACAAAAGCCAA CTGACCCGGGAATTAGGGGGGACTTTGGAATATCGCCACGGTCAGTGGGTAAATCACCGCACTGCCATCGAAAACTTTGCCTTGACCTTG AAGACCACTGCCCAGATGCTGCAGACGTTTGGGTCCTGCCTGGCCACAGCAGAGCTGCCCAGAAGCATGCTATCCACGGAAGACCTTCTC ATGTCCCACACAAGGCAGCGGGACAAGCTGCAGGATGAGCTGAAATTACTTGGAAAGCAGGGGACCACATTGCTGTCATGCATCCAAGAA CCAGCAACCAAATGTCCCAACAGCAAACTCAATCTCAACCAACTTGAGAATGTAACTACCATGGAAAGGTTATTAGTTCAACTGGATGAA ACAGAAAAAGCCTTTAGTCACTTTTGGTCTGAGCATCATCTGAAGCTTAACCAGTGCCTACAACTACAGCATTTTGAGCACGATTTTTGT AAGGCTAAGCTTGCCCTGGATAATTTGCTGGAAGAGCAAGCAGAGTTTACAGGCATTGGAGACAGCGTGATGCACGTGGAGCAGATTCTT AAGGAACACAAAAAACTGGAGGAAAAAAGCCAGGAGCCCCTGGAAAAGGCCCAGCTGCTGGCACTGGTTGGGGACCAGCTCATCCAAAGC CACCATTATGCAGCAGATGCCATCAGGCCCCGGTGTGTGGAGCTCAGGCACCTCTGTGACGATTTCATCAATGGAAACAAGAAAAAATGG GACATTTTAGGAAAGTCCTTAGAGTTTCATAGACAGCTGGACAAGGTCAGCCAATGGTGTGAGGCAGGAATCTACCTCTTGGCTTCCCAA GCTGTAGACAAGTGCCAGTCTCGAGAAGGGGTTGATATCGCCTTGAACGACATTGCGACATTCCTGGGCACAGTCAAGGAGTACCCGTTG CTCAGCCCCAAGGAGTTTTACAACGAGTTTGAGTTGCTGCTCACCCTCGATGCAAAGGCCAAAGCCCAGAAAGTTTTGCAGAGGCTGGAT GATGTCCAGGAAATATTTCACAAGAGGCAAGTGAGTCTGATGAAACTGGCAGCCAAACAGACTCGTCCAGTGCAACCTGTGGCCCCACAT CCTGAGTCTTCACCAAAATGGGTGTCATCAAAAACCAGCCAGCCCTCCACCTCGGTCCCTCTAGCTCGTCCTCTGAGAACGTCTGAGGAA CCTTATACGGAGACAGAGTTGAACTCCCGGGGAAAGGAAGATGATGAGACTAAATTTGAAGTCAAGAGTGAAGAAATCTTTGAAAGCCAT CATGAAAGGGGGAACCCTGAGCTGGAGCAGCAGGCCAGGCTCGGAGACCTTTCCCCCCGCAGTATTCTCGCCGCCCAGCAGTCAAGATTC TGGCTTGGTTTCCCTCTCGAGCAGCTCTCCTATTAGTAACAAGAGCACAAAGGCAGTGCTTGAATGTGAGCCTGCGTCGGAGCCCAGCAG CTTCACAGTCACTCCCGTCATCGAGGAGGACGAGTGAGCAGTGCCTGCTGCCGATGGCGGTTCACTTGGAGTAACAGGCTTATTCCACTT TCCATGGGGTTTGTTAATATTTTGCATTGACTCATACTATCTTAACTGTTGAGAACGTATTTGGTTTATATTCCTTAGAGTTTAGTCCAG AGGCTGTAACACATTTGTAATACTTTAGGGTCCGTGACTACCATCTGCATGGTTTAAGTGCTTTACTCACGGAGTTTAAATAATAGTGTT CATTTTTTTAATGACACTGGTTTCATGTAGTTTTCAAGAAATAAAAGAATTCATTCAAGTGAAGCCATTTGTGTGCCTCTAAATGAGTCA >52171_52171_4_MCF2L2-DMRT1_MCF2L2_chr3_182994660_ENST00000473233_DMRT1_chr9_967985_ENST00000569227_length(amino acids)=641AA_BP=617 MLSCLKEEMPPQELTRRLATVITHVDEIMQQEVRPLMAVEIIEQLHRQFAILSGGRGEDGAPIITFPEFSGFKHIPDEDFLNVMTYLTSI PSVEAASIGFIVVIDRRRDKWSSVKASLTRIAVAFPGNLQLIFILRPSRFIQRTFTDIGIKYYRNEFKTKVPIIMVNSVSDLHGYIDKSQ LTRELGGTLEYRHGQWVNHRTAIENFALTLKTTAQMLQTFGSCLATAELPRSMLSTEDLLMSHTRQRDKLQDELKLLGKQGTTLLSCIQE PATKCPNSKLNLNQLENVTTMERLLVQLDETEKAFSHFWSEHHLKLNQCLQLQHFEHDFCKAKLALDNLLEEQAEFTGIGDSVMHVEQIL KEHKKLEEKSQEPLEKAQLLALVGDQLIQSHHYAADAIRPRCVELRHLCDDFINGNKKKWDILGKSLEFHRQLDKVSQWCEAGIYLLASQ AVDKCQSREGVDIALNDIATFLGTVKEYPLLSPKEFYNEFELLLTLDAKAKAQKVLQRLDDVQEIFHKRQVSLMKLAAKQTRPVQPVAPH PESSPKWVSSKTSQPSTSVPLARPLRTSEEPYTETELNSRGKEDDETKFEVKSEEIFESHHERGNPELEQQARLGDLSPRSILAAQQSRF -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for MCF2L2-DMRT1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for MCF2L2-DMRT1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for MCF2L2-DMRT1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
