
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:MED12L-GOLIM4 (FusionGDB2 ID:52538) |
Fusion Gene Summary for MED12L-GOLIM4 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: MED12L-GOLIM4 | Fusion gene ID: 52538 | Hgene | Tgene | Gene symbol | MED12L | GOLIM4 | Gene ID | 116931 | 27333 |
| Gene name | mediator complex subunit 12L | golgi integral membrane protein 4 | |
| Synonyms | NOPAR|TNRC11L|TRALP|TRALPUSH | GIMPC|GOLPH4|GPP130|P138 | |
| Cytomap | 3q25.1 | 3q26.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | mediator of RNA polymerase II transcription subunit 12-like proteinmediator complex subunit 12 likemediator of RNA polymerase II transcription, subunit 12 homolog-likeno opposite paired repeat proteinthyroid hormone receptor-associated-like proteintr | Golgi integral membrane protein 4130 kDa golgi-localized phosphoproteincis Golgi-localized calcium-binding proteingolgi integral membrane protein, cisgolgi phosphoprotein 4golgi phosphoprotein of 130 kDagolgi-localized phosphoprotein of 130 kDatype | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q86YW9 | O00461 | |
| Ensembl transtripts involved in fusion gene | ENST00000309237, ENST00000422248, ENST00000273432, ENST00000474524, ENST00000491549, | ENST00000309027, ENST00000470487, | |
| Fusion gene scores | * DoF score | 9 X 9 X 5=405 | 7 X 6 X 5=210 |
| # samples | 9 | 7 | |
| ** MAII score | log2(9/405*10)=-2.16992500144231 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/210*10)=-1.58496250072116 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: MED12L [Title/Abstract] AND GOLIM4 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | MED12L(150911453)-GOLIM4(167742883), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | MED12L-GOLIM4 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. MED12L-GOLIM4 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across MED12L (5'-gene) Fusion gene breakpoints across MED12L (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
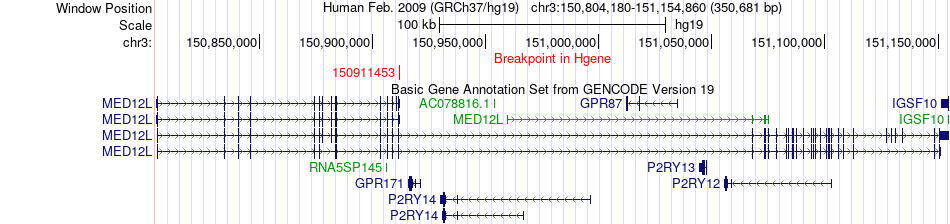 |
 Fusion gene breakpoints across GOLIM4 (3'-gene) Fusion gene breakpoints across GOLIM4 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
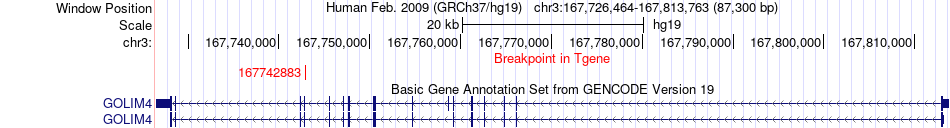 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUSC | TCGA-63-A5M9-01A | MED12L | chr3 | 150911453 | - | GOLIM4 | chr3 | 167742883 | - |
| ChimerDB4 | LUSC | TCGA-63-A5M9-01A | MED12L | chr3 | 150911453 | + | GOLIM4 | chr3 | 167742883 | - |
| ChimerDB4 | LUSC | TCGA-63-A5M9 | MED12L | chr3 | 150911453 | + | GOLIM4 | chr3 | 167742883 | - |
Top |
Fusion Gene ORF analysis for MED12L-GOLIM4 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| Frame-shift | ENST00000309237 | ENST00000309027 | MED12L | chr3 | 150911453 | + | GOLIM4 | chr3 | 167742883 | - |
| Frame-shift | ENST00000309237 | ENST00000470487 | MED12L | chr3 | 150911453 | + | GOLIM4 | chr3 | 167742883 | - |
| Frame-shift | ENST00000422248 | ENST00000309027 | MED12L | chr3 | 150911453 | + | GOLIM4 | chr3 | 167742883 | - |
| Frame-shift | ENST00000422248 | ENST00000470487 | MED12L | chr3 | 150911453 | + | GOLIM4 | chr3 | 167742883 | - |
| In-frame | ENST00000273432 | ENST00000309027 | MED12L | chr3 | 150911453 | + | GOLIM4 | chr3 | 167742883 | - |
| In-frame | ENST00000273432 | ENST00000470487 | MED12L | chr3 | 150911453 | + | GOLIM4 | chr3 | 167742883 | - |
| In-frame | ENST00000474524 | ENST00000309027 | MED12L | chr3 | 150911453 | + | GOLIM4 | chr3 | 167742883 | - |
| In-frame | ENST00000474524 | ENST00000470487 | MED12L | chr3 | 150911453 | + | GOLIM4 | chr3 | 167742883 | - |
| intron-3CDS | ENST00000491549 | ENST00000309027 | MED12L | chr3 | 150911453 | + | GOLIM4 | chr3 | 167742883 | - |
| intron-3CDS | ENST00000491549 | ENST00000470487 | MED12L | chr3 | 150911453 | + | GOLIM4 | chr3 | 167742883 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000474524 | MED12L | chr3 | 150911453 | + | ENST00000470487 | GOLIM4 | chr3 | 167742883 | - | 4243 | 2183 | 38 | 2650 | 870 |
| ENST00000474524 | MED12L | chr3 | 150911453 | + | ENST00000309027 | GOLIM4 | chr3 | 167742883 | - | 2685 | 2183 | 38 | 2650 | 870 |
| ENST00000273432 | MED12L | chr3 | 150911453 | + | ENST00000470487 | GOLIM4 | chr3 | 167742883 | - | 3818 | 1758 | 33 | 2225 | 730 |
| ENST00000273432 | MED12L | chr3 | 150911453 | + | ENST00000309027 | GOLIM4 | chr3 | 167742883 | - | 2260 | 1758 | 33 | 2225 | 730 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000474524 | ENST00000470487 | MED12L | chr3 | 150911453 | + | GOLIM4 | chr3 | 167742883 | - | 0.000607806 | 0.99939215 |
| ENST00000474524 | ENST00000309027 | MED12L | chr3 | 150911453 | + | GOLIM4 | chr3 | 167742883 | - | 0.005014546 | 0.99498546 |
| ENST00000273432 | ENST00000470487 | MED12L | chr3 | 150911453 | + | GOLIM4 | chr3 | 167742883 | - | 0.000780483 | 0.99921954 |
| ENST00000273432 | ENST00000309027 | MED12L | chr3 | 150911453 | + | GOLIM4 | chr3 | 167742883 | - | 0.006608471 | 0.9933915 |
Top |
Fusion Genomic Features for MED12L-GOLIM4 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
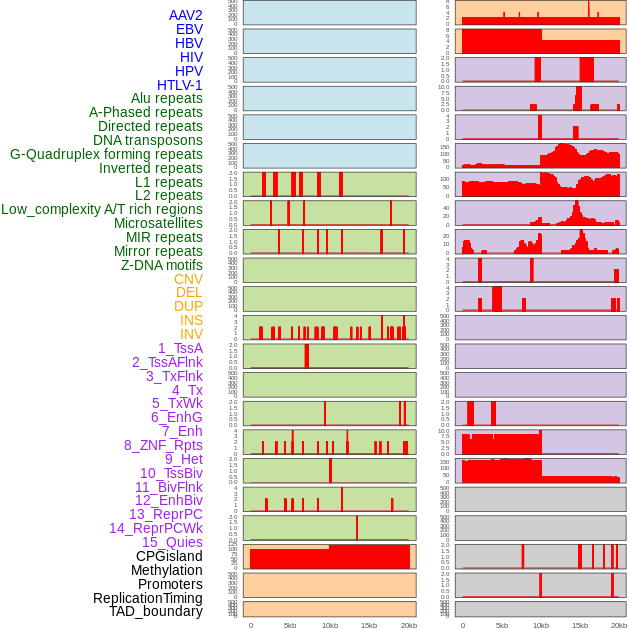 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for MED12L-GOLIM4 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:150911453/chr3:167742883) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MED12L | GOLIM4 |
| FUNCTION: May be a component of the Mediator complex, a coactivator involved in the regulated transcription of nearly all RNA polymerase II-dependent genes. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. Mediator is recruited to promoters by direct interactions with regulatory proteins and serves as a scaffold for the assembly of a functional preinitiation complex with RNA polymerase II and the general transcription factors (By similarity). {ECO:0000250}. | FUNCTION: Plays a role in endosome to Golgi protein trafficking; mediates protein transport along the late endosome-bypass pathway from the early endosome to the Golgi. {ECO:0000269|PubMed:15331763}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MED12L | chr3:150911453 | chr3:167742883 | ENST00000309237 | + | 1 | 15 | 1877_2134 | 0 | 757.0 | Compositional bias | Note=Gln-rich |
| Hgene | MED12L | chr3:150911453 | chr3:167742883 | ENST00000422248 | + | 1 | 14 | 1877_2134 | 0 | 722.0 | Compositional bias | Note=Gln-rich |
| Hgene | MED12L | chr3:150911453 | chr3:167742883 | ENST00000474524 | + | 14 | 43 | 1877_2134 | 715 | 2146.0 | Compositional bias | Note=Gln-rich |
| Tgene | GOLIM4 | chr3:150911453 | chr3:167742883 | ENST00000470487 | 11 | 16 | 35_244 | 541 | 697.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | GOLIM4 | chr3:150911453 | chr3:167742883 | ENST00000470487 | 11 | 16 | 311_681 | 541 | 697.0 | Compositional bias | Note=Glu-rich | |
| Tgene | GOLIM4 | chr3:150911453 | chr3:167742883 | ENST00000470487 | 11 | 16 | 404_513 | 541 | 697.0 | Compositional bias | Note=Gln-rich | |
| Tgene | GOLIM4 | chr3:150911453 | chr3:167742883 | ENST00000470487 | 11 | 16 | 176_248 | 541 | 697.0 | Region | Note=Golgi targeting | |
| Tgene | GOLIM4 | chr3:150911453 | chr3:167742883 | ENST00000470487 | 11 | 16 | 38_107 | 541 | 697.0 | Region | Note=Golgi targeting | |
| Tgene | GOLIM4 | chr3:150911453 | chr3:167742883 | ENST00000470487 | 11 | 16 | 80_175 | 541 | 697.0 | Region | Note=Endosome targeting | |
| Tgene | GOLIM4 | chr3:150911453 | chr3:167742883 | ENST00000470487 | 11 | 16 | 2_12 | 541 | 697.0 | Topological domain | Cytoplasmic | |
| Tgene | GOLIM4 | chr3:150911453 | chr3:167742883 | ENST00000470487 | 11 | 16 | 34_696 | 541 | 697.0 | Topological domain | Lumenal | |
| Tgene | GOLIM4 | chr3:150911453 | chr3:167742883 | ENST00000470487 | 11 | 16 | 13_33 | 541 | 697.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
Top |
Fusion Gene Sequence for MED12L-GOLIM4 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >52538_52538_1_MED12L-GOLIM4_MED12L_chr3_150911453_ENST00000273432_GOLIM4_chr3_167742883_ENST00000309027_length(transcript)=2260nt_BP=1758nt TCTCATTCATTTCGCCGGTTAACATGAGAGATCATGGCCGCCTTCGGGCTTCTCAGCTATGAGCAGAGACCGCTGAAGCGCCCCCGGCTC GGGCCGCCCGACGTCTACCCACAGGACCCCAAGCAGAAGGAGGATGAACTTACTGCTGTGAATGTAAAGCAAGGCTTCAATAATCAGCCA GCCTTCACTGGAGATGAACATGGCTCAGCCAGAAATATTGTAATTAACCCATCAAAGATTGGAGCTTATTTTAGCAGCATATTAGCTGAG AAACTGAAGCTTAACACTTTCCAGGACACGGGAAAGAAGAAACCACAAGTTAATGCTAAAGATAATTATTGGCTGGTTACTGCTCGATCC CAGAGTGCAATTCATAGTTGGTTTTCTGACTTAGCAGGAAATAAGCCACTTTCTATTTTGGCAAAAAAGGTTCCTATCCTTAGTAAAAAA GAGGATGTTTTTGCATATTTAGCTAAATATTCTGTGCCAATGGTTCGAGCAACGTGGCTGATCAAGATGACTTGTGCCTATTATTCTGCT ATATCTGAAGCTAAAATTAAGAAACGTCAGGCTCCTGATCCGAATTTGGAGTGGACACAGATATCTACCAGATATCTTCGAGAGCAGTTG GCCAAGATTTCTGACTTTTACCACATGGCCTCCAGCACGGGCGATGGCCCTGTCCCTGTGCCACCAGAGGTGGAGCAAGCCATGAAGCAA TGGGAATACAACGAAAAGCTAGCATTTCACATGTTCCAGGAAGGAATGTTAGAAAAACACGAATATTTGACATGGATCCTGGATGTTTTA GAAAAGATCAGACCAATGGATGATGATCTTCTTAAACTCTTGCTACCACTAATGCTGCAGGTTCGGGCAAGGATTTATGAAGTAGAACAA CAGATAAAACAAAGAGGCCGTGCAGTGGAAGTTCGGTGGTCATTTGACAAGTGCCAAGAATCCACAGCAGGGGTGACTATTAGTCGGGTT TTGCACACGTTGGAAGTTTTGGATCGTCACTGTTTTGACCGAACTGATTCCAGCAATTCCATGGAGACACTTTATCATAAGATTTTCTGG GCAAACCAAAACAAAGATAACCAAGAGGTTGCGCCCAACGATGAAGCTGTGGTGACGCTGTTATGTGAATGGGCCGTGAGCTGCAAACGG TCTGGCAAGCACAGGGCCATGGCTGTGGCAAAACTCCTGGAGAAGAGGCAAGCAGAAATTGAGGCAGAGAGATGTGGTGAATCAGAAGTC TTAGATGAGAAGGAGTCTATTTCTTCATCCTCTCTTGCTGGATCCAGTTTGCCTGTTTTCCAGAATGTGCTGTTAAGGTTTTTAGATACA CAGGCCCCCTCTTTGTCGGACCCAAACAGTGAATGTGAAAAGGTGGAATTTGTGAACCTGGTGCTGCTCTTCTGCGAGTTCATCCGCCAT GATGTCTTCTCCCATGACGCATACATGTGTACCCTCATATCTCGAGGAGATTTGTCAGTCACTGCCTCAACTCGGCCGCGGTCACCAGTA GGGGAAAATGCAGATGAACACTATTCAAAGGACCATGATGTGAAAATGGAGATTTTTTCTCCTATGCCTGGAGAATCCTGTGAGAATGCC AACACTTCGTTGGGCAGAAGAATGTCAGTTAATTGTGAGAAGTTGGTGAAGAGGGAAAAGCCAAGGGAATTAATTTTTCCATCTAATTAT GACCTCCTTCGCCACTTACAGTATGCAACACATTTTCCTATACCTCTGAGGGCAGCTGTAGAAGATATAAACCCAGCAGATGACCCTAAT AATCAAGGTGAGGATGAATTTGAAGAAGCCGAGCAAGTGAGAGAAGAAAATTTGCCAGATGAAAATGAAGAGCAAAAACAAAGTAATCAA AAGCAAGAGAATACAGAAGTGGAGGAACATTTGGTGATGGCAGGAAATCCAGACCAGCAGGAGGACAATGTTGATGAACAGTACCAGGAA GAGGCAGAAGAGGAGGTTCAGGAAGATTTGACTGAAGAGAAAAAAAGGGAACTGGAGCATAATGCTGAAGAGACCTATGGTGAAAATGAT GAAAATACTGATGATAAAAATAATGATGGAGAAGAGCAAGAAGTTCGAGATGACAACCGCCCCAAAGGCCGAGAGGAACACTACGAGGAG GAAGAAGAGGAGGAAGAAGACGGGGCTGCAGTTGCTGAGAAATCACATCGAAGAGCTGAAATGTAGCGGCACCCAATTTCTAGACAACGC >52538_52538_1_MED12L-GOLIM4_MED12L_chr3_150911453_ENST00000273432_GOLIM4_chr3_167742883_ENST00000309027_length(amino acids)=730AA_BP=575 MAAFGLLSYEQRPLKRPRLGPPDVYPQDPKQKEDELTAVNVKQGFNNQPAFTGDEHGSARNIVINPSKIGAYFSSILAEKLKLNTFQDTG KKKPQVNAKDNYWLVTARSQSAIHSWFSDLAGNKPLSILAKKVPILSKKEDVFAYLAKYSVPMVRATWLIKMTCAYYSAISEAKIKKRQA PDPNLEWTQISTRYLREQLAKISDFYHMASSTGDGPVPVPPEVEQAMKQWEYNEKLAFHMFQEGMLEKHEYLTWILDVLEKIRPMDDDLL KLLLPLMLQVRARIYEVEQQIKQRGRAVEVRWSFDKCQESTAGVTISRVLHTLEVLDRHCFDRTDSSNSMETLYHKIFWANQNKDNQEVA PNDEAVVTLLCEWAVSCKRSGKHRAMAVAKLLEKRQAEIEAERCGESEVLDEKESISSSSLAGSSLPVFQNVLLRFLDTQAPSLSDPNSE CEKVEFVNLVLLFCEFIRHDVFSHDAYMCTLISRGDLSVTASTRPRSPVGENADEHYSKDHDVKMEIFSPMPGESCENANTSLGRRMSVN CEKLVKREKPRELIFPSNYDLLRHLQYATHFPIPLRAAVEDINPADDPNNQGEDEFEEAEQVREENLPDENEEQKQSNQKQENTEVEEHL VMAGNPDQQEDNVDEQYQEEAEEEVQEDLTEEKKRELEHNAEETYGENDENTDDKNNDGEEQEVRDDNRPKGREEHYEEEEEEEEDGAAV -------------------------------------------------------------- >52538_52538_2_MED12L-GOLIM4_MED12L_chr3_150911453_ENST00000273432_GOLIM4_chr3_167742883_ENST00000470487_length(transcript)=3818nt_BP=1758nt TCTCATTCATTTCGCCGGTTAACATGAGAGATCATGGCCGCCTTCGGGCTTCTCAGCTATGAGCAGAGACCGCTGAAGCGCCCCCGGCTC GGGCCGCCCGACGTCTACCCACAGGACCCCAAGCAGAAGGAGGATGAACTTACTGCTGTGAATGTAAAGCAAGGCTTCAATAATCAGCCA GCCTTCACTGGAGATGAACATGGCTCAGCCAGAAATATTGTAATTAACCCATCAAAGATTGGAGCTTATTTTAGCAGCATATTAGCTGAG AAACTGAAGCTTAACACTTTCCAGGACACGGGAAAGAAGAAACCACAAGTTAATGCTAAAGATAATTATTGGCTGGTTACTGCTCGATCC CAGAGTGCAATTCATAGTTGGTTTTCTGACTTAGCAGGAAATAAGCCACTTTCTATTTTGGCAAAAAAGGTTCCTATCCTTAGTAAAAAA GAGGATGTTTTTGCATATTTAGCTAAATATTCTGTGCCAATGGTTCGAGCAACGTGGCTGATCAAGATGACTTGTGCCTATTATTCTGCT ATATCTGAAGCTAAAATTAAGAAACGTCAGGCTCCTGATCCGAATTTGGAGTGGACACAGATATCTACCAGATATCTTCGAGAGCAGTTG GCCAAGATTTCTGACTTTTACCACATGGCCTCCAGCACGGGCGATGGCCCTGTCCCTGTGCCACCAGAGGTGGAGCAAGCCATGAAGCAA TGGGAATACAACGAAAAGCTAGCATTTCACATGTTCCAGGAAGGAATGTTAGAAAAACACGAATATTTGACATGGATCCTGGATGTTTTA GAAAAGATCAGACCAATGGATGATGATCTTCTTAAACTCTTGCTACCACTAATGCTGCAGGTTCGGGCAAGGATTTATGAAGTAGAACAA CAGATAAAACAAAGAGGCCGTGCAGTGGAAGTTCGGTGGTCATTTGACAAGTGCCAAGAATCCACAGCAGGGGTGACTATTAGTCGGGTT TTGCACACGTTGGAAGTTTTGGATCGTCACTGTTTTGACCGAACTGATTCCAGCAATTCCATGGAGACACTTTATCATAAGATTTTCTGG GCAAACCAAAACAAAGATAACCAAGAGGTTGCGCCCAACGATGAAGCTGTGGTGACGCTGTTATGTGAATGGGCCGTGAGCTGCAAACGG TCTGGCAAGCACAGGGCCATGGCTGTGGCAAAACTCCTGGAGAAGAGGCAAGCAGAAATTGAGGCAGAGAGATGTGGTGAATCAGAAGTC TTAGATGAGAAGGAGTCTATTTCTTCATCCTCTCTTGCTGGATCCAGTTTGCCTGTTTTCCAGAATGTGCTGTTAAGGTTTTTAGATACA CAGGCCCCCTCTTTGTCGGACCCAAACAGTGAATGTGAAAAGGTGGAATTTGTGAACCTGGTGCTGCTCTTCTGCGAGTTCATCCGCCAT GATGTCTTCTCCCATGACGCATACATGTGTACCCTCATATCTCGAGGAGATTTGTCAGTCACTGCCTCAACTCGGCCGCGGTCACCAGTA GGGGAAAATGCAGATGAACACTATTCAAAGGACCATGATGTGAAAATGGAGATTTTTTCTCCTATGCCTGGAGAATCCTGTGAGAATGCC AACACTTCGTTGGGCAGAAGAATGTCAGTTAATTGTGAGAAGTTGGTGAAGAGGGAAAAGCCAAGGGAATTAATTTTTCCATCTAATTAT GACCTCCTTCGCCACTTACAGTATGCAACACATTTTCCTATACCTCTGAGGGCAGCTGTAGAAGATATAAACCCAGCAGATGACCCTAAT AATCAAGGTGAGGATGAATTTGAAGAAGCCGAGCAAGTGAGAGAAGAAAATTTGCCAGATGAAAATGAAGAGCAAAAACAAAGTAATCAA AAGCAAGAGAATACAGAAGTGGAGGAACATTTGGTGATGGCAGGAAATCCAGACCAGCAGGAGGACAATGTTGATGAACAGTACCAGGAA GAGGCAGAAGAGGAGGTTCAGGAAGATTTGACTGAAGAGAAAAAAAGGGAACTGGAGCATAATGCTGAAGAGACCTATGGTGAAAATGAT GAAAATACTGATGATAAAAATAATGATGGAGAAGAGCAAGAAGTTCGAGATGACAACCGCCCCAAAGGCCGAGAGGAACACTACGAGGAG GAAGAAGAGGAGGAAGAAGACGGGGCTGCAGTTGCTGAGAAATCACATCGAAGAGCTGAAATGTAGCGGCACCCAATTTCTAGACAACGC TCAGCCAACGGATTCTTTTCAAGCTGCTCAAACATAAATCTGCCTACTGAACTCTAGGATATTTAATTACAAAAATTAAGAACTTAGACT TTTTTAAAACTTTTGTATTAGAAATGCGCATACATTTATATGAATATATTTTGATAACGTAGGTCTAGAGCTTCTTTTATATTCAAGCTA AACATGAAAAAGAAGAAAAACAATAAAGTAAACCTGAGCCCCCACGTCCCAATTTTTTTAATAGATTATGTGATGTTGGAAAGCTCATTG ATTTTGTATATGTTTCAGCGTGTTACCTTTCTGGCTTCCAGTTCCCAGGTGTTCTTTGTTTGCCTTTGATAAAATACAGGATTTAAGAAC AGAGAGTACTGCAAAATGCCATGCAGACTTTAAAGAGAATGGCCTGTTTACTAATTGCTGCCCTTCTGATGTCTTTATGTATAGCTCTGA TAGAATTTTCACCAGTCTATGTATCTCTGGAGTGAGATCTATGTACAAAGTGACATACAATTGGAAATCCATTTTTGTTGTAAAGACATT GTTTTTCAGACTTTTCAGATCAATTAGAAAAATGTCATTGCTTTAAAATCATAGCTGTTCTGTTTAAGAGGAATTGAATTTAAAAATGAG AGAGTATTAAAACTCATGTGGCAGTATCCTGGTCTTAATCAGGTATTGCAGTGTTCAAACAAGGTATGGACACATCAACATATTCACCTT TTGGAAGGACTAGTGACTTGGTTAAAAACTAATGAGTGTTTGCGTGTGGTGTTTCTATGCGAGCACCCATTCTCAATTTTTTTTTTTTTT TTTTGTTTGAAGCCATTGATTGATATTCTTGTTTCAGTGTCTGGTTCCCTATAAGTAATATATATTTCCTCTTTAAGCTCTGTGCCTCAA TATGCAATCACTTTGATAATAAGTATAAATATGAAGACATACCAAACTGCAAATAGGATTATTTTTCAAAACTATATGGTGAAACATATG GTGATATGATAGACTTATGGTTTTTCTGTTTGCTGGGATTGGGTCTGCTTTTTGCCTATGTGTAAGGCATTTTAATCATTGTGAATATAA TAGATTCCTTAAAATAAGAAAATTAAAAATCAGAAACTTCCCTTGGAGTAGAGACATTTCAGTTGCAAGCATAAAGATTTTTAAATATAT AACACTAGGTTATATAATTCATATATGTGTGGTTTTGTATTAATAATCAAATTAATAAAAACATTTCATTTTCTACCATTGTATGAACGT AATTTTATTTTTTAATAACCACACTCATAGATGGTTTTGATTGTTCCAGTTGAGTGATTGTGTATGTACGCATAGGAATGTTTTGTTCAG TTGTACTTTGTATTTCCAATAATCAGAAATCTTACCTTTATGTATAAAAACCCATTATGTAATGTAAAATGTATAATATTGTACATAATA TTCAGAATACAATTATTTTGGTAAATTAGCTTGTATTTTTAATGTCCCAGTTGCAGTATTTAATTATTTGAACCATACTGAAAGTTGGTA >52538_52538_2_MED12L-GOLIM4_MED12L_chr3_150911453_ENST00000273432_GOLIM4_chr3_167742883_ENST00000470487_length(amino acids)=730AA_BP=575 MAAFGLLSYEQRPLKRPRLGPPDVYPQDPKQKEDELTAVNVKQGFNNQPAFTGDEHGSARNIVINPSKIGAYFSSILAEKLKLNTFQDTG KKKPQVNAKDNYWLVTARSQSAIHSWFSDLAGNKPLSILAKKVPILSKKEDVFAYLAKYSVPMVRATWLIKMTCAYYSAISEAKIKKRQA PDPNLEWTQISTRYLREQLAKISDFYHMASSTGDGPVPVPPEVEQAMKQWEYNEKLAFHMFQEGMLEKHEYLTWILDVLEKIRPMDDDLL KLLLPLMLQVRARIYEVEQQIKQRGRAVEVRWSFDKCQESTAGVTISRVLHTLEVLDRHCFDRTDSSNSMETLYHKIFWANQNKDNQEVA PNDEAVVTLLCEWAVSCKRSGKHRAMAVAKLLEKRQAEIEAERCGESEVLDEKESISSSSLAGSSLPVFQNVLLRFLDTQAPSLSDPNSE CEKVEFVNLVLLFCEFIRHDVFSHDAYMCTLISRGDLSVTASTRPRSPVGENADEHYSKDHDVKMEIFSPMPGESCENANTSLGRRMSVN CEKLVKREKPRELIFPSNYDLLRHLQYATHFPIPLRAAVEDINPADDPNNQGEDEFEEAEQVREENLPDENEEQKQSNQKQENTEVEEHL VMAGNPDQQEDNVDEQYQEEAEEEVQEDLTEEKKRELEHNAEETYGENDENTDDKNNDGEEQEVRDDNRPKGREEHYEEEEEEEEDGAAV -------------------------------------------------------------- >52538_52538_3_MED12L-GOLIM4_MED12L_chr3_150911453_ENST00000474524_GOLIM4_chr3_167742883_ENST00000309027_length(transcript)=2685nt_BP=2183nt CCAACTCTCATTCATTTCGCCGGTTAACATGAGAGATCATGGCCGCCTTCGGGCTTCTCAGCTATGAGCAGAGACCGCTGAAGCGCCCCC GGCTCGGGCCGCCCGACGTCTACCCACAGGACCCCAAGCAGAAGGAGGATGAACTTACTGCTGTGAATGTAAAGCAAGGCTTCAATAATC AGCCAGCCTTCACTGGAGATGAACATGGCTCAGCCAGAAATATTGTAATTAACCCATCAAAGATTGGAGCTTATTTTAGCAGCATATTAG CTGAGAAACTGAAGCTTAACACTTTCCAGGACACGGGAAAGAAGAAACCACAAGTTAATGCTAAAGATAATTATTGGCTGGTTACTGCTC GATCCCAGAGTGCAATTCATAGTTGGTTTTCTGACTTAGCAGGAAATAAGCCACTTTCTATTTTGGCAAAAAAGGTTCCTATCCTTAGTA AAAAAGAGGATGTTTTTGCATATTTAGCTAAATATTCTGTGCCAATGGTTCGAGCAACGTGGCTGATCAAGATGACTTGTGCCTATTATT CTGCTATATCTGAAGCTAAAATTAAGAAACGTCAGGCTCCTGATCCGAATTTGGAGTGGACACAGATATCTACCAGATATCTTCGAGAGC AGTTGGCCAAGATTTCTGACTTTTACCACATGGCCTCCAGCACGGGCGATGGCCCTGTCCCTGTGCCACCAGAGGTGGAGCAAGCCATGA AGCAATGGGAATACAACGAAAAGCTAGCATTTCACATGTTCCAGGAAGGAATGTTAGAAAAACACGAATATTTGACATGGATCCTGGATG TTTTAGAAAAGATCAGACCAATGGATGATGATCTTCTTAAACTCTTGCTACCACTAATGCTGCAGTATTCAGATGAGTTTGTTCAGTCGG CCTACCTGTCTCGTCGTCTTGCCTACTTTTGTGCCCGGCGTCTTTCCTTGCTGCTGAGCGATAGCCCCAACCTCCTTGCTGCCCACTCAC CCCACATGATGATAGGACCAAACAACTCGAGTATCGGGGCCCCCAGCCCTGGCCCCCCCGGCCCTGGCATGAGCCCCGTGCAGCTGGCCT TCTCAGATTTTCTTTCCTGTGCACAGCATGGTCCCCTGGTTTATGGACTTAGTTGTATGTTGCAGACTGTCACTCTCTGTTGCCCAAGTG CCTTGGTGTGGAATTATTCCACAAATGAAAATAAGAGCGCAAACCCAGGCTCACCCCTGGATCTGCTGCAGGTGGCCCCGTCCAGCCTCC CCATGCCGGGTGGGAACACGGCTTTCAATCAGCAGGTTCGGGCAAGGATTTATGAAGTAGAACAACAGATAAAACAAAGAGGCCGTGCAG TGGAAGTTCGGTGGTCATTTGACAAGTGCCAAGAATCCACAGCAGGGGTGACTATTAGTCGGGTTTTGCACACGTTGGAAGTTTTGGATC GTCACTGTTTTGACCGAACTGATTCCAGCAATTCCATGGAGACACTTTATCATAAGATTTTCTGGGCAAACCAAAACAAAGATAACCAAG AGGTTGCGCCCAACGATGAAGCTGTGGTGACGCTGTTATGTGAATGGGCCGTGAGCTGCAAACGGTCTGGCAAGCACAGGGCCATGGCTG TGGCAAAACTCCTGGAGAAGAGGCAAGCAGAAATTGAGGCAGAGAGATGTGGTGAATCAGAAGTCTTAGATGAGAAGGAGTCTATTTCTT CATCCTCTCTTGCTGGATCCAGTTTGCCTGTTTTCCAGAATGTGCTGTTAAGGTTTTTAGATACACAGGCCCCCTCTTTGTCGGACCCAA ACAGTGAATGTGAAAAGGTGGAATTTGTGAACCTGGTGCTGCTCTTCTGCGAGTTCATCCGCCATGATGTCTTCTCCCATGACGCATACA TGTGTACCCTCATATCTCGAGGAGATTTGTCAGTCACTGCCTCAACTCGGCCGCGGTCACCAGTAGGGGAAAATGCAGATGAACACTATT CAAAGGACCATGATGTGAAAATGGAGATTTTTTCTCCTATGCCTGGAGAATCCTGTGAGAATGCCAACACTTCGTTGGGCAGAAGAATGT CAGTTAATTGTGAGAAGTTGGTGAAGAGGGAAAAGCCAAGGGAATTAATTTTTCCATCTAATTATGACCTCCTTCGCCACTTACAGTATG CAACACATTTTCCTATACCTCTGAGGGCAGCTGTAGAAGATATAAACCCAGCAGATGACCCTAATAATCAAGGTGAGGATGAATTTGAAG AAGCCGAGCAAGTGAGAGAAGAAAATTTGCCAGATGAAAATGAAGAGCAAAAACAAAGTAATCAAAAGCAAGAGAATACAGAAGTGGAGG AACATTTGGTGATGGCAGGAAATCCAGACCAGCAGGAGGACAATGTTGATGAACAGTACCAGGAAGAGGCAGAAGAGGAGGTTCAGGAAG ATTTGACTGAAGAGAAAAAAAGGGAACTGGAGCATAATGCTGAAGAGACCTATGGTGAAAATGATGAAAATACTGATGATAAAAATAATG ATGGAGAAGAGCAAGAAGTTCGAGATGACAACCGCCCCAAAGGCCGAGAGGAACACTACGAGGAGGAAGAAGAGGAGGAAGAAGACGGGG >52538_52538_3_MED12L-GOLIM4_MED12L_chr3_150911453_ENST00000474524_GOLIM4_chr3_167742883_ENST00000309027_length(amino acids)=870AA_BP=715 MAAFGLLSYEQRPLKRPRLGPPDVYPQDPKQKEDELTAVNVKQGFNNQPAFTGDEHGSARNIVINPSKIGAYFSSILAEKLKLNTFQDTG KKKPQVNAKDNYWLVTARSQSAIHSWFSDLAGNKPLSILAKKVPILSKKEDVFAYLAKYSVPMVRATWLIKMTCAYYSAISEAKIKKRQA PDPNLEWTQISTRYLREQLAKISDFYHMASSTGDGPVPVPPEVEQAMKQWEYNEKLAFHMFQEGMLEKHEYLTWILDVLEKIRPMDDDLL KLLLPLMLQYSDEFVQSAYLSRRLAYFCARRLSLLLSDSPNLLAAHSPHMMIGPNNSSIGAPSPGPPGPGMSPVQLAFSDFLSCAQHGPL VYGLSCMLQTVTLCCPSALVWNYSTNENKSANPGSPLDLLQVAPSSLPMPGGNTAFNQQVRARIYEVEQQIKQRGRAVEVRWSFDKCQES TAGVTISRVLHTLEVLDRHCFDRTDSSNSMETLYHKIFWANQNKDNQEVAPNDEAVVTLLCEWAVSCKRSGKHRAMAVAKLLEKRQAEIE AERCGESEVLDEKESISSSSLAGSSLPVFQNVLLRFLDTQAPSLSDPNSECEKVEFVNLVLLFCEFIRHDVFSHDAYMCTLISRGDLSVT ASTRPRSPVGENADEHYSKDHDVKMEIFSPMPGESCENANTSLGRRMSVNCEKLVKREKPRELIFPSNYDLLRHLQYATHFPIPLRAAVE DINPADDPNNQGEDEFEEAEQVREENLPDENEEQKQSNQKQENTEVEEHLVMAGNPDQQEDNVDEQYQEEAEEEVQEDLTEEKKRELEHN -------------------------------------------------------------- >52538_52538_4_MED12L-GOLIM4_MED12L_chr3_150911453_ENST00000474524_GOLIM4_chr3_167742883_ENST00000470487_length(transcript)=4243nt_BP=2183nt CCAACTCTCATTCATTTCGCCGGTTAACATGAGAGATCATGGCCGCCTTCGGGCTTCTCAGCTATGAGCAGAGACCGCTGAAGCGCCCCC GGCTCGGGCCGCCCGACGTCTACCCACAGGACCCCAAGCAGAAGGAGGATGAACTTACTGCTGTGAATGTAAAGCAAGGCTTCAATAATC AGCCAGCCTTCACTGGAGATGAACATGGCTCAGCCAGAAATATTGTAATTAACCCATCAAAGATTGGAGCTTATTTTAGCAGCATATTAG CTGAGAAACTGAAGCTTAACACTTTCCAGGACACGGGAAAGAAGAAACCACAAGTTAATGCTAAAGATAATTATTGGCTGGTTACTGCTC GATCCCAGAGTGCAATTCATAGTTGGTTTTCTGACTTAGCAGGAAATAAGCCACTTTCTATTTTGGCAAAAAAGGTTCCTATCCTTAGTA AAAAAGAGGATGTTTTTGCATATTTAGCTAAATATTCTGTGCCAATGGTTCGAGCAACGTGGCTGATCAAGATGACTTGTGCCTATTATT CTGCTATATCTGAAGCTAAAATTAAGAAACGTCAGGCTCCTGATCCGAATTTGGAGTGGACACAGATATCTACCAGATATCTTCGAGAGC AGTTGGCCAAGATTTCTGACTTTTACCACATGGCCTCCAGCACGGGCGATGGCCCTGTCCCTGTGCCACCAGAGGTGGAGCAAGCCATGA AGCAATGGGAATACAACGAAAAGCTAGCATTTCACATGTTCCAGGAAGGAATGTTAGAAAAACACGAATATTTGACATGGATCCTGGATG TTTTAGAAAAGATCAGACCAATGGATGATGATCTTCTTAAACTCTTGCTACCACTAATGCTGCAGTATTCAGATGAGTTTGTTCAGTCGG CCTACCTGTCTCGTCGTCTTGCCTACTTTTGTGCCCGGCGTCTTTCCTTGCTGCTGAGCGATAGCCCCAACCTCCTTGCTGCCCACTCAC CCCACATGATGATAGGACCAAACAACTCGAGTATCGGGGCCCCCAGCCCTGGCCCCCCCGGCCCTGGCATGAGCCCCGTGCAGCTGGCCT TCTCAGATTTTCTTTCCTGTGCACAGCATGGTCCCCTGGTTTATGGACTTAGTTGTATGTTGCAGACTGTCACTCTCTGTTGCCCAAGTG CCTTGGTGTGGAATTATTCCACAAATGAAAATAAGAGCGCAAACCCAGGCTCACCCCTGGATCTGCTGCAGGTGGCCCCGTCCAGCCTCC CCATGCCGGGTGGGAACACGGCTTTCAATCAGCAGGTTCGGGCAAGGATTTATGAAGTAGAACAACAGATAAAACAAAGAGGCCGTGCAG TGGAAGTTCGGTGGTCATTTGACAAGTGCCAAGAATCCACAGCAGGGGTGACTATTAGTCGGGTTTTGCACACGTTGGAAGTTTTGGATC GTCACTGTTTTGACCGAACTGATTCCAGCAATTCCATGGAGACACTTTATCATAAGATTTTCTGGGCAAACCAAAACAAAGATAACCAAG AGGTTGCGCCCAACGATGAAGCTGTGGTGACGCTGTTATGTGAATGGGCCGTGAGCTGCAAACGGTCTGGCAAGCACAGGGCCATGGCTG TGGCAAAACTCCTGGAGAAGAGGCAAGCAGAAATTGAGGCAGAGAGATGTGGTGAATCAGAAGTCTTAGATGAGAAGGAGTCTATTTCTT CATCCTCTCTTGCTGGATCCAGTTTGCCTGTTTTCCAGAATGTGCTGTTAAGGTTTTTAGATACACAGGCCCCCTCTTTGTCGGACCCAA ACAGTGAATGTGAAAAGGTGGAATTTGTGAACCTGGTGCTGCTCTTCTGCGAGTTCATCCGCCATGATGTCTTCTCCCATGACGCATACA TGTGTACCCTCATATCTCGAGGAGATTTGTCAGTCACTGCCTCAACTCGGCCGCGGTCACCAGTAGGGGAAAATGCAGATGAACACTATT CAAAGGACCATGATGTGAAAATGGAGATTTTTTCTCCTATGCCTGGAGAATCCTGTGAGAATGCCAACACTTCGTTGGGCAGAAGAATGT CAGTTAATTGTGAGAAGTTGGTGAAGAGGGAAAAGCCAAGGGAATTAATTTTTCCATCTAATTATGACCTCCTTCGCCACTTACAGTATG CAACACATTTTCCTATACCTCTGAGGGCAGCTGTAGAAGATATAAACCCAGCAGATGACCCTAATAATCAAGGTGAGGATGAATTTGAAG AAGCCGAGCAAGTGAGAGAAGAAAATTTGCCAGATGAAAATGAAGAGCAAAAACAAAGTAATCAAAAGCAAGAGAATACAGAAGTGGAGG AACATTTGGTGATGGCAGGAAATCCAGACCAGCAGGAGGACAATGTTGATGAACAGTACCAGGAAGAGGCAGAAGAGGAGGTTCAGGAAG ATTTGACTGAAGAGAAAAAAAGGGAACTGGAGCATAATGCTGAAGAGACCTATGGTGAAAATGATGAAAATACTGATGATAAAAATAATG ATGGAGAAGAGCAAGAAGTTCGAGATGACAACCGCCCCAAAGGCCGAGAGGAACACTACGAGGAGGAAGAAGAGGAGGAAGAAGACGGGG CTGCAGTTGCTGAGAAATCACATCGAAGAGCTGAAATGTAGCGGCACCCAATTTCTAGACAACGCTCAGCCAACGGATTCTTTTCAAGCT GCTCAAACATAAATCTGCCTACTGAACTCTAGGATATTTAATTACAAAAATTAAGAACTTAGACTTTTTTAAAACTTTTGTATTAGAAAT GCGCATACATTTATATGAATATATTTTGATAACGTAGGTCTAGAGCTTCTTTTATATTCAAGCTAAACATGAAAAAGAAGAAAAACAATA AAGTAAACCTGAGCCCCCACGTCCCAATTTTTTTAATAGATTATGTGATGTTGGAAAGCTCATTGATTTTGTATATGTTTCAGCGTGTTA CCTTTCTGGCTTCCAGTTCCCAGGTGTTCTTTGTTTGCCTTTGATAAAATACAGGATTTAAGAACAGAGAGTACTGCAAAATGCCATGCA GACTTTAAAGAGAATGGCCTGTTTACTAATTGCTGCCCTTCTGATGTCTTTATGTATAGCTCTGATAGAATTTTCACCAGTCTATGTATC TCTGGAGTGAGATCTATGTACAAAGTGACATACAATTGGAAATCCATTTTTGTTGTAAAGACATTGTTTTTCAGACTTTTCAGATCAATT AGAAAAATGTCATTGCTTTAAAATCATAGCTGTTCTGTTTAAGAGGAATTGAATTTAAAAATGAGAGAGTATTAAAACTCATGTGGCAGT ATCCTGGTCTTAATCAGGTATTGCAGTGTTCAAACAAGGTATGGACACATCAACATATTCACCTTTTGGAAGGACTAGTGACTTGGTTAA AAACTAATGAGTGTTTGCGTGTGGTGTTTCTATGCGAGCACCCATTCTCAATTTTTTTTTTTTTTTTTTGTTTGAAGCCATTGATTGATA TTCTTGTTTCAGTGTCTGGTTCCCTATAAGTAATATATATTTCCTCTTTAAGCTCTGTGCCTCAATATGCAATCACTTTGATAATAAGTA TAAATATGAAGACATACCAAACTGCAAATAGGATTATTTTTCAAAACTATATGGTGAAACATATGGTGATATGATAGACTTATGGTTTTT CTGTTTGCTGGGATTGGGTCTGCTTTTTGCCTATGTGTAAGGCATTTTAATCATTGTGAATATAATAGATTCCTTAAAATAAGAAAATTA AAAATCAGAAACTTCCCTTGGAGTAGAGACATTTCAGTTGCAAGCATAAAGATTTTTAAATATATAACACTAGGTTATATAATTCATATA TGTGTGGTTTTGTATTAATAATCAAATTAATAAAAACATTTCATTTTCTACCATTGTATGAACGTAATTTTATTTTTTAATAACCACACT CATAGATGGTTTTGATTGTTCCAGTTGAGTGATTGTGTATGTACGCATAGGAATGTTTTGTTCAGTTGTACTTTGTATTTCCAATAATCA GAAATCTTACCTTTATGTATAAAAACCCATTATGTAATGTAAAATGTATAATATTGTACATAATATTCAGAATACAATTATTTTGGTAAA TTAGCTTGTATTTTTAATGTCCCAGTTGCAGTATTTAATTATTTGAACCATACTGAAAGTTGGTAATAGTCAATAAAGCTAAAAAAAAAA >52538_52538_4_MED12L-GOLIM4_MED12L_chr3_150911453_ENST00000474524_GOLIM4_chr3_167742883_ENST00000470487_length(amino acids)=870AA_BP=715 MAAFGLLSYEQRPLKRPRLGPPDVYPQDPKQKEDELTAVNVKQGFNNQPAFTGDEHGSARNIVINPSKIGAYFSSILAEKLKLNTFQDTG KKKPQVNAKDNYWLVTARSQSAIHSWFSDLAGNKPLSILAKKVPILSKKEDVFAYLAKYSVPMVRATWLIKMTCAYYSAISEAKIKKRQA PDPNLEWTQISTRYLREQLAKISDFYHMASSTGDGPVPVPPEVEQAMKQWEYNEKLAFHMFQEGMLEKHEYLTWILDVLEKIRPMDDDLL KLLLPLMLQYSDEFVQSAYLSRRLAYFCARRLSLLLSDSPNLLAAHSPHMMIGPNNSSIGAPSPGPPGPGMSPVQLAFSDFLSCAQHGPL VYGLSCMLQTVTLCCPSALVWNYSTNENKSANPGSPLDLLQVAPSSLPMPGGNTAFNQQVRARIYEVEQQIKQRGRAVEVRWSFDKCQES TAGVTISRVLHTLEVLDRHCFDRTDSSNSMETLYHKIFWANQNKDNQEVAPNDEAVVTLLCEWAVSCKRSGKHRAMAVAKLLEKRQAEIE AERCGESEVLDEKESISSSSLAGSSLPVFQNVLLRFLDTQAPSLSDPNSECEKVEFVNLVLLFCEFIRHDVFSHDAYMCTLISRGDLSVT ASTRPRSPVGENADEHYSKDHDVKMEIFSPMPGESCENANTSLGRRMSVNCEKLVKREKPRELIFPSNYDLLRHLQYATHFPIPLRAAVE DINPADDPNNQGEDEFEEAEQVREENLPDENEEQKQSNQKQENTEVEEHLVMAGNPDQQEDNVDEQYQEEAEEEVQEDLTEEKKRELEHN -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for MED12L-GOLIM4 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for MED12L-GOLIM4 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for MED12L-GOLIM4 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
