
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:MED15-LYZ (FusionGDB2 ID:52617) |
Fusion Gene Summary for MED15-LYZ |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: MED15-LYZ | Fusion gene ID: 52617 | Hgene | Tgene | Gene symbol | MED15 | LYZ | Gene ID | 51586 | 4069 |
| Gene name | mediator complex subunit 15 | lysozyme | |
| Synonyms | ARC105|CAG7A|CTG7A|PCQAP|TIG-1|TIG1|TNRC7 | LYZF1|LZM | |
| Cytomap | 22q11.21 | 12q15 | |
| Type of gene | protein-coding | protein-coding | |
| Description | mediator of RNA polymerase II transcription subunit 15CTG repeat protein 7aPC2 (positive cofactor 2, multiprotein complex) glutamine/Q-rich-associated proteinPC2 glutamine/Q-rich-associated proteinPC2-glutamine-rich-associated proteinTPA inducible ge | lysozyme C1,4-beta-N-acetylmuramidase Cc-type lysozymelysozyme F1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q96RN5 | P61626 | |
| Ensembl transtripts involved in fusion gene | ENST00000263205, ENST00000292733, ENST00000382974, ENST00000406969, ENST00000425759, ENST00000541476, ENST00000478831, ENST00000542773, | ENST00000548839, ENST00000261267, ENST00000549690, | |
| Fusion gene scores | * DoF score | 17 X 11 X 9=1683 | 51 X 27 X 13=17901 |
| # samples | 20 | 60 | |
| ** MAII score | log2(20/1683*10)=-3.07296327155522 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(60/17901*10)=-4.89893387178018 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: MED15 [Title/Abstract] AND LYZ [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | MED15(20905774)-LYZ(69743888), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | MED15-LYZ seems lost the major protein functional domain in Hgene partner, which is a cell metabolism gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | LYZ | GO:0031640 | killing of cells of other organism | 9727055 |
| Tgene | LYZ | GO:0042742 | defense response to bacterium | 21093056 |
| Tgene | LYZ | GO:0050830 | defense response to Gram-positive bacterium | 21093056 |
 Fusion gene breakpoints across MED15 (5'-gene) Fusion gene breakpoints across MED15 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
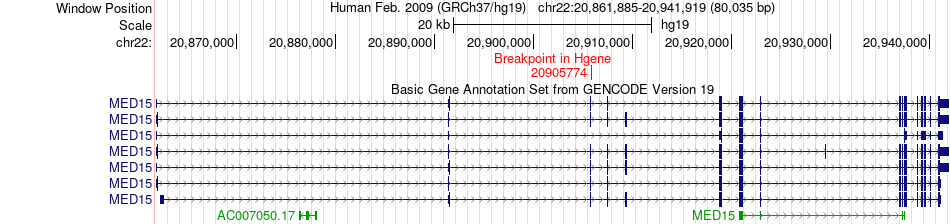 |
 Fusion gene breakpoints across LYZ (3'-gene) Fusion gene breakpoints across LYZ (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
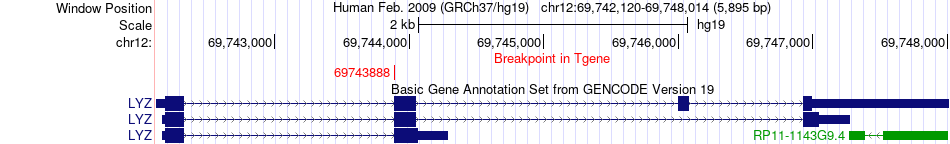 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-HU-A4GH-01A | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + |
Top |
Fusion Gene ORF analysis for MED15-LYZ |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| Frame-shift | ENST00000263205 | ENST00000548839 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + |
| Frame-shift | ENST00000292733 | ENST00000548839 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + |
| Frame-shift | ENST00000382974 | ENST00000548839 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + |
| Frame-shift | ENST00000406969 | ENST00000548839 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + |
| Frame-shift | ENST00000425759 | ENST00000548839 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + |
| Frame-shift | ENST00000541476 | ENST00000548839 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + |
| In-frame | ENST00000263205 | ENST00000261267 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + |
| In-frame | ENST00000263205 | ENST00000549690 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + |
| In-frame | ENST00000292733 | ENST00000261267 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + |
| In-frame | ENST00000292733 | ENST00000549690 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + |
| In-frame | ENST00000382974 | ENST00000261267 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + |
| In-frame | ENST00000382974 | ENST00000549690 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + |
| In-frame | ENST00000406969 | ENST00000261267 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + |
| In-frame | ENST00000406969 | ENST00000549690 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + |
| In-frame | ENST00000425759 | ENST00000261267 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + |
| In-frame | ENST00000425759 | ENST00000549690 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + |
| In-frame | ENST00000541476 | ENST00000261267 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + |
| In-frame | ENST00000541476 | ENST00000549690 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + |
| intron-3CDS | ENST00000478831 | ENST00000261267 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + |
| intron-3CDS | ENST00000478831 | ENST00000548839 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + |
| intron-3CDS | ENST00000478831 | ENST00000549690 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + |
| intron-3CDS | ENST00000542773 | ENST00000261267 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + |
| intron-3CDS | ENST00000542773 | ENST00000548839 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + |
| intron-3CDS | ENST00000542773 | ENST00000549690 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000425759 | MED15 | chr22 | 20905774 | + | ENST00000261267 | LYZ | chr12 | 69743888 | + | 1614 | 288 | 80 | 598 | 172 |
| ENST00000425759 | MED15 | chr22 | 20905774 | + | ENST00000549690 | LYZ | chr12 | 69743888 | + | 796 | 288 | 80 | 565 | 161 |
| ENST00000292733 | MED15 | chr22 | 20905774 | + | ENST00000261267 | LYZ | chr12 | 69743888 | + | 1611 | 285 | 77 | 595 | 172 |
| ENST00000292733 | MED15 | chr22 | 20905774 | + | ENST00000549690 | LYZ | chr12 | 69743888 | + | 793 | 285 | 77 | 562 | 161 |
| ENST00000263205 | MED15 | chr22 | 20905774 | + | ENST00000261267 | LYZ | chr12 | 69743888 | + | 1603 | 277 | 69 | 587 | 172 |
| ENST00000263205 | MED15 | chr22 | 20905774 | + | ENST00000549690 | LYZ | chr12 | 69743888 | + | 785 | 277 | 69 | 554 | 161 |
| ENST00000406969 | MED15 | chr22 | 20905774 | + | ENST00000261267 | LYZ | chr12 | 69743888 | + | 1584 | 258 | 2 | 568 | 188 |
| ENST00000406969 | MED15 | chr22 | 20905774 | + | ENST00000549690 | LYZ | chr12 | 69743888 | + | 766 | 258 | 2 | 535 | 177 |
| ENST00000382974 | MED15 | chr22 | 20905774 | + | ENST00000261267 | LYZ | chr12 | 69743888 | + | 1572 | 246 | 38 | 556 | 172 |
| ENST00000382974 | MED15 | chr22 | 20905774 | + | ENST00000549690 | LYZ | chr12 | 69743888 | + | 754 | 246 | 38 | 523 | 161 |
| ENST00000541476 | MED15 | chr22 | 20905774 | + | ENST00000261267 | LYZ | chr12 | 69743888 | + | 1859 | 533 | 403 | 843 | 146 |
| ENST00000541476 | MED15 | chr22 | 20905774 | + | ENST00000549690 | LYZ | chr12 | 69743888 | + | 1041 | 533 | 403 | 810 | 135 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000425759 | ENST00000261267 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + | 0.007862827 | 0.99213713 |
| ENST00000425759 | ENST00000549690 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + | 0.028583305 | 0.9714167 |
| ENST00000292733 | ENST00000261267 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + | 0.007562958 | 0.99243706 |
| ENST00000292733 | ENST00000549690 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + | 0.02739554 | 0.9726044 |
| ENST00000263205 | ENST00000261267 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + | 0.007315783 | 0.9926842 |
| ENST00000263205 | ENST00000549690 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + | 0.02819218 | 0.9718078 |
| ENST00000406969 | ENST00000261267 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + | 0.011697226 | 0.98830277 |
| ENST00000406969 | ENST00000549690 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + | 0.045514546 | 0.9544855 |
| ENST00000382974 | ENST00000261267 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + | 0.007737746 | 0.99226224 |
| ENST00000382974 | ENST00000549690 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + | 0.03125221 | 0.9687478 |
| ENST00000541476 | ENST00000261267 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + | 0.017584732 | 0.98241526 |
| ENST00000541476 | ENST00000549690 | MED15 | chr22 | 20905774 | + | LYZ | chr12 | 69743888 | + | 0.077965915 | 0.9220341 |
Top |
Fusion Genomic Features for MED15-LYZ |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for MED15-LYZ |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr22:20905774/chr12:69743888) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MED15 | LYZ |
| FUNCTION: Component of the Mediator complex, a coactivator involved in the regulated transcription of nearly all RNA polymerase II-dependent genes. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. Mediator is recruited to promoters by direct interactions with regulatory proteins and serves as a scaffold for the assembly of a functional preinitiation complex with RNA polymerase II and the general transcription factors. Required for cholesterol-dependent gene regulation. Positively regulates the Nodal signaling pathway. {ECO:0000269|PubMed:12167862, ECO:0000269|PubMed:16630888, ECO:0000269|PubMed:16799563}. | FUNCTION: Lysozymes have primarily a bacteriolytic function; those in tissues and body fluids are associated with the monocyte-macrophage system and enhance the activity of immunoagents. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000263205 | + | 3 | 18 | 161_174 | 69 | 789.0 | Compositional bias | Note=Poly-Gln |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000263205 | + | 3 | 18 | 178_193 | 69 | 789.0 | Compositional bias | Note=Poly-Gln |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000263205 | + | 3 | 18 | 205_218 | 69 | 789.0 | Compositional bias | Note=Poly-Gln |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000263205 | + | 3 | 18 | 226_239 | 69 | 789.0 | Compositional bias | Note=Poly-Gln |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000263205 | + | 3 | 18 | 243_262 | 69 | 789.0 | Compositional bias | Note=Poly-Gln |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000263205 | + | 3 | 18 | 266_515 | 69 | 789.0 | Compositional bias | Note=Pro-rich |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000263205 | + | 3 | 18 | 360_367 | 69 | 789.0 | Compositional bias | Note=Poly-Gln |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000263205 | + | 3 | 18 | 449_456 | 69 | 789.0 | Compositional bias | Note=Poly-Pro |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000263205 | + | 3 | 18 | 602_611 | 69 | 789.0 | Compositional bias | Note=Poly-Pro |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000292733 | + | 3 | 17 | 161_174 | 69 | 749.0 | Compositional bias | Note=Poly-Gln |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000292733 | + | 3 | 17 | 178_193 | 69 | 749.0 | Compositional bias | Note=Poly-Gln |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000292733 | + | 3 | 17 | 205_218 | 69 | 749.0 | Compositional bias | Note=Poly-Gln |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000292733 | + | 3 | 17 | 226_239 | 69 | 749.0 | Compositional bias | Note=Poly-Gln |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000292733 | + | 3 | 17 | 243_262 | 69 | 749.0 | Compositional bias | Note=Poly-Gln |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000292733 | + | 3 | 17 | 266_515 | 69 | 749.0 | Compositional bias | Note=Pro-rich |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000292733 | + | 3 | 17 | 360_367 | 69 | 749.0 | Compositional bias | Note=Poly-Gln |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000292733 | + | 3 | 17 | 449_456 | 69 | 749.0 | Compositional bias | Note=Poly-Pro |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000292733 | + | 3 | 17 | 602_611 | 69 | 749.0 | Compositional bias | Note=Poly-Pro |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000382974 | + | 3 | 16 | 161_174 | 69 | 678.0 | Compositional bias | Note=Poly-Gln |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000382974 | + | 3 | 16 | 178_193 | 69 | 678.0 | Compositional bias | Note=Poly-Gln |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000382974 | + | 3 | 16 | 205_218 | 69 | 678.0 | Compositional bias | Note=Poly-Gln |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000382974 | + | 3 | 16 | 226_239 | 69 | 678.0 | Compositional bias | Note=Poly-Gln |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000382974 | + | 3 | 16 | 243_262 | 69 | 678.0 | Compositional bias | Note=Poly-Gln |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000382974 | + | 3 | 16 | 266_515 | 69 | 678.0 | Compositional bias | Note=Pro-rich |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000382974 | + | 3 | 16 | 360_367 | 69 | 678.0 | Compositional bias | Note=Poly-Gln |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000382974 | + | 3 | 16 | 449_456 | 69 | 678.0 | Compositional bias | Note=Poly-Pro |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000382974 | + | 3 | 16 | 602_611 | 69 | 678.0 | Compositional bias | Note=Poly-Pro |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000263205 | + | 3 | 18 | 547_564 | 69 | 789.0 | Motif | Nuclear localization signal |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000292733 | + | 3 | 17 | 547_564 | 69 | 749.0 | Motif | Nuclear localization signal |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000382974 | + | 3 | 16 | 547_564 | 69 | 678.0 | Motif | Nuclear localization signal |
| Tgene | LYZ | chr22:20905774 | chr12:69743888 | ENST00000261267 | 0 | 4 | 19_148 | 45 | 149.0 | Domain | C-type lysozyme |
Top |
Fusion Gene Sequence for MED15-LYZ |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >52617_52617_1_MED15-LYZ_MED15_chr22_20905774_ENST00000263205_LYZ_chr12_69743888_ENST00000261267_length(transcript)=1603nt_BP=277nt GGCTCTGTGACTGAGGCGGCGGCGGTGGCGGCCAAGCGGGATACGGGCGGCGGGAGCTGGGGAACAGGCATGGACGTTTCCGGGCAAGAG ACCGACTGGCGGAGCACCGCCTTCCGGCAGAAGCTGGTCAGTCAAATCGAGGATGCCATGAGGAAAGCTGGTGTGGCACACAGTAAATCC AGCAAGGATATGGAGAGCCATGTTTTCCTGAAGGCCAAGACCCGGGACGAATACCTTTCTCTCGTGGCCAGGCTCATTATCCATTTTCGA GACATTCGGATGTGTTTGGCCAAATGGGAGAGTGGTTACAACACACGAGCTACAAACTACAATGCTGGAGACAGAAGCACTGATTATGGG ATATTTCAGATCAATAGCCGCTACTGGTGTAATGATGGCAAAACCCCAGGAGCAGTTAATGCCTGTCATTTATCCTGCAGTGCTTTGCTG CAAGATAACATCGCTGATGCTGTAGCTTGTGCAAAGAGGGTTGTCCGTGATCCACAAGGCATTAGAGCATGGGTGGCATGGAGAAATCGT TGTCAAAACAGAGATGTCCGTCAGTATGTTCAAGGTTGTGGAGTGTAACTCCAGAATTTTCCTTCTTCAGCTCATTTTGTCTCTCTCACA TTAAGGGAGTAGGAATTAAGTGAAAGGTCACACTACCATTATTTCCCCTTCAAACAAATAATATTTTTACAGAAGCAGGAGCAAAATATG GCCTTTCTTCTAAGAGATATAATGTTCACTAATGTGGTTATTTTACATTAAGCCTACAACATTTTTCAGTTTGCAAATAGAACTAATACT GGTGAAAATTTACCTAAAACCTTGGTTATCAAATACATCTCCAGTACATTCCGTTCTTTTTTTTTTTGAGACAGTCTCGCTCTGTCGCCC AGGCTGGAGTGCAGTGGCGCAATCTCGGCTCACTGCAACCTCCACCTCCCGGGTTCACGCCATTCTCCTGCCTCAGCCTCCCGAGTAGCT GGGATTACGGGCGCCCGCCACCACGCCCGGCTAATTTTTTGTATTTTTAGTAGAGACAGGGTTTCACCGTGTTAGCCAGGATGGTCTCGA TCTCCTGACCTTGTGATCCACCCACCTCGGCCTCCCAAAGTGCTGGGATTACAGGCGTGAGCCACTGCGCCCGGCCACATTCAGTTCTTA TCAAAGAAATAACCCAGACTTAATCTTGAATGATACGATTATGCCCAATATTAAGTAAAAAATATAAGAAAAGGTTATCTTAAATAGATC TTAGGCAAAATACCAGCTGATGAAGGCATCTGATGCCTTCATCTGTTCAGTCATCTCCAAAAACAGTAAAAATAACCACTTTTTGTTGGG CAATATGAAATTTTTAAAGGAGTAGAATACCAAATGATAGAAACAGACTGCCTGAATTGAGAATTTTGATTTCTTAAAGTGTGTTTCTTT CTAAATTGCTGTTCCTTAATTTGATTAATTTAATTCATGTATTATGATTAAATCTGAGGCAGATGAGCTTACAAGTATTGAAATAATTAC >52617_52617_1_MED15-LYZ_MED15_chr22_20905774_ENST00000263205_LYZ_chr12_69743888_ENST00000261267_length(amino acids)=172AA_BP=70 MDVSGQETDWRSTAFRQKLVSQIEDAMRKAGVAHSKSSKDMESHVFLKAKTRDEYLSLVARLIIHFRDIRMCLAKWESGYNTRATNYNAG -------------------------------------------------------------- >52617_52617_2_MED15-LYZ_MED15_chr22_20905774_ENST00000263205_LYZ_chr12_69743888_ENST00000549690_length(transcript)=785nt_BP=277nt GGCTCTGTGACTGAGGCGGCGGCGGTGGCGGCCAAGCGGGATACGGGCGGCGGGAGCTGGGGAACAGGCATGGACGTTTCCGGGCAAGAG ACCGACTGGCGGAGCACCGCCTTCCGGCAGAAGCTGGTCAGTCAAATCGAGGATGCCATGAGGAAAGCTGGTGTGGCACACAGTAAATCC AGCAAGGATATGGAGAGCCATGTTTTCCTGAAGGCCAAGACCCGGGACGAATACCTTTCTCTCGTGGCCAGGCTCATTATCCATTTTCGA GACATTCGGATGTGTTTGGCCAAATGGGAGAGTGGTTACAACACACGAGCTACAAACTACAATGCTGGAGACAGAAGCACTGATTATGGG ATATTTCAGATCAATAGCCGCTACTGGTGTAATGATGGCAAAACCCCAGGAGCAGTTAATGCCTGTCATTTATCCTGCAGTGGGTGGCAT GGAGAAATCGTTGTCAAAACAGAGATGTCCGTCAGTATGTTCAAGGTTGTGGAGTGTAACTCCAGAATTTTCCTTCTTCAGCTCATTTTG TCTCTCTCACATTAAGGGAGTAGGAATTAAGTGAAAGGTCACACTACCATTATTTCCCCTTCAAACAAATAATATTTTTACAGAAGCAGG AGCAAAATATGGCCTTTCTTCTAAGAGATATAATGTTCACTAATGTGGTTATTTTACATTAAGCCTACAACATTTTTCAGTTTGCAAATA >52617_52617_2_MED15-LYZ_MED15_chr22_20905774_ENST00000263205_LYZ_chr12_69743888_ENST00000549690_length(amino acids)=161AA_BP=70 MDVSGQETDWRSTAFRQKLVSQIEDAMRKAGVAHSKSSKDMESHVFLKAKTRDEYLSLVARLIIHFRDIRMCLAKWESGYNTRATNYNAG -------------------------------------------------------------- >52617_52617_3_MED15-LYZ_MED15_chr22_20905774_ENST00000292733_LYZ_chr12_69743888_ENST00000261267_length(transcript)=1611nt_BP=285nt TTGGGCCTGGCTCTGTGACTGAGGCGGCGGCGGTGGCGGCCAAGCGGGATACGGGCGGCGGGAGCTGGGGAACAGGCATGGACGTTTCCG GGCAAGAGACCGACTGGCGGAGCACCGCCTTCCGGCAGAAGCTGGTCAGTCAAATCGAGGATGCCATGAGGAAAGCTGGTGTGGCACACA GTAAATCCAGCAAGGATATGGAGAGCCATGTTTTCCTGAAGGCCAAGACCCGGGACGAATACCTTTCTCTCGTGGCCAGGCTCATTATCC ATTTTCGAGACATTCGGATGTGTTTGGCCAAATGGGAGAGTGGTTACAACACACGAGCTACAAACTACAATGCTGGAGACAGAAGCACTG ATTATGGGATATTTCAGATCAATAGCCGCTACTGGTGTAATGATGGCAAAACCCCAGGAGCAGTTAATGCCTGTCATTTATCCTGCAGTG CTTTGCTGCAAGATAACATCGCTGATGCTGTAGCTTGTGCAAAGAGGGTTGTCCGTGATCCACAAGGCATTAGAGCATGGGTGGCATGGA GAAATCGTTGTCAAAACAGAGATGTCCGTCAGTATGTTCAAGGTTGTGGAGTGTAACTCCAGAATTTTCCTTCTTCAGCTCATTTTGTCT CTCTCACATTAAGGGAGTAGGAATTAAGTGAAAGGTCACACTACCATTATTTCCCCTTCAAACAAATAATATTTTTACAGAAGCAGGAGC AAAATATGGCCTTTCTTCTAAGAGATATAATGTTCACTAATGTGGTTATTTTACATTAAGCCTACAACATTTTTCAGTTTGCAAATAGAA CTAATACTGGTGAAAATTTACCTAAAACCTTGGTTATCAAATACATCTCCAGTACATTCCGTTCTTTTTTTTTTTGAGACAGTCTCGCTC TGTCGCCCAGGCTGGAGTGCAGTGGCGCAATCTCGGCTCACTGCAACCTCCACCTCCCGGGTTCACGCCATTCTCCTGCCTCAGCCTCCC GAGTAGCTGGGATTACGGGCGCCCGCCACCACGCCCGGCTAATTTTTTGTATTTTTAGTAGAGACAGGGTTTCACCGTGTTAGCCAGGAT GGTCTCGATCTCCTGACCTTGTGATCCACCCACCTCGGCCTCCCAAAGTGCTGGGATTACAGGCGTGAGCCACTGCGCCCGGCCACATTC AGTTCTTATCAAAGAAATAACCCAGACTTAATCTTGAATGATACGATTATGCCCAATATTAAGTAAAAAATATAAGAAAAGGTTATCTTA AATAGATCTTAGGCAAAATACCAGCTGATGAAGGCATCTGATGCCTTCATCTGTTCAGTCATCTCCAAAAACAGTAAAAATAACCACTTT TTGTTGGGCAATATGAAATTTTTAAAGGAGTAGAATACCAAATGATAGAAACAGACTGCCTGAATTGAGAATTTTGATTTCTTAAAGTGT GTTTCTTTCTAAATTGCTGTTCCTTAATTTGATTAATTTAATTCATGTATTATGATTAAATCTGAGGCAGATGAGCTTACAAGTATTGAA >52617_52617_3_MED15-LYZ_MED15_chr22_20905774_ENST00000292733_LYZ_chr12_69743888_ENST00000261267_length(amino acids)=172AA_BP=70 MDVSGQETDWRSTAFRQKLVSQIEDAMRKAGVAHSKSSKDMESHVFLKAKTRDEYLSLVARLIIHFRDIRMCLAKWESGYNTRATNYNAG -------------------------------------------------------------- >52617_52617_4_MED15-LYZ_MED15_chr22_20905774_ENST00000292733_LYZ_chr12_69743888_ENST00000549690_length(transcript)=793nt_BP=285nt TTGGGCCTGGCTCTGTGACTGAGGCGGCGGCGGTGGCGGCCAAGCGGGATACGGGCGGCGGGAGCTGGGGAACAGGCATGGACGTTTCCG GGCAAGAGACCGACTGGCGGAGCACCGCCTTCCGGCAGAAGCTGGTCAGTCAAATCGAGGATGCCATGAGGAAAGCTGGTGTGGCACACA GTAAATCCAGCAAGGATATGGAGAGCCATGTTTTCCTGAAGGCCAAGACCCGGGACGAATACCTTTCTCTCGTGGCCAGGCTCATTATCC ATTTTCGAGACATTCGGATGTGTTTGGCCAAATGGGAGAGTGGTTACAACACACGAGCTACAAACTACAATGCTGGAGACAGAAGCACTG ATTATGGGATATTTCAGATCAATAGCCGCTACTGGTGTAATGATGGCAAAACCCCAGGAGCAGTTAATGCCTGTCATTTATCCTGCAGTG GGTGGCATGGAGAAATCGTTGTCAAAACAGAGATGTCCGTCAGTATGTTCAAGGTTGTGGAGTGTAACTCCAGAATTTTCCTTCTTCAGC TCATTTTGTCTCTCTCACATTAAGGGAGTAGGAATTAAGTGAAAGGTCACACTACCATTATTTCCCCTTCAAACAAATAATATTTTTACA GAAGCAGGAGCAAAATATGGCCTTTCTTCTAAGAGATATAATGTTCACTAATGTGGTTATTTTACATTAAGCCTACAACATTTTTCAGTT >52617_52617_4_MED15-LYZ_MED15_chr22_20905774_ENST00000292733_LYZ_chr12_69743888_ENST00000549690_length(amino acids)=161AA_BP=70 MDVSGQETDWRSTAFRQKLVSQIEDAMRKAGVAHSKSSKDMESHVFLKAKTRDEYLSLVARLIIHFRDIRMCLAKWESGYNTRATNYNAG -------------------------------------------------------------- >52617_52617_5_MED15-LYZ_MED15_chr22_20905774_ENST00000382974_LYZ_chr12_69743888_ENST00000261267_length(transcript)=1572nt_BP=246nt CCAAGCGGGATACGGGCGGCGGGAGCTGGGGAACAGGCATGGACGTTTCCGGGCAAGAGACCGACTGGCGGAGCACCGCCTTCCGGCAGA AGCTGGTCAGTCAAATCGAGGATGCCATGAGGAAAGCTGGTGTGGCACACAGTAAATCCAGCAAGGATATGGAGAGCCATGTTTTCCTGA AGGCCAAGACCCGGGACGAATACCTTTCTCTCGTGGCCAGGCTCATTATCCATTTTCGAGACATTCGGATGTGTTTGGCCAAATGGGAGA GTGGTTACAACACACGAGCTACAAACTACAATGCTGGAGACAGAAGCACTGATTATGGGATATTTCAGATCAATAGCCGCTACTGGTGTA ATGATGGCAAAACCCCAGGAGCAGTTAATGCCTGTCATTTATCCTGCAGTGCTTTGCTGCAAGATAACATCGCTGATGCTGTAGCTTGTG CAAAGAGGGTTGTCCGTGATCCACAAGGCATTAGAGCATGGGTGGCATGGAGAAATCGTTGTCAAAACAGAGATGTCCGTCAGTATGTTC AAGGTTGTGGAGTGTAACTCCAGAATTTTCCTTCTTCAGCTCATTTTGTCTCTCTCACATTAAGGGAGTAGGAATTAAGTGAAAGGTCAC ACTACCATTATTTCCCCTTCAAACAAATAATATTTTTACAGAAGCAGGAGCAAAATATGGCCTTTCTTCTAAGAGATATAATGTTCACTA ATGTGGTTATTTTACATTAAGCCTACAACATTTTTCAGTTTGCAAATAGAACTAATACTGGTGAAAATTTACCTAAAACCTTGGTTATCA AATACATCTCCAGTACATTCCGTTCTTTTTTTTTTTGAGACAGTCTCGCTCTGTCGCCCAGGCTGGAGTGCAGTGGCGCAATCTCGGCTC ACTGCAACCTCCACCTCCCGGGTTCACGCCATTCTCCTGCCTCAGCCTCCCGAGTAGCTGGGATTACGGGCGCCCGCCACCACGCCCGGC TAATTTTTTGTATTTTTAGTAGAGACAGGGTTTCACCGTGTTAGCCAGGATGGTCTCGATCTCCTGACCTTGTGATCCACCCACCTCGGC CTCCCAAAGTGCTGGGATTACAGGCGTGAGCCACTGCGCCCGGCCACATTCAGTTCTTATCAAAGAAATAACCCAGACTTAATCTTGAAT GATACGATTATGCCCAATATTAAGTAAAAAATATAAGAAAAGGTTATCTTAAATAGATCTTAGGCAAAATACCAGCTGATGAAGGCATCT GATGCCTTCATCTGTTCAGTCATCTCCAAAAACAGTAAAAATAACCACTTTTTGTTGGGCAATATGAAATTTTTAAAGGAGTAGAATACC AAATGATAGAAACAGACTGCCTGAATTGAGAATTTTGATTTCTTAAAGTGTGTTTCTTTCTAAATTGCTGTTCCTTAATTTGATTAATTT AATTCATGTATTATGATTAAATCTGAGGCAGATGAGCTTACAAGTATTGAAATAATTACTAATTAATCACAAATGTGAAGTTATGCATGA >52617_52617_5_MED15-LYZ_MED15_chr22_20905774_ENST00000382974_LYZ_chr12_69743888_ENST00000261267_length(amino acids)=172AA_BP=70 MDVSGQETDWRSTAFRQKLVSQIEDAMRKAGVAHSKSSKDMESHVFLKAKTRDEYLSLVARLIIHFRDIRMCLAKWESGYNTRATNYNAG -------------------------------------------------------------- >52617_52617_6_MED15-LYZ_MED15_chr22_20905774_ENST00000382974_LYZ_chr12_69743888_ENST00000549690_length(transcript)=754nt_BP=246nt CCAAGCGGGATACGGGCGGCGGGAGCTGGGGAACAGGCATGGACGTTTCCGGGCAAGAGACCGACTGGCGGAGCACCGCCTTCCGGCAGA AGCTGGTCAGTCAAATCGAGGATGCCATGAGGAAAGCTGGTGTGGCACACAGTAAATCCAGCAAGGATATGGAGAGCCATGTTTTCCTGA AGGCCAAGACCCGGGACGAATACCTTTCTCTCGTGGCCAGGCTCATTATCCATTTTCGAGACATTCGGATGTGTTTGGCCAAATGGGAGA GTGGTTACAACACACGAGCTACAAACTACAATGCTGGAGACAGAAGCACTGATTATGGGATATTTCAGATCAATAGCCGCTACTGGTGTA ATGATGGCAAAACCCCAGGAGCAGTTAATGCCTGTCATTTATCCTGCAGTGGGTGGCATGGAGAAATCGTTGTCAAAACAGAGATGTCCG TCAGTATGTTCAAGGTTGTGGAGTGTAACTCCAGAATTTTCCTTCTTCAGCTCATTTTGTCTCTCTCACATTAAGGGAGTAGGAATTAAG TGAAAGGTCACACTACCATTATTTCCCCTTCAAACAAATAATATTTTTACAGAAGCAGGAGCAAAATATGGCCTTTCTTCTAAGAGATAT AATGTTCACTAATGTGGTTATTTTACATTAAGCCTACAACATTTTTCAGTTTGCAAATAGAACTAATACTGGTGAAAATTTACCTAAAAC >52617_52617_6_MED15-LYZ_MED15_chr22_20905774_ENST00000382974_LYZ_chr12_69743888_ENST00000549690_length(amino acids)=161AA_BP=70 MDVSGQETDWRSTAFRQKLVSQIEDAMRKAGVAHSKSSKDMESHVFLKAKTRDEYLSLVARLIIHFRDIRMCLAKWESGYNTRATNYNAG -------------------------------------------------------------- >52617_52617_7_MED15-LYZ_MED15_chr22_20905774_ENST00000406969_LYZ_chr12_69743888_ENST00000261267_length(transcript)=1584nt_BP=258nt GACTGAGGCGGCGGCGGTGGCGGCCAAGCGGGATACGGGCGGCGGGAGCTGGGGAACAGGCATGGACGTTTCCGGGCAAGAGACCGACTG GCGGAGCACCGCCTTCCGGCAGAAGCTGCGAGGATGCCATGAGGAAAGCTGGTGTGGCACACAGTAAATCCAGCAAGGATATGGAGAGCC ATGTTTTCCTGAAGGCCAAGACCCGGGACGAATACCTTTCTCTCGTGGCCAGGCTCATTATCCATTTTCGAGACATTCGGATGTGTTTGG CCAAATGGGAGAGTGGTTACAACACACGAGCTACAAACTACAATGCTGGAGACAGAAGCACTGATTATGGGATATTTCAGATCAATAGCC GCTACTGGTGTAATGATGGCAAAACCCCAGGAGCAGTTAATGCCTGTCATTTATCCTGCAGTGCTTTGCTGCAAGATAACATCGCTGATG CTGTAGCTTGTGCAAAGAGGGTTGTCCGTGATCCACAAGGCATTAGAGCATGGGTGGCATGGAGAAATCGTTGTCAAAACAGAGATGTCC GTCAGTATGTTCAAGGTTGTGGAGTGTAACTCCAGAATTTTCCTTCTTCAGCTCATTTTGTCTCTCTCACATTAAGGGAGTAGGAATTAA GTGAAAGGTCACACTACCATTATTTCCCCTTCAAACAAATAATATTTTTACAGAAGCAGGAGCAAAATATGGCCTTTCTTCTAAGAGATA TAATGTTCACTAATGTGGTTATTTTACATTAAGCCTACAACATTTTTCAGTTTGCAAATAGAACTAATACTGGTGAAAATTTACCTAAAA CCTTGGTTATCAAATACATCTCCAGTACATTCCGTTCTTTTTTTTTTTGAGACAGTCTCGCTCTGTCGCCCAGGCTGGAGTGCAGTGGCG CAATCTCGGCTCACTGCAACCTCCACCTCCCGGGTTCACGCCATTCTCCTGCCTCAGCCTCCCGAGTAGCTGGGATTACGGGCGCCCGCC ACCACGCCCGGCTAATTTTTTGTATTTTTAGTAGAGACAGGGTTTCACCGTGTTAGCCAGGATGGTCTCGATCTCCTGACCTTGTGATCC ACCCACCTCGGCCTCCCAAAGTGCTGGGATTACAGGCGTGAGCCACTGCGCCCGGCCACATTCAGTTCTTATCAAAGAAATAACCCAGAC TTAATCTTGAATGATACGATTATGCCCAATATTAAGTAAAAAATATAAGAAAAGGTTATCTTAAATAGATCTTAGGCAAAATACCAGCTG ATGAAGGCATCTGATGCCTTCATCTGTTCAGTCATCTCCAAAAACAGTAAAAATAACCACTTTTTGTTGGGCAATATGAAATTTTTAAAG GAGTAGAATACCAAATGATAGAAACAGACTGCCTGAATTGAGAATTTTGATTTCTTAAAGTGTGTTTCTTTCTAAATTGCTGTTCCTTAA TTTGATTAATTTAATTCATGTATTATGATTAAATCTGAGGCAGATGAGCTTACAAGTATTGAAATAATTACTAATTAATCACAAATGTGA >52617_52617_7_MED15-LYZ_MED15_chr22_20905774_ENST00000406969_LYZ_chr12_69743888_ENST00000261267_length(amino acids)=188AA_BP=86 LRRRRWRPSGIRAAGAGEQAWTFPGKRPTGGAPPSGRSCEDAMRKAGVAHSKSSKDMESHVFLKAKTRDEYLSLVARLIIHFRDIRMCLA KWESGYNTRATNYNAGDRSTDYGIFQINSRYWCNDGKTPGAVNACHLSCSALLQDNIADAVACAKRVVRDPQGIRAWVAWRNRCQNRDVR -------------------------------------------------------------- >52617_52617_8_MED15-LYZ_MED15_chr22_20905774_ENST00000406969_LYZ_chr12_69743888_ENST00000549690_length(transcript)=766nt_BP=258nt GACTGAGGCGGCGGCGGTGGCGGCCAAGCGGGATACGGGCGGCGGGAGCTGGGGAACAGGCATGGACGTTTCCGGGCAAGAGACCGACTG GCGGAGCACCGCCTTCCGGCAGAAGCTGCGAGGATGCCATGAGGAAAGCTGGTGTGGCACACAGTAAATCCAGCAAGGATATGGAGAGCC ATGTTTTCCTGAAGGCCAAGACCCGGGACGAATACCTTTCTCTCGTGGCCAGGCTCATTATCCATTTTCGAGACATTCGGATGTGTTTGG CCAAATGGGAGAGTGGTTACAACACACGAGCTACAAACTACAATGCTGGAGACAGAAGCACTGATTATGGGATATTTCAGATCAATAGCC GCTACTGGTGTAATGATGGCAAAACCCCAGGAGCAGTTAATGCCTGTCATTTATCCTGCAGTGGGTGGCATGGAGAAATCGTTGTCAAAA CAGAGATGTCCGTCAGTATGTTCAAGGTTGTGGAGTGTAACTCCAGAATTTTCCTTCTTCAGCTCATTTTGTCTCTCTCACATTAAGGGA GTAGGAATTAAGTGAAAGGTCACACTACCATTATTTCCCCTTCAAACAAATAATATTTTTACAGAAGCAGGAGCAAAATATGGCCTTTCT TCTAAGAGATATAATGTTCACTAATGTGGTTATTTTACATTAAGCCTACAACATTTTTCAGTTTGCAAATAGAACTAATACTGGTGAAAA >52617_52617_8_MED15-LYZ_MED15_chr22_20905774_ENST00000406969_LYZ_chr12_69743888_ENST00000549690_length(amino acids)=177AA_BP=86 LRRRRWRPSGIRAAGAGEQAWTFPGKRPTGGAPPSGRSCEDAMRKAGVAHSKSSKDMESHVFLKAKTRDEYLSLVARLIIHFRDIRMCLA -------------------------------------------------------------- >52617_52617_9_MED15-LYZ_MED15_chr22_20905774_ENST00000425759_LYZ_chr12_69743888_ENST00000261267_length(transcript)=1614nt_BP=288nt GGTTTGGGCCTGGCTCTGTGACTGAGGCGGCGGCGGTGGCGGCCAAGCGGGATACGGGCGGCGGGAGCTGGGGAACAGGCATGGACGTTT CCGGGCAAGAGACCGACTGGCGGAGCACCGCCTTCCGGCAGAAGCTGGTCAGTCAAATCGAGGATGCCATGAGGAAAGCTGGTGTGGCAC ACAGTAAATCCAGCAAGGATATGGAGAGCCATGTTTTCCTGAAGGCCAAGACCCGGGACGAATACCTTTCTCTCGTGGCCAGGCTCATTA TCCATTTTCGAGACATTCGGATGTGTTTGGCCAAATGGGAGAGTGGTTACAACACACGAGCTACAAACTACAATGCTGGAGACAGAAGCA CTGATTATGGGATATTTCAGATCAATAGCCGCTACTGGTGTAATGATGGCAAAACCCCAGGAGCAGTTAATGCCTGTCATTTATCCTGCA GTGCTTTGCTGCAAGATAACATCGCTGATGCTGTAGCTTGTGCAAAGAGGGTTGTCCGTGATCCACAAGGCATTAGAGCATGGGTGGCAT GGAGAAATCGTTGTCAAAACAGAGATGTCCGTCAGTATGTTCAAGGTTGTGGAGTGTAACTCCAGAATTTTCCTTCTTCAGCTCATTTTG TCTCTCTCACATTAAGGGAGTAGGAATTAAGTGAAAGGTCACACTACCATTATTTCCCCTTCAAACAAATAATATTTTTACAGAAGCAGG AGCAAAATATGGCCTTTCTTCTAAGAGATATAATGTTCACTAATGTGGTTATTTTACATTAAGCCTACAACATTTTTCAGTTTGCAAATA GAACTAATACTGGTGAAAATTTACCTAAAACCTTGGTTATCAAATACATCTCCAGTACATTCCGTTCTTTTTTTTTTTGAGACAGTCTCG CTCTGTCGCCCAGGCTGGAGTGCAGTGGCGCAATCTCGGCTCACTGCAACCTCCACCTCCCGGGTTCACGCCATTCTCCTGCCTCAGCCT CCCGAGTAGCTGGGATTACGGGCGCCCGCCACCACGCCCGGCTAATTTTTTGTATTTTTAGTAGAGACAGGGTTTCACCGTGTTAGCCAG GATGGTCTCGATCTCCTGACCTTGTGATCCACCCACCTCGGCCTCCCAAAGTGCTGGGATTACAGGCGTGAGCCACTGCGCCCGGCCACA TTCAGTTCTTATCAAAGAAATAACCCAGACTTAATCTTGAATGATACGATTATGCCCAATATTAAGTAAAAAATATAAGAAAAGGTTATC TTAAATAGATCTTAGGCAAAATACCAGCTGATGAAGGCATCTGATGCCTTCATCTGTTCAGTCATCTCCAAAAACAGTAAAAATAACCAC TTTTTGTTGGGCAATATGAAATTTTTAAAGGAGTAGAATACCAAATGATAGAAACAGACTGCCTGAATTGAGAATTTTGATTTCTTAAAG TGTGTTTCTTTCTAAATTGCTGTTCCTTAATTTGATTAATTTAATTCATGTATTATGATTAAATCTGAGGCAGATGAGCTTACAAGTATT >52617_52617_9_MED15-LYZ_MED15_chr22_20905774_ENST00000425759_LYZ_chr12_69743888_ENST00000261267_length(amino acids)=172AA_BP=70 MDVSGQETDWRSTAFRQKLVSQIEDAMRKAGVAHSKSSKDMESHVFLKAKTRDEYLSLVARLIIHFRDIRMCLAKWESGYNTRATNYNAG -------------------------------------------------------------- >52617_52617_10_MED15-LYZ_MED15_chr22_20905774_ENST00000425759_LYZ_chr12_69743888_ENST00000549690_length(transcript)=796nt_BP=288nt GGTTTGGGCCTGGCTCTGTGACTGAGGCGGCGGCGGTGGCGGCCAAGCGGGATACGGGCGGCGGGAGCTGGGGAACAGGCATGGACGTTT CCGGGCAAGAGACCGACTGGCGGAGCACCGCCTTCCGGCAGAAGCTGGTCAGTCAAATCGAGGATGCCATGAGGAAAGCTGGTGTGGCAC ACAGTAAATCCAGCAAGGATATGGAGAGCCATGTTTTCCTGAAGGCCAAGACCCGGGACGAATACCTTTCTCTCGTGGCCAGGCTCATTA TCCATTTTCGAGACATTCGGATGTGTTTGGCCAAATGGGAGAGTGGTTACAACACACGAGCTACAAACTACAATGCTGGAGACAGAAGCA CTGATTATGGGATATTTCAGATCAATAGCCGCTACTGGTGTAATGATGGCAAAACCCCAGGAGCAGTTAATGCCTGTCATTTATCCTGCA GTGGGTGGCATGGAGAAATCGTTGTCAAAACAGAGATGTCCGTCAGTATGTTCAAGGTTGTGGAGTGTAACTCCAGAATTTTCCTTCTTC AGCTCATTTTGTCTCTCTCACATTAAGGGAGTAGGAATTAAGTGAAAGGTCACACTACCATTATTTCCCCTTCAAACAAATAATATTTTT ACAGAAGCAGGAGCAAAATATGGCCTTTCTTCTAAGAGATATAATGTTCACTAATGTGGTTATTTTACATTAAGCCTACAACATTTTTCA >52617_52617_10_MED15-LYZ_MED15_chr22_20905774_ENST00000425759_LYZ_chr12_69743888_ENST00000549690_length(amino acids)=161AA_BP=70 MDVSGQETDWRSTAFRQKLVSQIEDAMRKAGVAHSKSSKDMESHVFLKAKTRDEYLSLVARLIIHFRDIRMCLAKWESGYNTRATNYNAG -------------------------------------------------------------- >52617_52617_11_MED15-LYZ_MED15_chr22_20905774_ENST00000541476_LYZ_chr12_69743888_ENST00000261267_length(transcript)=1859nt_BP=533nt ACACTGAATAGGGAAAGGGAGAGAAGCAGCCGGCTGTAGTGGGTTTAGCAGGGCTGGGTGCTTCATGGTTTCTCTTCCAGAAAAGCGCAT CAACGGTGAAAGAAAGTGATGGGAGGAGGGTGGATGTAAGATGAGATTGGGAAGGTAGTTGTTTGGGGTGTGGGCGGTGGGACGGAGGAT CTGAGGGTGACCCCCCGAAGGAATGTGGACTTTGCCGAAGGATGGCAGGAAGCCGCTGTCAGGAAGCGATCGTCGTTTCTGGAGGAGAGC GTGAAGAGCTGGGGTCAGTGGCCCCGCTCAGAGGAACTCTAGCCCCTCACCACCGACTGCTTGTTTCTGTTAGCAGTTTTTTTCATGCGC GTCCGTGTGAAGAGACCACCAAACAGGCTTTGTCGAGGATGCCATGAGGAAAGCTGGTGTGGCACACAGTAAATCCAGCAAGGATATGGA GAGCCATGTTTTCCTGAAGGCCAAGACCCGGGACGAATACCTTTCTCTCGTGGCCAGGCTCATTATCCATTTTCGAGACATTCGGATGTG TTTGGCCAAATGGGAGAGTGGTTACAACACACGAGCTACAAACTACAATGCTGGAGACAGAAGCACTGATTATGGGATATTTCAGATCAA TAGCCGCTACTGGTGTAATGATGGCAAAACCCCAGGAGCAGTTAATGCCTGTCATTTATCCTGCAGTGCTTTGCTGCAAGATAACATCGC TGATGCTGTAGCTTGTGCAAAGAGGGTTGTCCGTGATCCACAAGGCATTAGAGCATGGGTGGCATGGAGAAATCGTTGTCAAAACAGAGA TGTCCGTCAGTATGTTCAAGGTTGTGGAGTGTAACTCCAGAATTTTCCTTCTTCAGCTCATTTTGTCTCTCTCACATTAAGGGAGTAGGA ATTAAGTGAAAGGTCACACTACCATTATTTCCCCTTCAAACAAATAATATTTTTACAGAAGCAGGAGCAAAATATGGCCTTTCTTCTAAG AGATATAATGTTCACTAATGTGGTTATTTTACATTAAGCCTACAACATTTTTCAGTTTGCAAATAGAACTAATACTGGTGAAAATTTACC TAAAACCTTGGTTATCAAATACATCTCCAGTACATTCCGTTCTTTTTTTTTTTGAGACAGTCTCGCTCTGTCGCCCAGGCTGGAGTGCAG TGGCGCAATCTCGGCTCACTGCAACCTCCACCTCCCGGGTTCACGCCATTCTCCTGCCTCAGCCTCCCGAGTAGCTGGGATTACGGGCGC CCGCCACCACGCCCGGCTAATTTTTTGTATTTTTAGTAGAGACAGGGTTTCACCGTGTTAGCCAGGATGGTCTCGATCTCCTGACCTTGT GATCCACCCACCTCGGCCTCCCAAAGTGCTGGGATTACAGGCGTGAGCCACTGCGCCCGGCCACATTCAGTTCTTATCAAAGAAATAACC CAGACTTAATCTTGAATGATACGATTATGCCCAATATTAAGTAAAAAATATAAGAAAAGGTTATCTTAAATAGATCTTAGGCAAAATACC AGCTGATGAAGGCATCTGATGCCTTCATCTGTTCAGTCATCTCCAAAAACAGTAAAAATAACCACTTTTTGTTGGGCAATATGAAATTTT TAAAGGAGTAGAATACCAAATGATAGAAACAGACTGCCTGAATTGAGAATTTTGATTTCTTAAAGTGTGTTTCTTTCTAAATTGCTGTTC CTTAATTTGATTAATTTAATTCATGTATTATGATTAAATCTGAGGCAGATGAGCTTACAAGTATTGAAATAATTACTAATTAATCACAAA >52617_52617_11_MED15-LYZ_MED15_chr22_20905774_ENST00000541476_LYZ_chr12_69743888_ENST00000261267_length(amino acids)=146AA_BP=44 MRKAGVAHSKSSKDMESHVFLKAKTRDEYLSLVARLIIHFRDIRMCLAKWESGYNTRATNYNAGDRSTDYGIFQINSRYWCNDGKTPGAV -------------------------------------------------------------- >52617_52617_12_MED15-LYZ_MED15_chr22_20905774_ENST00000541476_LYZ_chr12_69743888_ENST00000549690_length(transcript)=1041nt_BP=533nt ACACTGAATAGGGAAAGGGAGAGAAGCAGCCGGCTGTAGTGGGTTTAGCAGGGCTGGGTGCTTCATGGTTTCTCTTCCAGAAAAGCGCAT CAACGGTGAAAGAAAGTGATGGGAGGAGGGTGGATGTAAGATGAGATTGGGAAGGTAGTTGTTTGGGGTGTGGGCGGTGGGACGGAGGAT CTGAGGGTGACCCCCCGAAGGAATGTGGACTTTGCCGAAGGATGGCAGGAAGCCGCTGTCAGGAAGCGATCGTCGTTTCTGGAGGAGAGC GTGAAGAGCTGGGGTCAGTGGCCCCGCTCAGAGGAACTCTAGCCCCTCACCACCGACTGCTTGTTTCTGTTAGCAGTTTTTTTCATGCGC GTCCGTGTGAAGAGACCACCAAACAGGCTTTGTCGAGGATGCCATGAGGAAAGCTGGTGTGGCACACAGTAAATCCAGCAAGGATATGGA GAGCCATGTTTTCCTGAAGGCCAAGACCCGGGACGAATACCTTTCTCTCGTGGCCAGGCTCATTATCCATTTTCGAGACATTCGGATGTG TTTGGCCAAATGGGAGAGTGGTTACAACACACGAGCTACAAACTACAATGCTGGAGACAGAAGCACTGATTATGGGATATTTCAGATCAA TAGCCGCTACTGGTGTAATGATGGCAAAACCCCAGGAGCAGTTAATGCCTGTCATTTATCCTGCAGTGGGTGGCATGGAGAAATCGTTGT CAAAACAGAGATGTCCGTCAGTATGTTCAAGGTTGTGGAGTGTAACTCCAGAATTTTCCTTCTTCAGCTCATTTTGTCTCTCTCACATTA AGGGAGTAGGAATTAAGTGAAAGGTCACACTACCATTATTTCCCCTTCAAACAAATAATATTTTTACAGAAGCAGGAGCAAAATATGGCC TTTCTTCTAAGAGATATAATGTTCACTAATGTGGTTATTTTACATTAAGCCTACAACATTTTTCAGTTTGCAAATAGAACTAATACTGGT >52617_52617_12_MED15-LYZ_MED15_chr22_20905774_ENST00000541476_LYZ_chr12_69743888_ENST00000549690_length(amino acids)=135AA_BP=44 MRKAGVAHSKSSKDMESHVFLKAKTRDEYLSLVARLIIHFRDIRMCLAKWESGYNTRATNYNAGDRSTDYGIFQINSRYWCNDGKTPGAV -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for MED15-LYZ |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000263205 | + | 3 | 18 | 9_73 | 69.33333333333333 | 789.0 | SREBF1 |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000292733 | + | 3 | 17 | 9_73 | 69.33333333333333 | 749.0 | SREBF1 |
| Hgene | MED15 | chr22:20905774 | chr12:69743888 | ENST00000382974 | + | 3 | 16 | 9_73 | 69.33333333333333 | 678.0 | SREBF1 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for MED15-LYZ |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for MED15-LYZ |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
