
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:MGAT5-SP100 (FusionGDB2 ID:53360) |
Fusion Gene Summary for MGAT5-SP100 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: MGAT5-SP100 | Fusion gene ID: 53360 | Hgene | Tgene | Gene symbol | MGAT5 | SP100 | Gene ID | 4249 | 6672 |
| Gene name | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase | SP100 nuclear antigen | |
| Synonyms | GNT-V|GNT-VA|MGAT5A|glcNAc-T V | lysp100b | |
| Cytomap | 2q21.2-q21.3 | 2q37.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase AN-acetylglucosaminyl-transferase Valpha-mannoside beta-1,6-N-acetylglucosaminyltransferasealpha-mannoside beta-1,6-N-acetylglucosaminyltransferase Vmannoside acetylglucosaminyltran | nuclear autoantigen Sp-100SP100-HMG nuclear autoantigennuclear dot-associated Sp100 proteinspeckled 100 kDa | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q09328 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000468758, ENST00000281923, ENST00000409645, | ENST00000264052, ENST00000340126, ENST00000341950, ENST00000409112, ENST00000409341, ENST00000409824, ENST00000409897, ENST00000427101, | |
| Fusion gene scores | * DoF score | 11 X 12 X 3=396 | 3 X 3 X 2=18 |
| # samples | 14 | 4 | |
| ** MAII score | log2(14/396*10)=-1.50007360313464 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(4/18*10)=1.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: MGAT5 [Title/Abstract] AND SP100 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | MGAT5(135012215)-SP100(231327150), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | MGAT5-SP100 seems lost the major protein functional domain in Hgene partner, which is a cell metabolism gene due to the frame-shifted ORF. MGAT5-SP100 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. MGAT5-SP100 seems lost the major protein functional domain in Tgene partner, which is a epigenetic factor due to the frame-shifted ORF. MGAT5-SP100 seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. MGAT5-SP100 seems lost the major protein functional domain in Tgene partner, which is a transcription factor due to the frame-shifted ORF. MGAT5-SP100 seems lost the major protein functional domain in Tgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | MGAT5 | GO:0006487 | protein N-linked glycosylation | 24846175 |
| Hgene | MGAT5 | GO:0018279 | protein N-linked glycosylation via asparagine | 10395745|30140003 |
| Hgene | MGAT5 | GO:0030335 | positive regulation of cell migration | 24846175 |
| Hgene | MGAT5 | GO:1903614 | negative regulation of protein tyrosine phosphatase activity | 24846175 |
| Hgene | MGAT5 | GO:1904894 | positive regulation of STAT cascade | 24846175 |
| Tgene | SP100 | GO:0000122 | negative regulation of transcription by RNA polymerase II | 15247905 |
| Tgene | SP100 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | 14647468 |
| Tgene | SP100 | GO:0034340 | response to type I interferon | 9230084|15247905 |
| Tgene | SP100 | GO:0034341 | response to interferon-gamma | 9230084 |
| Tgene | SP100 | GO:0043392 | negative regulation of DNA binding | 15247905 |
| Tgene | SP100 | GO:0043433 | negative regulation of DNA-binding transcription factor activity | 15247905 |
| Tgene | SP100 | GO:0045185 | maintenance of protein location | 12470659 |
 Fusion gene breakpoints across MGAT5 (5'-gene) Fusion gene breakpoints across MGAT5 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
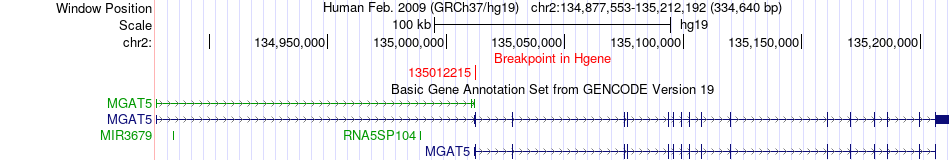 |
 Fusion gene breakpoints across SP100 (3'-gene) Fusion gene breakpoints across SP100 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
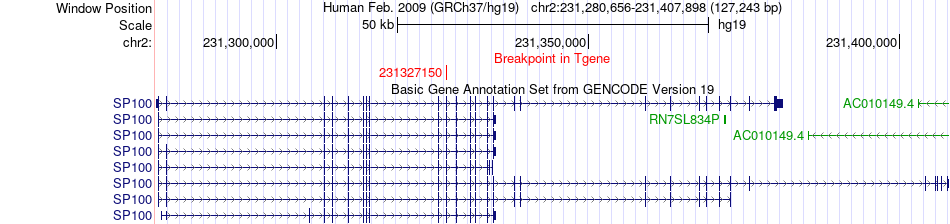 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-HU-A4GJ-01A | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + |
Top |
Fusion Gene ORF analysis for MGAT5-SP100 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| Frame-shift | ENST00000468758 | ENST00000264052 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + |
| Frame-shift | ENST00000468758 | ENST00000340126 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + |
| Frame-shift | ENST00000468758 | ENST00000341950 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + |
| Frame-shift | ENST00000468758 | ENST00000409112 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + |
| Frame-shift | ENST00000468758 | ENST00000409341 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + |
| Frame-shift | ENST00000468758 | ENST00000409824 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + |
| Frame-shift | ENST00000468758 | ENST00000409897 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + |
| Frame-shift | ENST00000468758 | ENST00000427101 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + |
| In-frame | ENST00000281923 | ENST00000264052 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + |
| In-frame | ENST00000281923 | ENST00000340126 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + |
| In-frame | ENST00000281923 | ENST00000341950 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + |
| In-frame | ENST00000281923 | ENST00000409112 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + |
| In-frame | ENST00000281923 | ENST00000409341 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + |
| In-frame | ENST00000281923 | ENST00000409824 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + |
| In-frame | ENST00000281923 | ENST00000409897 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + |
| In-frame | ENST00000281923 | ENST00000427101 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + |
| In-frame | ENST00000409645 | ENST00000264052 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + |
| In-frame | ENST00000409645 | ENST00000340126 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + |
| In-frame | ENST00000409645 | ENST00000341950 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + |
| In-frame | ENST00000409645 | ENST00000409112 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + |
| In-frame | ENST00000409645 | ENST00000409341 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + |
| In-frame | ENST00000409645 | ENST00000409824 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + |
| In-frame | ENST00000409645 | ENST00000409897 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + |
| In-frame | ENST00000409645 | ENST00000427101 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000409645 | MGAT5 | chr2 | 135012215 | + | ENST00000264052 | SP100 | chr2 | 231327150 | + | 3047 | 493 | 252 | 2159 | 635 |
| ENST00000409645 | MGAT5 | chr2 | 135012215 | + | ENST00000409112 | SP100 | chr2 | 231327150 | + | 1687 | 493 | 252 | 1586 | 444 |
| ENST00000409645 | MGAT5 | chr2 | 135012215 | + | ENST00000340126 | SP100 | chr2 | 231327150 | + | 2415 | 493 | 252 | 2177 | 641 |
| ENST00000409645 | MGAT5 | chr2 | 135012215 | + | ENST00000427101 | SP100 | chr2 | 231327150 | + | 1363 | 493 | 252 | 953 | 233 |
| ENST00000409645 | MGAT5 | chr2 | 135012215 | + | ENST00000409824 | SP100 | chr2 | 231327150 | + | 1340 | 493 | 252 | 962 | 236 |
| ENST00000409645 | MGAT5 | chr2 | 135012215 | + | ENST00000409341 | SP100 | chr2 | 231327150 | + | 1368 | 493 | 252 | 962 | 236 |
| ENST00000409645 | MGAT5 | chr2 | 135012215 | + | ENST00000341950 | SP100 | chr2 | 231327150 | + | 942 | 493 | 252 | 941 | 229 |
| ENST00000409645 | MGAT5 | chr2 | 135012215 | + | ENST00000409897 | SP100 | chr2 | 231327150 | + | 1381 | 493 | 252 | 962 | 236 |
| ENST00000281923 | MGAT5 | chr2 | 135012215 | + | ENST00000264052 | SP100 | chr2 | 231327150 | + | 2940 | 386 | 145 | 2052 | 635 |
| ENST00000281923 | MGAT5 | chr2 | 135012215 | + | ENST00000409112 | SP100 | chr2 | 231327150 | + | 1580 | 386 | 145 | 1479 | 444 |
| ENST00000281923 | MGAT5 | chr2 | 135012215 | + | ENST00000340126 | SP100 | chr2 | 231327150 | + | 2308 | 386 | 145 | 2070 | 641 |
| ENST00000281923 | MGAT5 | chr2 | 135012215 | + | ENST00000427101 | SP100 | chr2 | 231327150 | + | 1256 | 386 | 145 | 846 | 233 |
| ENST00000281923 | MGAT5 | chr2 | 135012215 | + | ENST00000409824 | SP100 | chr2 | 231327150 | + | 1233 | 386 | 145 | 855 | 236 |
| ENST00000281923 | MGAT5 | chr2 | 135012215 | + | ENST00000409341 | SP100 | chr2 | 231327150 | + | 1261 | 386 | 145 | 855 | 236 |
| ENST00000281923 | MGAT5 | chr2 | 135012215 | + | ENST00000341950 | SP100 | chr2 | 231327150 | + | 835 | 386 | 145 | 834 | 230 |
| ENST00000281923 | MGAT5 | chr2 | 135012215 | + | ENST00000409897 | SP100 | chr2 | 231327150 | + | 1274 | 386 | 145 | 855 | 236 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000409645 | ENST00000264052 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + | 0.0009897 | 0.9990103 |
| ENST00000409645 | ENST00000409112 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + | 0.008976928 | 0.99102306 |
| ENST00000409645 | ENST00000340126 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + | 0.003545954 | 0.9964541 |
| ENST00000409645 | ENST00000427101 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + | 0.003296366 | 0.9967037 |
| ENST00000409645 | ENST00000409824 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + | 0.004365132 | 0.99563485 |
| ENST00000409645 | ENST00000409341 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + | 0.004185962 | 0.995814 |
| ENST00000409645 | ENST00000341950 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + | 0.007266083 | 0.99273396 |
| ENST00000409645 | ENST00000409897 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + | 0.004068144 | 0.9959319 |
| ENST00000281923 | ENST00000264052 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + | 0.000852339 | 0.99914765 |
| ENST00000281923 | ENST00000409112 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + | 0.007357612 | 0.99264234 |
| ENST00000281923 | ENST00000340126 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + | 0.003314668 | 0.9966853 |
| ENST00000281923 | ENST00000427101 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + | 0.00368984 | 0.9963102 |
| ENST00000281923 | ENST00000409824 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + | 0.005155684 | 0.9948443 |
| ENST00000281923 | ENST00000409341 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + | 0.005113911 | 0.9948861 |
| ENST00000281923 | ENST00000341950 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + | 0.009890941 | 0.9901091 |
| ENST00000281923 | ENST00000409897 | MGAT5 | chr2 | 135012215 | + | SP100 | chr2 | 231327150 | + | 0.005144892 | 0.99485505 |
Top |
Fusion Genomic Features for MGAT5-SP100 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
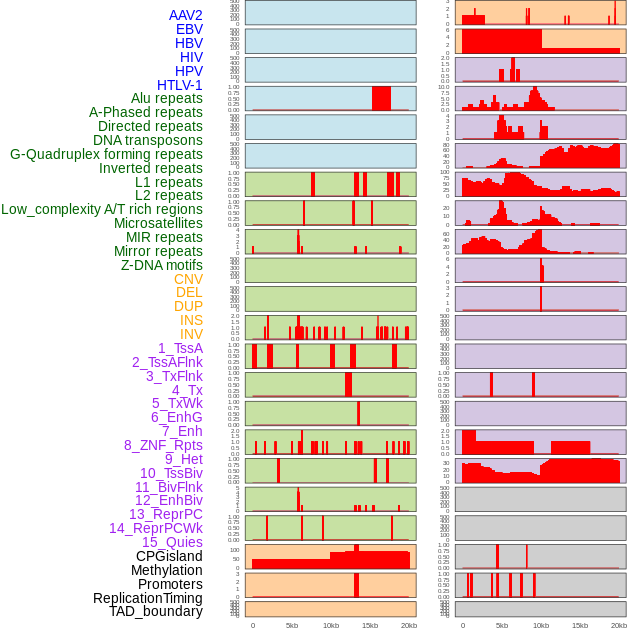 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
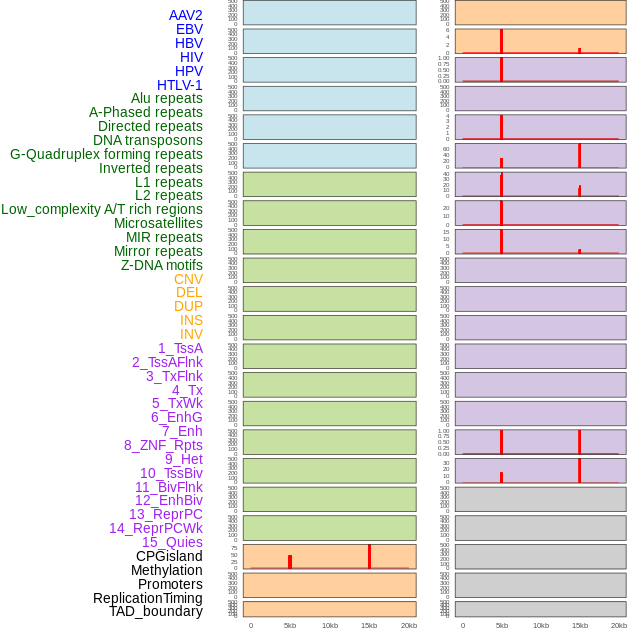 |
Top |
Fusion Protein Features for MGAT5-SP100 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:135012215/chr2:231327150) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MGAT5 | . |
| FUNCTION: Catalyzes the addition of N-acetylglucosamine (GlcNAc) in beta 1-6 linkage to the alpha-linked mannose of biantennary N-linked oligosaccharides (PubMed:10395745, PubMed:30140003). Catalyzes an important step in the biosynthesis of branched, complex-type N-glycans, such as those found on EGFR, TGFR (TGF-beta receptor) and CDH2 (PubMed:10395745, PubMed:22614033, PubMed:30140003). Via its role in the biosynthesis of complex N-glycans, plays an important role in the activation of cellular signaling pathways, reorganization of the actin cytoskeleton, cell-cell adhesion and cell migration. MGAT5-dependent EGFR N-glycosylation enhances the interaction between EGFR and LGALS3 and thereby prevents rapid EGFR endocytosis and prolongs EGFR signaling. Required for efficient interaction between TGFB1 and its receptor. Enhances activation of intracellular signaling pathways by several types of growth factors, including FGF2, PDGF, IGF, TGFB1 and EGF. MGAT5-dependent CDH2 N-glycosylation inhibits CDH2-mediated homotypic cell-cell adhesion and contributes to the regulation of downstream signaling pathways. Promotes cell migration. Contributes to the regulation of the inflammatory response. MGAT5-dependent TCR N-glycosylation enhances the interaction between TCR and LGALS3, limits agonist-induced TCR clustering, and thereby dampens TCR-mediated responses to antigens. Required for normal leukocyte evasation and accumulation at sites of inflammation (By similarity). Inhibits attachment of monocytes to the vascular endothelium and subsequent monocyte diapedesis (PubMed:22614033). {ECO:0000250|UniProtKB:Q8R4G6, ECO:0000269|PubMed:10395745, ECO:0000269|PubMed:22614033, ECO:0000269|PubMed:30140003}.; FUNCTION: [Secreted alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A]: Promotes proliferation of umbilical vein endothelial cells and angiogenesis, at least in part by promoting the release of the growth factor FGF2 from the extracellular matrix. {ECO:0000269|PubMed:11872751}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MGAT5 | chr2:135012215 | chr2:231327150 | ENST00000281923 | + | 1 | 16 | 1_13 | 80 | 742.0 | Topological domain | Cytoplasmic |
| Hgene | MGAT5 | chr2:135012215 | chr2:231327150 | ENST00000409645 | + | 2 | 17 | 1_13 | 80 | 742.0 | Topological domain | Cytoplasmic |
| Hgene | MGAT5 | chr2:135012215 | chr2:231327150 | ENST00000281923 | + | 1 | 16 | 14_30 | 80 | 742.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Hgene | MGAT5 | chr2:135012215 | chr2:231327150 | ENST00000409645 | + | 2 | 17 | 14_30 | 80 | 742.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000264052 | 8 | 25 | 759_764 | 324 | 880.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000264052 | 8 | 25 | 854_859 | 324 | 880.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000264052 | 8 | 25 | 860_868 | 324 | 880.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000340126 | 8 | 29 | 759_764 | 324 | 886.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000340126 | 8 | 29 | 854_859 | 324 | 886.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000340126 | 8 | 29 | 860_868 | 324 | 886.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409341 | 8 | 15 | 759_764 | 324 | 481.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409341 | 8 | 15 | 854_859 | 324 | 481.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409341 | 8 | 15 | 860_868 | 324 | 481.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409897 | 9 | 16 | 759_764 | 289 | 446.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409897 | 9 | 16 | 854_859 | 289 | 446.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409897 | 9 | 16 | 860_868 | 289 | 446.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000427101 | 7 | 14 | 759_764 | 299 | 453.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000427101 | 7 | 14 | 854_859 | 299 | 453.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000427101 | 7 | 14 | 860_868 | 299 | 453.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000264052 | 8 | 25 | 677_753 | 324 | 880.0 | DNA binding | HMG box 1 | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000264052 | 8 | 25 | 769_837 | 324 | 880.0 | DNA binding | HMG box 2 | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000340126 | 8 | 29 | 677_753 | 324 | 886.0 | DNA binding | HMG box 1 | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000340126 | 8 | 29 | 769_837 | 324 | 886.0 | DNA binding | HMG box 2 | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409341 | 8 | 15 | 677_753 | 324 | 481.0 | DNA binding | HMG box 1 | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409341 | 8 | 15 | 769_837 | 324 | 481.0 | DNA binding | HMG box 2 | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409897 | 9 | 16 | 677_753 | 289 | 446.0 | DNA binding | HMG box 1 | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409897 | 9 | 16 | 769_837 | 289 | 446.0 | DNA binding | HMG box 2 | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000427101 | 7 | 14 | 677_753 | 299 | 453.0 | DNA binding | HMG box 1 | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000427101 | 7 | 14 | 769_837 | 299 | 453.0 | DNA binding | HMG box 2 | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000264052 | 8 | 25 | 595_676 | 324 | 880.0 | Domain | SAND | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000340126 | 8 | 29 | 595_676 | 324 | 886.0 | Domain | SAND | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409341 | 8 | 15 | 595_676 | 324 | 481.0 | Domain | SAND | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409897 | 9 | 16 | 595_676 | 289 | 446.0 | Domain | SAND | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000427101 | 7 | 14 | 595_676 | 299 | 453.0 | Domain | SAND | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000264052 | 8 | 25 | 536_553 | 324 | 880.0 | Motif | Nuclear localization signal | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000264052 | 8 | 25 | 568_592 | 324 | 880.0 | Motif | Nuclear localization signal | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000264052 | 8 | 25 | 717_734 | 324 | 880.0 | Motif | Nuclear localization signal | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000340126 | 8 | 29 | 536_553 | 324 | 886.0 | Motif | Nuclear localization signal | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000340126 | 8 | 29 | 568_592 | 324 | 886.0 | Motif | Nuclear localization signal | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000340126 | 8 | 29 | 717_734 | 324 | 886.0 | Motif | Nuclear localization signal | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409341 | 8 | 15 | 536_553 | 324 | 481.0 | Motif | Nuclear localization signal | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409341 | 8 | 15 | 568_592 | 324 | 481.0 | Motif | Nuclear localization signal | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409341 | 8 | 15 | 717_734 | 324 | 481.0 | Motif | Nuclear localization signal | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409897 | 9 | 16 | 536_553 | 289 | 446.0 | Motif | Nuclear localization signal | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409897 | 9 | 16 | 568_592 | 289 | 446.0 | Motif | Nuclear localization signal | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409897 | 9 | 16 | 717_734 | 289 | 446.0 | Motif | Nuclear localization signal | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000427101 | 7 | 14 | 536_553 | 299 | 453.0 | Motif | Nuclear localization signal | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000427101 | 7 | 14 | 568_592 | 299 | 453.0 | Motif | Nuclear localization signal | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000427101 | 7 | 14 | 717_734 | 299 | 453.0 | Motif | Nuclear localization signal |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MGAT5 | chr2:135012215 | chr2:231327150 | ENST00000281923 | + | 1 | 16 | 213_741 | 80 | 742.0 | Region | Sufficient for catalytic activity |
| Hgene | MGAT5 | chr2:135012215 | chr2:231327150 | ENST00000281923 | + | 1 | 16 | 264_269 | 80 | 742.0 | Region | Important for activity in FGF2 release |
| Hgene | MGAT5 | chr2:135012215 | chr2:231327150 | ENST00000281923 | + | 1 | 16 | 378_379 | 80 | 742.0 | Region | Substrate binding |
| Hgene | MGAT5 | chr2:135012215 | chr2:231327150 | ENST00000409645 | + | 2 | 17 | 213_741 | 80 | 742.0 | Region | Sufficient for catalytic activity |
| Hgene | MGAT5 | chr2:135012215 | chr2:231327150 | ENST00000409645 | + | 2 | 17 | 264_269 | 80 | 742.0 | Region | Important for activity in FGF2 release |
| Hgene | MGAT5 | chr2:135012215 | chr2:231327150 | ENST00000409645 | + | 2 | 17 | 378_379 | 80 | 742.0 | Region | Substrate binding |
| Hgene | MGAT5 | chr2:135012215 | chr2:231327150 | ENST00000281923 | + | 1 | 16 | 31_741 | 80 | 742.0 | Topological domain | Lumenal |
| Hgene | MGAT5 | chr2:135012215 | chr2:231327150 | ENST00000409645 | + | 2 | 17 | 31_741 | 80 | 742.0 | Topological domain | Lumenal |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000264052 | 8 | 25 | 156_164 | 324 | 880.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000264052 | 8 | 25 | 3_6 | 324 | 880.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000340126 | 8 | 29 | 156_164 | 324 | 886.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000340126 | 8 | 29 | 3_6 | 324 | 886.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409341 | 8 | 15 | 156_164 | 324 | 481.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409341 | 8 | 15 | 3_6 | 324 | 481.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409897 | 9 | 16 | 156_164 | 289 | 446.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409897 | 9 | 16 | 3_6 | 289 | 446.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000427101 | 7 | 14 | 156_164 | 299 | 453.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000427101 | 7 | 14 | 3_6 | 299 | 453.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000264052 | 8 | 25 | 33_149 | 324 | 880.0 | Domain | HSR | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000340126 | 8 | 29 | 33_149 | 324 | 886.0 | Domain | HSR | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409341 | 8 | 15 | 33_149 | 324 | 481.0 | Domain | HSR | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409897 | 9 | 16 | 33_149 | 289 | 446.0 | Domain | HSR | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000427101 | 7 | 14 | 33_149 | 299 | 453.0 | Domain | HSR | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000264052 | 8 | 25 | 165_168 | 324 | 880.0 | Motif | Note=D-box%3B recognition signal for CDC20-mediated degradation | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000264052 | 8 | 25 | 284_297 | 324 | 880.0 | Motif | Note=PxVxL motif | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000340126 | 8 | 29 | 165_168 | 324 | 886.0 | Motif | Note=D-box%3B recognition signal for CDC20-mediated degradation | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000340126 | 8 | 29 | 284_297 | 324 | 886.0 | Motif | Note=PxVxL motif | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409341 | 8 | 15 | 165_168 | 324 | 481.0 | Motif | Note=D-box%3B recognition signal for CDC20-mediated degradation | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409341 | 8 | 15 | 284_297 | 324 | 481.0 | Motif | Note=PxVxL motif | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409897 | 9 | 16 | 165_168 | 289 | 446.0 | Motif | Note=D-box%3B recognition signal for CDC20-mediated degradation | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000409897 | 9 | 16 | 284_297 | 289 | 446.0 | Motif | Note=PxVxL motif | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000427101 | 7 | 14 | 165_168 | 299 | 453.0 | Motif | Note=D-box%3B recognition signal for CDC20-mediated degradation | |
| Tgene | SP100 | chr2:135012215 | chr2:231327150 | ENST00000427101 | 7 | 14 | 284_297 | 299 | 453.0 | Motif | Note=PxVxL motif |
Top |
Fusion Gene Sequence for MGAT5-SP100 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >53360_53360_1_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000281923_SP100_chr2_231327150_ENST00000264052_length(transcript)=2940nt_BP=386nt CAGCAGAATGGAAGTGAGGAAAGGCAACCAGCTGACACAGGAGCCAGAGTGAGACCAGCAGACTCTCACACTCAACCTACACCATGAATT TGTGTCTATCTTCTACGCGTTAAGAGCCAAGGACAGGTGAAGTTGCCAGAGAGCAATGGCTCTCTTCACTCCGTGGAAGTTGTCCTCTCA GAAGCTGGGCTTTTTCCTGGTGACTTTTGGCTTCATTTGGGGTATGATGCTTCTGCACTTTACCATCCAGCAGCGAACTCAGCCTGAAAG CAGCTCCATGCTGCGCGAGCAGATCCTGGACCTCAGCAAAAGGTACATCAAGGCACTGGCAGAAGAAAACAGGAATGTGGTGGATGGGCC ATACGCTGGAGTCATGACAGCTTATGTCATCAGCAGTGAGGACTCTGAAGGATCCACTGACGTTGATGAGCCCTTAGAAGTCTTCATCTC AGCACCGAGAAGTGAGCCTGTGATCAATAATGACAACCCTTTAGAATCAAATGATGAAAAGGAGGGCCAAGAAGCCACTTGCTCACGACC CCAGATTGTACCAGAGCCCATGGATTTCAGAAAATTATCTACATTCAGAGAAAGTTTTAAGAAAAGAGTGATAGGACAAGACCACGACTT TTCAGAATCCAGTGAGGAGGAGGCGCCCGCAGAAGCCTCGAGCGGGGCACTGAGAAGCAAGCATGGTGAGAAGGCTCCTATGACTTCTAG AAGTACATCTACTTGGAGAATACCCAGCAGGAAGAGACGTTTCAGCAGTAGTGACTTTTCAGACCTGAGTAATGGAGAAGAGCTTCAGGA AACCTGCAGCTCATCCCTAAGAAGAGGGTCAGGATCACAGCCACAAGAACCTGAAAATAAGAAGTGCTCCTGTGTCATGTGTTTTCCAAA AGGTGTGCCAAGAAGCCAAGAAGCAAGGACTGAAAGTAGTCAAGCATCTGACATGATGGATACCATGGATGTTGAAAACAATTCTACTTT GGAAAAACACAGTGGGAAAAGAAGAAAAAAGAGAAGGCATAGATCTAAAGTAAATGGTCTCCAAAGAGGGAGAAAGAAAGACAGACCTAG AAAACATTTAACTCTGAATAACAAAGTCCAAAAGAAAAGATGGCAACAAAGAGGAAGAAAAGCCAACACTAGACCTTTGAAAAGAAGAAG AAAAAGAGGTCCAAGAATTCCCAAAGATGAAAATATTAATTTTAAACAATCTGAACTTCCTGTGACCTGTGGTGAGGTGAAGGGCACTCT ATATAAGGAGCGATTCAAACAAGGAACCTCAAAGAAGTGTATACAGAGTGAGGATAAAAAGTGGTTCACTCCCAGGGAATTTGAAATTGA AGGAGACCGCGGAGCATCCAAGAACTGGAAGCTAAGTATACGCTGCGGTGGATATACCCTGAAAGTCCTGATGGAGAACAAATTTCTGCC AGAACCACCAAGCACAAGAAAAAAGAGAATACTGGAATCTCACAACAATACCTTAGTTGACCCTTGTGAGGAGCATAAGAAGAAGAACCC AGATGCTTCAGTCAAGTTCTCAGAGTTTTTAAAGAAGTGCTCAGAGACATGGAAGACCATTTTTGCTAAAGAGAAAGGAAAATTTGAAGA TATGGCAAAGGCGGACAAGGCCCATTATGAAAGAGAAATGAAAACCTATATCCCTCCTAAAGGGGAGAAAAAAAAGAAGTTCAAGGATCC CAATGCACCCAAGAGGCCTCCTTTGGCCTTTTTCCTGTTCTGCTCTGAGTATCGCCCAAAAATCAAAGGAGAACATCCTGGCCTGTCCAT TGATGATGTTGTGAAGAAACTGGCAGGGATGTGGAATAACACCGCTGCAGCTGACAAGCAGTTTTATGAAAAGAAGGCTGCAAAGCTGAA GGAAAAATACAAAAAGGATATTGCTGCATATCGAGCTAAAGGAAAGCCTAATTCAGCAAAAAAGAGAGTTGTCAAGGCTGAAAAAAGCAA GAAAAAGAAGGAAGAGGAAGAAGATGAAGAGGATGAACAAGAGGAGGAAAATGAAGAAGATGATGATAAATAAGTTGCTTCTAGTGCAGT TTTTTTCTTGTCTATAAAGCATTTAAGCTGCCTGTACACAACTCACTCCTTTTAAAGAAAAAAACTTCAACGTAAGACTGTGTAAGATTT GTTTTTAAACCGTACACTGTGTTTTTTTGTATAGTTAACCACTACCGAATGTGTCTTCAGATAGCCCTGTCCTGGTGGTATTTAGCCACT AACCTTTGCCTGGTACAGTATGGGGGTTGTAAATTGGCATGGAAATTTAAAGCAGGTTCTTGTTAGTGCACAGCACAAATTAGTTGTATA TGAGGATGGTAGTTTTTTCACCTTCAGTTGTCTCTGATGTAGCTTATACAAAACATTTGTTGTTCTGTTAACTGAATGCCACTCTGTAAT TGCAAAAAAAAAAAACAGTTGCAGCTGTTTTGTTGACATTCTGAATGCTTCTAAGTAAATACAATTTTTAAAAAACCGTATGAGGGAACT GTGTAGACAAGGTACCAGGTCAGTCTTCTTCCATGTCTATTAGCTCCACAAAGCCAATCTCAATCCCTCAAAACAATCTTGTCATACTTG AAAATATGACACTCTAGTCAAAGCCTTGGTAAAATAATCAGTGTTTCCAATCTGTCCTGTTACAAAAGAAACAGATTATTATTGAACTTA TGCAAATAACCATTGTCATAAGAATGTTTATGAATAGTTTCCAAATTATGGCAAATTCATGTAGAGAGAGAAAAGTAACTGTTTTGGTTT TGCTCACAAAAGTCTACTTTACCTAAGGGCTGTCAGATATAAGTAACTTAAAAGAAAGAGAAGTTTTCTTGACTTTTGAAAACAAAATAT >53360_53360_1_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000281923_SP100_chr2_231327150_ENST00000264052_length(amino acids)=635AA_BP=80 MALFTPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQPESSSMLREQILDLSKRYIKALAEENRNVVDGPYAGVMTAYVISSEDSEGS TDVDEPLEVFISAPRSEPVINNDNPLESNDEKEGQEATCSRPQIVPEPMDFRKLSTFRESFKKRVIGQDHDFSESSEEEAPAEASSGALR SKHGEKAPMTSRSTSTWRIPSRKRRFSSSDFSDLSNGEELQETCSSSLRRGSGSQPQEPENKKCSCVMCFPKGVPRSQEARTESSQASDM MDTMDVENNSTLEKHSGKRRKKRRHRSKVNGLQRGRKKDRPRKHLTLNNKVQKKRWQQRGRKANTRPLKRRRKRGPRIPKDENINFKQSE LPVTCGEVKGTLYKERFKQGTSKKCIQSEDKKWFTPREFEIEGDRGASKNWKLSIRCGGYTLKVLMENKFLPEPPSTRKKRILESHNNTL VDPCEEHKKKNPDASVKFSEFLKKCSETWKTIFAKEKGKFEDMAKADKAHYEREMKTYIPPKGEKKKKFKDPNAPKRPPLAFFLFCSEYR PKIKGEHPGLSIDDVVKKLAGMWNNTAAADKQFYEKKAAKLKEKYKKDIAAYRAKGKPNSAKKRVVKAEKSKKKKEEEEDEEDEQEEENE -------------------------------------------------------------- >53360_53360_2_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000281923_SP100_chr2_231327150_ENST00000340126_length(transcript)=2308nt_BP=386nt CAGCAGAATGGAAGTGAGGAAAGGCAACCAGCTGACACAGGAGCCAGAGTGAGACCAGCAGACTCTCACACTCAACCTACACCATGAATT TGTGTCTATCTTCTACGCGTTAAGAGCCAAGGACAGGTGAAGTTGCCAGAGAGCAATGGCTCTCTTCACTCCGTGGAAGTTGTCCTCTCA GAAGCTGGGCTTTTTCCTGGTGACTTTTGGCTTCATTTGGGGTATGATGCTTCTGCACTTTACCATCCAGCAGCGAACTCAGCCTGAAAG CAGCTCCATGCTGCGCGAGCAGATCCTGGACCTCAGCAAAAGGTACATCAAGGCACTGGCAGAAGAAAACAGGAATGTGGTGGATGGGCC ATACGCTGGAGTCATGACAGCTTATGTCATCAGCAGTGAGGACTCTGAAGGATCCACTGACGTTGATGAGCCCTTAGAAGTCTTCATCTC AGCACCGAGAAGTGAGCCTGTGATCAATAATGACAACCCTTTAGAATCAAATGATGAAAAGGAGGGCCAAGAAGCCACTTGCTCACGACC CCAGATTGTACCAGAGCCCATGGATTTCAGAAAATTATCTACATTCAGAGAAAGTTTTAAGAAAAGAGTGATAGGACAAGACCACGACTT TTCAGAATCCAGTGAGGAGGAGGCGCCCGCAGAAGCCTCGAGCGGGGCACTGAGAAGCAAGCATGGTGAGAAGGCTCCTATGACTTCTAG AAGTACATCTACTTGGAGAATACCCAGCAGGAAGAGACGTTTCAGCAGTAGTGACTTTTCAGACCTGAGTAATGGAGAAGAGCTTCAGGA AACCTGCAGCTCATCCCTAAGAAGAGGGTCAGGATCACAGCCACAAGAACCTGAAAATAAGAAGTGCTCCTGTGTCATGTGTTTTCCAAA AGGTGTGCCAAGAAGCCAAGAAGCAAGGACTGAAAGTAGTCAAGCATCTGACATGATGGATACCATGGATGTTGAAAACAATTCTACTTT GGAAAAACACAGTGGGAAAAGAAGAAAAAAGAGAAGGCATAGATCTAAAGTAAATGGTCTCCAAAGAGGGAGAAAGAAAGACAGACCTAG AAAACATTTAACTCTGAATAACAAAGTCCAAAAGAAAAGATGGCAACAAAGAGGAAGAAAAGCCAACACTAGACCTTTGAAAAGAAGAAG AAAAAGAGGTCCAAGAATTCCCAAAGATGAAAATATTAATTTTAAACAATCTGAACTTCCTGTGACCTGTGGTGAGGTGAAGGGCACTCT ATATAAGGAGCGATTCAAACAAGGAACCTCAAAGAAGTGTATACAGAGTGAGGATAAAAAGTGGTTCACTCCCAGGGAATTTGAAATTGA AGGAGACCGCGGAGCATCCAAGAACTGGAAGCTAAGTATACGCTGCGGTGGATATACCCTGAAAGTCCTGATGGAGAACAAATTTCTGCC AGAACCACCAAGCACAAGAAAAAAGAGAATACTGGAATCTCACAACAATACCTTAGTTGACCCTTGTCCGGAAAACTCAAATATATGTGA GGTGTGCAACAAATGGGGACGGCTGTTCTGCTGCGACACTTGTCCAAGATCCTTTCATGAGCACTGCCACATCCCATCCGTGGAAGCTAA CAAGAACCCGTGGAGTTGCATCTTCTGCAGGATAAAGACTATTCAGGAAAGATGCCCAGAAAGCCAATCAGGTCATCAGGAATCTGAAGT CCTGATGAGGCAGATGCTGCCTGAGGAGCAGTTGAAATGTGAATTCCTCCTCTTGAAGGTCTACTGTGATTCGAAAAGCTGCTTTTTCGC CTCAGAACCGTATTATAACAGAGAGGGGTCTCAGGGCCCACAGAAGCCCATGTGGTTAAACAAAGTCAAGACAAGTTTGAATGAGCAGAT GTACACCCGAGTAGAAGGGTTTGTGCAGGACATGCGTCTCATCTTTCATAACCACAAGGAATTTTACAGGGAAGATAAATTCACCAGACT GGGAATTCAAGTACAGGACATCTTTGAGAAGAATTTCAGAAACATTTTTGCAATTCAGGAAACAAGCAAGAACATTATAATGTTTATTTA GCCATTCTTATCTCCTCCCTTCAGATCCTCTGGCAGCTAGCTACGCAATGTGCCTGTGGTCCCACTAATCTGTGACTGCTCCTGTGGAAA CTCCACATCACAATTCTCCAAAATTTATCATTGCCATTTTAAAACCGTCTTTTCAGCTTTCAATAAAATTCAACACCCCTTCATGTTAAA >53360_53360_2_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000281923_SP100_chr2_231327150_ENST00000340126_length(amino acids)=641AA_BP=80 MALFTPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQPESSSMLREQILDLSKRYIKALAEENRNVVDGPYAGVMTAYVISSEDSEGS TDVDEPLEVFISAPRSEPVINNDNPLESNDEKEGQEATCSRPQIVPEPMDFRKLSTFRESFKKRVIGQDHDFSESSEEEAPAEASSGALR SKHGEKAPMTSRSTSTWRIPSRKRRFSSSDFSDLSNGEELQETCSSSLRRGSGSQPQEPENKKCSCVMCFPKGVPRSQEARTESSQASDM MDTMDVENNSTLEKHSGKRRKKRRHRSKVNGLQRGRKKDRPRKHLTLNNKVQKKRWQQRGRKANTRPLKRRRKRGPRIPKDENINFKQSE LPVTCGEVKGTLYKERFKQGTSKKCIQSEDKKWFTPREFEIEGDRGASKNWKLSIRCGGYTLKVLMENKFLPEPPSTRKKRILESHNNTL VDPCPENSNICEVCNKWGRLFCCDTCPRSFHEHCHIPSVEANKNPWSCIFCRIKTIQERCPESQSGHQESEVLMRQMLPEEQLKCEFLLL KVYCDSKSCFFASEPYYNREGSQGPQKPMWLNKVKTSLNEQMYTRVEGFVQDMRLIFHNHKEFYREDKFTRLGIQVQDIFEKNFRNIFAI -------------------------------------------------------------- >53360_53360_3_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000281923_SP100_chr2_231327150_ENST00000341950_length(transcript)=835nt_BP=386nt CAGCAGAATGGAAGTGAGGAAAGGCAACCAGCTGACACAGGAGCCAGAGTGAGACCAGCAGACTCTCACACTCAACCTACACCATGAATT TGTGTCTATCTTCTACGCGTTAAGAGCCAAGGACAGGTGAAGTTGCCAGAGAGCAATGGCTCTCTTCACTCCGTGGAAGTTGTCCTCTCA GAAGCTGGGCTTTTTCCTGGTGACTTTTGGCTTCATTTGGGGTATGATGCTTCTGCACTTTACCATCCAGCAGCGAACTCAGCCTGAAAG CAGCTCCATGCTGCGCGAGCAGATCCTGGACCTCAGCAAAAGGTACATCAAGGCACTGGCAGAAGAAAACAGGAATGTGGTGGATGGGCC ATACGCTGGAGTCATGACAGCTTATGTCATCAGCAGTGAGGACTCTGAAGGATCCACTGACGTTGATGAGCCCTTAGAAGTCTTCATCTC AGCACCGAGAAGTGAGCCTGTGATCAATAATGACAACCCTTTAGAATCAAATGATGAAAAGGAGGGCCAAGAAGCCACTTGCTCACGACC CCAGATTGTACCAGAGCCCATGGATTTCAGAAAATTATCTACATTCAGAGAAAGTTTTAAGAAAAGAGTGATAGGACAAGACCACGACTT TTCAGAATCCAGTGAGGAGGAGGCGCCCGCAGAAGCCTCGAGCGGGGCACTGAGAAGCAAGCATGGTGAGAAGGCTCCTATGACTTCTAG AAGTACATCTACTTGGAGAATACCCAGCAGGAAGAGACAATTAAAAAAAAAAAAGAAGAAGAAACAATGTCATCCCCAGCCCCAGCCTCA >53360_53360_3_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000281923_SP100_chr2_231327150_ENST00000341950_length(amino acids)=230AA_BP=80 MALFTPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQPESSSMLREQILDLSKRYIKALAEENRNVVDGPYAGVMTAYVISSEDSEGS TDVDEPLEVFISAPRSEPVINNDNPLESNDEKEGQEATCSRPQIVPEPMDFRKLSTFRESFKKRVIGQDHDFSESSEEEAPAEASSGALR -------------------------------------------------------------- >53360_53360_4_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000281923_SP100_chr2_231327150_ENST00000409112_length(transcript)=1580nt_BP=386nt CAGCAGAATGGAAGTGAGGAAAGGCAACCAGCTGACACAGGAGCCAGAGTGAGACCAGCAGACTCTCACACTCAACCTACACCATGAATT TGTGTCTATCTTCTACGCGTTAAGAGCCAAGGACAGGTGAAGTTGCCAGAGAGCAATGGCTCTCTTCACTCCGTGGAAGTTGTCCTCTCA GAAGCTGGGCTTTTTCCTGGTGACTTTTGGCTTCATTTGGGGTATGATGCTTCTGCACTTTACCATCCAGCAGCGAACTCAGCCTGAAAG CAGCTCCATGCTGCGCGAGCAGATCCTGGACCTCAGCAAAAGGTACATCAAGGCACTGGCAGAAGAAAACAGGAATGTGGTGGATGGGCC ATACGCTGGAGTCATGACAGCTTATGTCATCAGCAGTGAGGACTCTGAAGGATCCACTGACGTTGATGAGCCCTTAGAAGTCTTCATCTC AGCACCGAGAAGTGAGCCTGTGATCAATAATGACAACCCTTTAGAATCAAATGATGAAAAGGAGGGCCAAGAAGCCACTTGCTCACGACC CCAGATTGTACCAGAGCCCATGGATTTCAGAAAATTATCTACATTCAGAGAAAGTTTTAAGAAAAGAGTGATAGGACAAGACCACGACTT TTCAGAATCCAGTGAGGAGGAGGCGCCCGCAGAAGCCTCGAGCGGGGCACTGAGAAGCAAGCATGGTGAGAAGGCTCCTATGACTTCTAG AAGTACATCTACTTGGAGAATACCCAGCAGGAAGAGACGTTTCAGCAGTAGTGACTTTTCAGACCTGAGTAATGGAGAAGAGCTTCAGGA AACCTGCAGCTCATCCCTAAGAAGAGGGTCAGGATCACAGCCACAAGAACCTGAAAATAAGAAGTGCTCCTGTGTCATGTGTTTTCCAAA AGGTGTGCCAAGAAGCCAAGAAGCAAGGACTGAAAGTAGTCAAGCATCTGACATGATGGATACCATGGATGTTGAAAACAATTCTACTTT GGAAAAACACAGTGGGAAAAGAAGAAAAAAGAGAAGGCATAGATCTAAAGTAAATGGTCTCCAAAGAGGGAGAAAGAAAGACAGACCTAG AAAACATTTAACTCTGAATAACAAAGTCCAAAAGAAAAGATGGCAACAAAGAGGAAGAAAAGCCAACACTAGACCTTTGAAAAGAAGAAG AAAAAGAGGTCCAAGAATTCCCAAAGATGAAAATATTAATTTTAAACAATCTGAACTTCCTGTGACCTGTGGTGAGGTGAAGGGCACTCT ATATAAGGAGCGATTCAAACAAGGAACCTCAAAGAAGTGTATACAGAGTGAGGATAAAAAGTGGTTCACTCCCAGGGAATTTGAAATTGA AGGAGACCGCGGAGCATCCAAGAACTGGAAGCTAAGTATACGCTGCGGTGGATATACCCTGAAAGTCCTGATGGAGAACAAATTTCTGCC AGAACCACCAAGCACAAGAAAAAAGGTGATGATCAAGTGATCTTCTGCCAATGTCTCGTCTATTATGTTGTTGATTTTCTATCTCTGTGG >53360_53360_4_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000281923_SP100_chr2_231327150_ENST00000409112_length(amino acids)=444AA_BP=80 MALFTPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQPESSSMLREQILDLSKRYIKALAEENRNVVDGPYAGVMTAYVISSEDSEGS TDVDEPLEVFISAPRSEPVINNDNPLESNDEKEGQEATCSRPQIVPEPMDFRKLSTFRESFKKRVIGQDHDFSESSEEEAPAEASSGALR SKHGEKAPMTSRSTSTWRIPSRKRRFSSSDFSDLSNGEELQETCSSSLRRGSGSQPQEPENKKCSCVMCFPKGVPRSQEARTESSQASDM MDTMDVENNSTLEKHSGKRRKKRRHRSKVNGLQRGRKKDRPRKHLTLNNKVQKKRWQQRGRKANTRPLKRRRKRGPRIPKDENINFKQSE -------------------------------------------------------------- >53360_53360_5_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000281923_SP100_chr2_231327150_ENST00000409341_length(transcript)=1261nt_BP=386nt CAGCAGAATGGAAGTGAGGAAAGGCAACCAGCTGACACAGGAGCCAGAGTGAGACCAGCAGACTCTCACACTCAACCTACACCATGAATT TGTGTCTATCTTCTACGCGTTAAGAGCCAAGGACAGGTGAAGTTGCCAGAGAGCAATGGCTCTCTTCACTCCGTGGAAGTTGTCCTCTCA GAAGCTGGGCTTTTTCCTGGTGACTTTTGGCTTCATTTGGGGTATGATGCTTCTGCACTTTACCATCCAGCAGCGAACTCAGCCTGAAAG CAGCTCCATGCTGCGCGAGCAGATCCTGGACCTCAGCAAAAGGTACATCAAGGCACTGGCAGAAGAAAACAGGAATGTGGTGGATGGGCC ATACGCTGGAGTCATGACAGCTTATGTCATCAGCAGTGAGGACTCTGAAGGATCCACTGACGTTGATGAGCCCTTAGAAGTCTTCATCTC AGCACCGAGAAGTGAGCCTGTGATCAATAATGACAACCCTTTAGAATCAAATGATGAAAAGGAGGGCCAAGAAGCCACTTGCTCACGACC CCAGATTGTACCAGAGCCCATGGATTTCAGAAAATTATCTACATTCAGAGAAAGTTTTAAGAAAAGAGTGATAGGACAAGACCACGACTT TTCAGAATCCAGTGAGGAGGAGGCGCCCGCAGAAGCCTCGAGCGGGGCACTGAGAAGCAAGCATGGTGAGAAGGCTCCTATGACTTCTAG AAGTACATCTACTTGGAGAATACCCAGCAGGAAGAGACGTTTCAGCAGTAGTGACTTTTCAGACCTGAGTAATGGAGAAGAGCTTCAGGA AACCTGCAGCTCATCCCTAAGAAGAGGGTCAGGTAAAGAAGATTAGGATGCCAAGACTTGGCCTGCAGAATGTCAGGAATGTGAATTAAA AGCTGCTGTTTCCAGACGCTTTTTATTCTGAGCACCTTCACTACCTTGTATCCAGTTCATCTGGGAACTCCTTTTTGCATTTTAGAAAAT GGAAAGAGGCAGGAAATTATGATAAACTCATGTTTAACAGAAAGAGTTTCACTGACTAAATGTATGTAATTATATTTTGTTGTTGTAGAA GAAATAAATAGCAAATTTGTGGTATTCTTTTTTTTAAACCTGCTCTCATTCCTATTAACACTAAGATCTTAGATTTTTATAGTGATAAAT GGGTTGACATCATTGTCATTTGTAATTGTAAAGCCTCAAAAGACAACTGTTCCTACTATGTAATTATAGACAGAAATAAAAACTTCAGAT >53360_53360_5_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000281923_SP100_chr2_231327150_ENST00000409341_length(amino acids)=236AA_BP=80 MALFTPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQPESSSMLREQILDLSKRYIKALAEENRNVVDGPYAGVMTAYVISSEDSEGS TDVDEPLEVFISAPRSEPVINNDNPLESNDEKEGQEATCSRPQIVPEPMDFRKLSTFRESFKKRVIGQDHDFSESSEEEAPAEASSGALR -------------------------------------------------------------- >53360_53360_6_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000281923_SP100_chr2_231327150_ENST00000409824_length(transcript)=1233nt_BP=386nt CAGCAGAATGGAAGTGAGGAAAGGCAACCAGCTGACACAGGAGCCAGAGTGAGACCAGCAGACTCTCACACTCAACCTACACCATGAATT TGTGTCTATCTTCTACGCGTTAAGAGCCAAGGACAGGTGAAGTTGCCAGAGAGCAATGGCTCTCTTCACTCCGTGGAAGTTGTCCTCTCA GAAGCTGGGCTTTTTCCTGGTGACTTTTGGCTTCATTTGGGGTATGATGCTTCTGCACTTTACCATCCAGCAGCGAACTCAGCCTGAAAG CAGCTCCATGCTGCGCGAGCAGATCCTGGACCTCAGCAAAAGGTACATCAAGGCACTGGCAGAAGAAAACAGGAATGTGGTGGATGGGCC ATACGCTGGAGTCATGACAGCTTATGTCATCAGCAGTGAGGACTCTGAAGGATCCACTGACGTTGATGAGCCCTTAGAAGTCTTCATCTC AGCACCGAGAAGTGAGCCTGTGATCAATAATGACAACCCTTTAGAATCAAATGATGAAAAGGAGGGCCAAGAAGCCACTTGCTCACGACC CCAGATTGTACCAGAGCCCATGGATTTCAGAAAATTATCTACATTCAGAGAAAGTTTTAAGAAAAGAGTGATAGGACAAGACCACGACTT TTCAGAATCCAGTGAGGAGGAGGCGCCCGCAGAAGCCTCGAGCGGGGCACTGAGAAGCAAGCATGGTGAGAAGGCTCCTATGACTTCTAG AAGTACATCTACTTGGAGAATACCCAGCAGGAAGAGACGTTTCAGCAGTAGTGACTTTTCAGACCTGAGTAATGGAGAAGAGCTTCAGGA AACCTGCAGCTCATCCCTAAGAAGAGGGTCAGGTAAAGAAGATTAGGATGCCAAGACTTGGCCTGCAGAATGTCAGGAATGTGAATTAAA AGCTGCTGTTTCCAGACGCTTTTTATTCTGAGCACCTTCACTACCTTGTATCCAGTTCATCTGGGAACTCCTTTTTGCATTTTAGAAAAT GGAAAGAGGCAGGAAATTATGATAAACTCATGTTTAACAGAAAGAGTTTCACTGACTAAATGTATGTAATTATATTTTGTTGTTGTAGAA GAAATAAATAGCAAATTTGTGGTATTCTTTTTTTTAAACCTGCTCTCATTCCTATTAACACTAAGATCTTAGATTTTTATAGTGATAAAT >53360_53360_6_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000281923_SP100_chr2_231327150_ENST00000409824_length(amino acids)=236AA_BP=80 MALFTPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQPESSSMLREQILDLSKRYIKALAEENRNVVDGPYAGVMTAYVISSEDSEGS TDVDEPLEVFISAPRSEPVINNDNPLESNDEKEGQEATCSRPQIVPEPMDFRKLSTFRESFKKRVIGQDHDFSESSEEEAPAEASSGALR -------------------------------------------------------------- >53360_53360_7_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000281923_SP100_chr2_231327150_ENST00000409897_length(transcript)=1274nt_BP=386nt CAGCAGAATGGAAGTGAGGAAAGGCAACCAGCTGACACAGGAGCCAGAGTGAGACCAGCAGACTCTCACACTCAACCTACACCATGAATT TGTGTCTATCTTCTACGCGTTAAGAGCCAAGGACAGGTGAAGTTGCCAGAGAGCAATGGCTCTCTTCACTCCGTGGAAGTTGTCCTCTCA GAAGCTGGGCTTTTTCCTGGTGACTTTTGGCTTCATTTGGGGTATGATGCTTCTGCACTTTACCATCCAGCAGCGAACTCAGCCTGAAAG CAGCTCCATGCTGCGCGAGCAGATCCTGGACCTCAGCAAAAGGTACATCAAGGCACTGGCAGAAGAAAACAGGAATGTGGTGGATGGGCC ATACGCTGGAGTCATGACAGCTTATGTCATCAGCAGTGAGGACTCTGAAGGATCCACTGACGTTGATGAGCCCTTAGAAGTCTTCATCTC AGCACCGAGAAGTGAGCCTGTGATCAATAATGACAACCCTTTAGAATCAAATGATGAAAAGGAGGGCCAAGAAGCCACTTGCTCACGACC CCAGATTGTACCAGAGCCCATGGATTTCAGAAAATTATCTACATTCAGAGAAAGTTTTAAGAAAAGAGTGATAGGACAAGACCACGACTT TTCAGAATCCAGTGAGGAGGAGGCGCCCGCAGAAGCCTCGAGCGGGGCACTGAGAAGCAAGCATGGTGAGAAGGCTCCTATGACTTCTAG AAGTACATCTACTTGGAGAATACCCAGCAGGAAGAGACGTTTCAGCAGTAGTGACTTTTCAGACCTGAGTAATGGAGAAGAGCTTCAGGA AACCTGCAGCTCATCCCTAAGAAGAGGGTCAGGTAAAGAAGATTAGGATGCCAAGACTTGGCCTGCAGAATGTCAGGAATGTGAATTAAA AGCTGCTGTTTCCAGACGCTTTTTATTCTGAGCACCTTCACTACCTTGTATCCAGTTCATCTGGGAACTCCTTTTTGCATTTTAGAAAAT GGAAAGAGGCAGGAAATTATGATAAACTCATGTTTAACAGAAAGAGTTTCACTGACTAAATGTATGTAATTATATTTTGTTGTTGTAGAA GAAATAAATAGCAAATTTGTGGTATTCTTTTTTTTAAACCTGCTCTCATTCCTATTAACACTAAGATCTTAGATTTTTATAGTGATAAAT GGGTTGACATCATTGTCATTTGTAATTGTAAAGCCTCAAAAGACAACTGTTCCTACTATGTAATTATAGACAGAAATAAAAACTTCAGAT >53360_53360_7_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000281923_SP100_chr2_231327150_ENST00000409897_length(amino acids)=236AA_BP=80 MALFTPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQPESSSMLREQILDLSKRYIKALAEENRNVVDGPYAGVMTAYVISSEDSEGS TDVDEPLEVFISAPRSEPVINNDNPLESNDEKEGQEATCSRPQIVPEPMDFRKLSTFRESFKKRVIGQDHDFSESSEEEAPAEASSGALR -------------------------------------------------------------- >53360_53360_8_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000281923_SP100_chr2_231327150_ENST00000427101_length(transcript)=1256nt_BP=386nt CAGCAGAATGGAAGTGAGGAAAGGCAACCAGCTGACACAGGAGCCAGAGTGAGACCAGCAGACTCTCACACTCAACCTACACCATGAATT TGTGTCTATCTTCTACGCGTTAAGAGCCAAGGACAGGTGAAGTTGCCAGAGAGCAATGGCTCTCTTCACTCCGTGGAAGTTGTCCTCTCA GAAGCTGGGCTTTTTCCTGGTGACTTTTGGCTTCATTTGGGGTATGATGCTTCTGCACTTTACCATCCAGCAGCGAACTCAGCCTGAAAG CAGCTCCATGCTGCGCGAGCAGATCCTGGACCTCAGCAAAAGGTACATCAAGGCACTGGCAGAAGAAAACAGGAATGTGGTGGATGGGCC ATACGCTGGAGTCATGACAGCTTATGTCATCAGCAGTGAGGACTCTGAAGGATCCACTGACGTTGATGAGCCCTTAGAAGTCTTCATCTC AGCACCGAGAAGTGAGCCTGTGATCAATAATGACAACCCTTTAGAATCAAATGATGAAAAGGAGGGCCAAGAAGCCACTTGCTCACGACC CCAGATTGTACCAGAGCCCATGGATTTCAGAAAATTATCTACATTCAGAGAAAGTTTTAAGAAAAGAGTGATAGGACAAGACCACGACTT TTCAGAATCCAGTGAGGAGGAGGCGCCCGCAGAAGCCTCGAGCGGGGCACTGAGAAGCAAGCATGCTCCTATGACTTCTAGAAGTACATC TACTTGGAGAATACCCAGCAGGAAGAGACGTTTCAGCAGTAGTGACTTTTCAGACCTGAGTAATGGAGAAGAGCTTCAGGAAACCTGCAG CTCATCCCTAAGAAGAGGGTCAGGTAAAGAAGATTAGGATGCCAAGACTTGGCCTGCAGAATGTCAGGAATGTGAATTAAAAGCTGCTGT TTCCAGACGCTTTTTATTCTGAGCACCTTCACTACCTTGTATCCAGTTCATCTGGGAACTCCTTTTTGCATTTTAGAAAATGGAAAGAGG CAGGAAATTATGATAAACTCATGTTTAACAGAAAGAGTTTCACTGACTAAATGTATGTAATTATATTTTGTTGTTGTAGAAGAAATAAAT AGCAAATTTGTGGTATTCTTTTTTTTAAACCTGCTCTCATTCCTATTAACACTAAGATCTTAGATTTTTATAGTGATAAATGGGTTGACA >53360_53360_8_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000281923_SP100_chr2_231327150_ENST00000427101_length(amino acids)=233AA_BP=80 MALFTPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQPESSSMLREQILDLSKRYIKALAEENRNVVDGPYAGVMTAYVISSEDSEGS TDVDEPLEVFISAPRSEPVINNDNPLESNDEKEGQEATCSRPQIVPEPMDFRKLSTFRESFKKRVIGQDHDFSESSEEEAPAEASSGALR -------------------------------------------------------------- >53360_53360_9_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000409645_SP100_chr2_231327150_ENST00000264052_length(transcript)=3047nt_BP=493nt AGCCCAGGTAGCCGCGCCGCAGGCTCGGGGCGTCGCGACCTCGCGCCTCGGGCCGCGTGGGCACGGCGGCCGGCGGGTGCTCCCGGCTGC TGCTGCTGCCCAACAAGGAGCAGAATGGAAGTGAGGAAAGGCAACCAGCTGACACAGGAGCCAGAGTGAGACCAGCAGACTCTCACACTC AACCTACACCATGAATTTGTGTCTATCTTCTACGCGTTAAGAGCCAAGGACAGGTGAAGTTGCCAGAGAGCAATGGCTCTCTTCACTCCG TGGAAGTTGTCCTCTCAGAAGCTGGGCTTTTTCCTGGTGACTTTTGGCTTCATTTGGGGTATGATGCTTCTGCACTTTACCATCCAGCAG CGAACTCAGCCTGAAAGCAGCTCCATGCTGCGCGAGCAGATCCTGGACCTCAGCAAAAGGTACATCAAGGCACTGGCAGAAGAAAACAGG AATGTGGTGGATGGGCCATACGCTGGAGTCATGACAGCTTATGTCATCAGCAGTGAGGACTCTGAAGGATCCACTGACGTTGATGAGCCC TTAGAAGTCTTCATCTCAGCACCGAGAAGTGAGCCTGTGATCAATAATGACAACCCTTTAGAATCAAATGATGAAAAGGAGGGCCAAGAA GCCACTTGCTCACGACCCCAGATTGTACCAGAGCCCATGGATTTCAGAAAATTATCTACATTCAGAGAAAGTTTTAAGAAAAGAGTGATA GGACAAGACCACGACTTTTCAGAATCCAGTGAGGAGGAGGCGCCCGCAGAAGCCTCGAGCGGGGCACTGAGAAGCAAGCATGGTGAGAAG GCTCCTATGACTTCTAGAAGTACATCTACTTGGAGAATACCCAGCAGGAAGAGACGTTTCAGCAGTAGTGACTTTTCAGACCTGAGTAAT GGAGAAGAGCTTCAGGAAACCTGCAGCTCATCCCTAAGAAGAGGGTCAGGATCACAGCCACAAGAACCTGAAAATAAGAAGTGCTCCTGT GTCATGTGTTTTCCAAAAGGTGTGCCAAGAAGCCAAGAAGCAAGGACTGAAAGTAGTCAAGCATCTGACATGATGGATACCATGGATGTT GAAAACAATTCTACTTTGGAAAAACACAGTGGGAAAAGAAGAAAAAAGAGAAGGCATAGATCTAAAGTAAATGGTCTCCAAAGAGGGAGA AAGAAAGACAGACCTAGAAAACATTTAACTCTGAATAACAAAGTCCAAAAGAAAAGATGGCAACAAAGAGGAAGAAAAGCCAACACTAGA CCTTTGAAAAGAAGAAGAAAAAGAGGTCCAAGAATTCCCAAAGATGAAAATATTAATTTTAAACAATCTGAACTTCCTGTGACCTGTGGT GAGGTGAAGGGCACTCTATATAAGGAGCGATTCAAACAAGGAACCTCAAAGAAGTGTATACAGAGTGAGGATAAAAAGTGGTTCACTCCC AGGGAATTTGAAATTGAAGGAGACCGCGGAGCATCCAAGAACTGGAAGCTAAGTATACGCTGCGGTGGATATACCCTGAAAGTCCTGATG GAGAACAAATTTCTGCCAGAACCACCAAGCACAAGAAAAAAGAGAATACTGGAATCTCACAACAATACCTTAGTTGACCCTTGTGAGGAG CATAAGAAGAAGAACCCAGATGCTTCAGTCAAGTTCTCAGAGTTTTTAAAGAAGTGCTCAGAGACATGGAAGACCATTTTTGCTAAAGAG AAAGGAAAATTTGAAGATATGGCAAAGGCGGACAAGGCCCATTATGAAAGAGAAATGAAAACCTATATCCCTCCTAAAGGGGAGAAAAAA AAGAAGTTCAAGGATCCCAATGCACCCAAGAGGCCTCCTTTGGCCTTTTTCCTGTTCTGCTCTGAGTATCGCCCAAAAATCAAAGGAGAA CATCCTGGCCTGTCCATTGATGATGTTGTGAAGAAACTGGCAGGGATGTGGAATAACACCGCTGCAGCTGACAAGCAGTTTTATGAAAAG AAGGCTGCAAAGCTGAAGGAAAAATACAAAAAGGATATTGCTGCATATCGAGCTAAAGGAAAGCCTAATTCAGCAAAAAAGAGAGTTGTC AAGGCTGAAAAAAGCAAGAAAAAGAAGGAAGAGGAAGAAGATGAAGAGGATGAACAAGAGGAGGAAAATGAAGAAGATGATGATAAATAA GTTGCTTCTAGTGCAGTTTTTTTCTTGTCTATAAAGCATTTAAGCTGCCTGTACACAACTCACTCCTTTTAAAGAAAAAAACTTCAACGT AAGACTGTGTAAGATTTGTTTTTAAACCGTACACTGTGTTTTTTTGTATAGTTAACCACTACCGAATGTGTCTTCAGATAGCCCTGTCCT GGTGGTATTTAGCCACTAACCTTTGCCTGGTACAGTATGGGGGTTGTAAATTGGCATGGAAATTTAAAGCAGGTTCTTGTTAGTGCACAG CACAAATTAGTTGTATATGAGGATGGTAGTTTTTTCACCTTCAGTTGTCTCTGATGTAGCTTATACAAAACATTTGTTGTTCTGTTAACT GAATGCCACTCTGTAATTGCAAAAAAAAAAAACAGTTGCAGCTGTTTTGTTGACATTCTGAATGCTTCTAAGTAAATACAATTTTTAAAA AACCGTATGAGGGAACTGTGTAGACAAGGTACCAGGTCAGTCTTCTTCCATGTCTATTAGCTCCACAAAGCCAATCTCAATCCCTCAAAA CAATCTTGTCATACTTGAAAATATGACACTCTAGTCAAAGCCTTGGTAAAATAATCAGTGTTTCCAATCTGTCCTGTTACAAAAGAAACA GATTATTATTGAACTTATGCAAATAACCATTGTCATAAGAATGTTTATGAATAGTTTCCAAATTATGGCAAATTCATGTAGAGAGAGAAA AGTAACTGTTTTGGTTTTGCTCACAAAAGTCTACTTTACCTAAGGGCTGTCAGATATAAGTAACTTAAAAGAAAGAGAAGTTTTCTTGAC >53360_53360_9_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000409645_SP100_chr2_231327150_ENST00000264052_length(amino acids)=635AA_BP=80 MALFTPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQPESSSMLREQILDLSKRYIKALAEENRNVVDGPYAGVMTAYVISSEDSEGS TDVDEPLEVFISAPRSEPVINNDNPLESNDEKEGQEATCSRPQIVPEPMDFRKLSTFRESFKKRVIGQDHDFSESSEEEAPAEASSGALR SKHGEKAPMTSRSTSTWRIPSRKRRFSSSDFSDLSNGEELQETCSSSLRRGSGSQPQEPENKKCSCVMCFPKGVPRSQEARTESSQASDM MDTMDVENNSTLEKHSGKRRKKRRHRSKVNGLQRGRKKDRPRKHLTLNNKVQKKRWQQRGRKANTRPLKRRRKRGPRIPKDENINFKQSE LPVTCGEVKGTLYKERFKQGTSKKCIQSEDKKWFTPREFEIEGDRGASKNWKLSIRCGGYTLKVLMENKFLPEPPSTRKKRILESHNNTL VDPCEEHKKKNPDASVKFSEFLKKCSETWKTIFAKEKGKFEDMAKADKAHYEREMKTYIPPKGEKKKKFKDPNAPKRPPLAFFLFCSEYR PKIKGEHPGLSIDDVVKKLAGMWNNTAAADKQFYEKKAAKLKEKYKKDIAAYRAKGKPNSAKKRVVKAEKSKKKKEEEEDEEDEQEEENE -------------------------------------------------------------- >53360_53360_10_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000409645_SP100_chr2_231327150_ENST00000340126_length(transcript)=2415nt_BP=493nt AGCCCAGGTAGCCGCGCCGCAGGCTCGGGGCGTCGCGACCTCGCGCCTCGGGCCGCGTGGGCACGGCGGCCGGCGGGTGCTCCCGGCTGC TGCTGCTGCCCAACAAGGAGCAGAATGGAAGTGAGGAAAGGCAACCAGCTGACACAGGAGCCAGAGTGAGACCAGCAGACTCTCACACTC AACCTACACCATGAATTTGTGTCTATCTTCTACGCGTTAAGAGCCAAGGACAGGTGAAGTTGCCAGAGAGCAATGGCTCTCTTCACTCCG TGGAAGTTGTCCTCTCAGAAGCTGGGCTTTTTCCTGGTGACTTTTGGCTTCATTTGGGGTATGATGCTTCTGCACTTTACCATCCAGCAG CGAACTCAGCCTGAAAGCAGCTCCATGCTGCGCGAGCAGATCCTGGACCTCAGCAAAAGGTACATCAAGGCACTGGCAGAAGAAAACAGG AATGTGGTGGATGGGCCATACGCTGGAGTCATGACAGCTTATGTCATCAGCAGTGAGGACTCTGAAGGATCCACTGACGTTGATGAGCCC TTAGAAGTCTTCATCTCAGCACCGAGAAGTGAGCCTGTGATCAATAATGACAACCCTTTAGAATCAAATGATGAAAAGGAGGGCCAAGAA GCCACTTGCTCACGACCCCAGATTGTACCAGAGCCCATGGATTTCAGAAAATTATCTACATTCAGAGAAAGTTTTAAGAAAAGAGTGATA GGACAAGACCACGACTTTTCAGAATCCAGTGAGGAGGAGGCGCCCGCAGAAGCCTCGAGCGGGGCACTGAGAAGCAAGCATGGTGAGAAG GCTCCTATGACTTCTAGAAGTACATCTACTTGGAGAATACCCAGCAGGAAGAGACGTTTCAGCAGTAGTGACTTTTCAGACCTGAGTAAT GGAGAAGAGCTTCAGGAAACCTGCAGCTCATCCCTAAGAAGAGGGTCAGGATCACAGCCACAAGAACCTGAAAATAAGAAGTGCTCCTGT GTCATGTGTTTTCCAAAAGGTGTGCCAAGAAGCCAAGAAGCAAGGACTGAAAGTAGTCAAGCATCTGACATGATGGATACCATGGATGTT GAAAACAATTCTACTTTGGAAAAACACAGTGGGAAAAGAAGAAAAAAGAGAAGGCATAGATCTAAAGTAAATGGTCTCCAAAGAGGGAGA AAGAAAGACAGACCTAGAAAACATTTAACTCTGAATAACAAAGTCCAAAAGAAAAGATGGCAACAAAGAGGAAGAAAAGCCAACACTAGA CCTTTGAAAAGAAGAAGAAAAAGAGGTCCAAGAATTCCCAAAGATGAAAATATTAATTTTAAACAATCTGAACTTCCTGTGACCTGTGGT GAGGTGAAGGGCACTCTATATAAGGAGCGATTCAAACAAGGAACCTCAAAGAAGTGTATACAGAGTGAGGATAAAAAGTGGTTCACTCCC AGGGAATTTGAAATTGAAGGAGACCGCGGAGCATCCAAGAACTGGAAGCTAAGTATACGCTGCGGTGGATATACCCTGAAAGTCCTGATG GAGAACAAATTTCTGCCAGAACCACCAAGCACAAGAAAAAAGAGAATACTGGAATCTCACAACAATACCTTAGTTGACCCTTGTCCGGAA AACTCAAATATATGTGAGGTGTGCAACAAATGGGGACGGCTGTTCTGCTGCGACACTTGTCCAAGATCCTTTCATGAGCACTGCCACATC CCATCCGTGGAAGCTAACAAGAACCCGTGGAGTTGCATCTTCTGCAGGATAAAGACTATTCAGGAAAGATGCCCAGAAAGCCAATCAGGT CATCAGGAATCTGAAGTCCTGATGAGGCAGATGCTGCCTGAGGAGCAGTTGAAATGTGAATTCCTCCTCTTGAAGGTCTACTGTGATTCG AAAAGCTGCTTTTTCGCCTCAGAACCGTATTATAACAGAGAGGGGTCTCAGGGCCCACAGAAGCCCATGTGGTTAAACAAAGTCAAGACA AGTTTGAATGAGCAGATGTACACCCGAGTAGAAGGGTTTGTGCAGGACATGCGTCTCATCTTTCATAACCACAAGGAATTTTACAGGGAA GATAAATTCACCAGACTGGGAATTCAAGTACAGGACATCTTTGAGAAGAATTTCAGAAACATTTTTGCAATTCAGGAAACAAGCAAGAAC ATTATAATGTTTATTTAGCCATTCTTATCTCCTCCCTTCAGATCCTCTGGCAGCTAGCTACGCAATGTGCCTGTGGTCCCACTAATCTGT GACTGCTCCTGTGGAAACTCCACATCACAATTCTCCAAAATTTATCATTGCCATTTTAAAACCGTCTTTTCAGCTTTCAATAAAATTCAA >53360_53360_10_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000409645_SP100_chr2_231327150_ENST00000340126_length(amino acids)=641AA_BP=80 MALFTPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQPESSSMLREQILDLSKRYIKALAEENRNVVDGPYAGVMTAYVISSEDSEGS TDVDEPLEVFISAPRSEPVINNDNPLESNDEKEGQEATCSRPQIVPEPMDFRKLSTFRESFKKRVIGQDHDFSESSEEEAPAEASSGALR SKHGEKAPMTSRSTSTWRIPSRKRRFSSSDFSDLSNGEELQETCSSSLRRGSGSQPQEPENKKCSCVMCFPKGVPRSQEARTESSQASDM MDTMDVENNSTLEKHSGKRRKKRRHRSKVNGLQRGRKKDRPRKHLTLNNKVQKKRWQQRGRKANTRPLKRRRKRGPRIPKDENINFKQSE LPVTCGEVKGTLYKERFKQGTSKKCIQSEDKKWFTPREFEIEGDRGASKNWKLSIRCGGYTLKVLMENKFLPEPPSTRKKRILESHNNTL VDPCPENSNICEVCNKWGRLFCCDTCPRSFHEHCHIPSVEANKNPWSCIFCRIKTIQERCPESQSGHQESEVLMRQMLPEEQLKCEFLLL KVYCDSKSCFFASEPYYNREGSQGPQKPMWLNKVKTSLNEQMYTRVEGFVQDMRLIFHNHKEFYREDKFTRLGIQVQDIFEKNFRNIFAI -------------------------------------------------------------- >53360_53360_11_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000409645_SP100_chr2_231327150_ENST00000341950_length(transcript)=942nt_BP=493nt AGCCCAGGTAGCCGCGCCGCAGGCTCGGGGCGTCGCGACCTCGCGCCTCGGGCCGCGTGGGCACGGCGGCCGGCGGGTGCTCCCGGCTGC TGCTGCTGCCCAACAAGGAGCAGAATGGAAGTGAGGAAAGGCAACCAGCTGACACAGGAGCCAGAGTGAGACCAGCAGACTCTCACACTC AACCTACACCATGAATTTGTGTCTATCTTCTACGCGTTAAGAGCCAAGGACAGGTGAAGTTGCCAGAGAGCAATGGCTCTCTTCACTCCG TGGAAGTTGTCCTCTCAGAAGCTGGGCTTTTTCCTGGTGACTTTTGGCTTCATTTGGGGTATGATGCTTCTGCACTTTACCATCCAGCAG CGAACTCAGCCTGAAAGCAGCTCCATGCTGCGCGAGCAGATCCTGGACCTCAGCAAAAGGTACATCAAGGCACTGGCAGAAGAAAACAGG AATGTGGTGGATGGGCCATACGCTGGAGTCATGACAGCTTATGTCATCAGCAGTGAGGACTCTGAAGGATCCACTGACGTTGATGAGCCC TTAGAAGTCTTCATCTCAGCACCGAGAAGTGAGCCTGTGATCAATAATGACAACCCTTTAGAATCAAATGATGAAAAGGAGGGCCAAGAA GCCACTTGCTCACGACCCCAGATTGTACCAGAGCCCATGGATTTCAGAAAATTATCTACATTCAGAGAAAGTTTTAAGAAAAGAGTGATA GGACAAGACCACGACTTTTCAGAATCCAGTGAGGAGGAGGCGCCCGCAGAAGCCTCGAGCGGGGCACTGAGAAGCAAGCATGGTGAGAAG GCTCCTATGACTTCTAGAAGTACATCTACTTGGAGAATACCCAGCAGGAAGAGACAATTAAAAAAAAAAAAGAAGAAGAAACAATGTCAT >53360_53360_11_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000409645_SP100_chr2_231327150_ENST00000341950_length(amino acids)=229AA_BP=80 MALFTPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQPESSSMLREQILDLSKRYIKALAEENRNVVDGPYAGVMTAYVISSEDSEGS TDVDEPLEVFISAPRSEPVINNDNPLESNDEKEGQEATCSRPQIVPEPMDFRKLSTFRESFKKRVIGQDHDFSESSEEEAPAEASSGALR -------------------------------------------------------------- >53360_53360_12_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000409645_SP100_chr2_231327150_ENST00000409112_length(transcript)=1687nt_BP=493nt AGCCCAGGTAGCCGCGCCGCAGGCTCGGGGCGTCGCGACCTCGCGCCTCGGGCCGCGTGGGCACGGCGGCCGGCGGGTGCTCCCGGCTGC TGCTGCTGCCCAACAAGGAGCAGAATGGAAGTGAGGAAAGGCAACCAGCTGACACAGGAGCCAGAGTGAGACCAGCAGACTCTCACACTC AACCTACACCATGAATTTGTGTCTATCTTCTACGCGTTAAGAGCCAAGGACAGGTGAAGTTGCCAGAGAGCAATGGCTCTCTTCACTCCG TGGAAGTTGTCCTCTCAGAAGCTGGGCTTTTTCCTGGTGACTTTTGGCTTCATTTGGGGTATGATGCTTCTGCACTTTACCATCCAGCAG CGAACTCAGCCTGAAAGCAGCTCCATGCTGCGCGAGCAGATCCTGGACCTCAGCAAAAGGTACATCAAGGCACTGGCAGAAGAAAACAGG AATGTGGTGGATGGGCCATACGCTGGAGTCATGACAGCTTATGTCATCAGCAGTGAGGACTCTGAAGGATCCACTGACGTTGATGAGCCC TTAGAAGTCTTCATCTCAGCACCGAGAAGTGAGCCTGTGATCAATAATGACAACCCTTTAGAATCAAATGATGAAAAGGAGGGCCAAGAA GCCACTTGCTCACGACCCCAGATTGTACCAGAGCCCATGGATTTCAGAAAATTATCTACATTCAGAGAAAGTTTTAAGAAAAGAGTGATA GGACAAGACCACGACTTTTCAGAATCCAGTGAGGAGGAGGCGCCCGCAGAAGCCTCGAGCGGGGCACTGAGAAGCAAGCATGGTGAGAAG GCTCCTATGACTTCTAGAAGTACATCTACTTGGAGAATACCCAGCAGGAAGAGACGTTTCAGCAGTAGTGACTTTTCAGACCTGAGTAAT GGAGAAGAGCTTCAGGAAACCTGCAGCTCATCCCTAAGAAGAGGGTCAGGATCACAGCCACAAGAACCTGAAAATAAGAAGTGCTCCTGT GTCATGTGTTTTCCAAAAGGTGTGCCAAGAAGCCAAGAAGCAAGGACTGAAAGTAGTCAAGCATCTGACATGATGGATACCATGGATGTT GAAAACAATTCTACTTTGGAAAAACACAGTGGGAAAAGAAGAAAAAAGAGAAGGCATAGATCTAAAGTAAATGGTCTCCAAAGAGGGAGA AAGAAAGACAGACCTAGAAAACATTTAACTCTGAATAACAAAGTCCAAAAGAAAAGATGGCAACAAAGAGGAAGAAAAGCCAACACTAGA CCTTTGAAAAGAAGAAGAAAAAGAGGTCCAAGAATTCCCAAAGATGAAAATATTAATTTTAAACAATCTGAACTTCCTGTGACCTGTGGT GAGGTGAAGGGCACTCTATATAAGGAGCGATTCAAACAAGGAACCTCAAAGAAGTGTATACAGAGTGAGGATAAAAAGTGGTTCACTCCC AGGGAATTTGAAATTGAAGGAGACCGCGGAGCATCCAAGAACTGGAAGCTAAGTATACGCTGCGGTGGATATACCCTGAAAGTCCTGATG GAGAACAAATTTCTGCCAGAACCACCAAGCACAAGAAAAAAGGTGATGATCAAGTGATCTTCTGCCAATGTCTCGTCTATTATGTTGTTG >53360_53360_12_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000409645_SP100_chr2_231327150_ENST00000409112_length(amino acids)=444AA_BP=80 MALFTPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQPESSSMLREQILDLSKRYIKALAEENRNVVDGPYAGVMTAYVISSEDSEGS TDVDEPLEVFISAPRSEPVINNDNPLESNDEKEGQEATCSRPQIVPEPMDFRKLSTFRESFKKRVIGQDHDFSESSEEEAPAEASSGALR SKHGEKAPMTSRSTSTWRIPSRKRRFSSSDFSDLSNGEELQETCSSSLRRGSGSQPQEPENKKCSCVMCFPKGVPRSQEARTESSQASDM MDTMDVENNSTLEKHSGKRRKKRRHRSKVNGLQRGRKKDRPRKHLTLNNKVQKKRWQQRGRKANTRPLKRRRKRGPRIPKDENINFKQSE -------------------------------------------------------------- >53360_53360_13_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000409645_SP100_chr2_231327150_ENST00000409341_length(transcript)=1368nt_BP=493nt AGCCCAGGTAGCCGCGCCGCAGGCTCGGGGCGTCGCGACCTCGCGCCTCGGGCCGCGTGGGCACGGCGGCCGGCGGGTGCTCCCGGCTGC TGCTGCTGCCCAACAAGGAGCAGAATGGAAGTGAGGAAAGGCAACCAGCTGACACAGGAGCCAGAGTGAGACCAGCAGACTCTCACACTC AACCTACACCATGAATTTGTGTCTATCTTCTACGCGTTAAGAGCCAAGGACAGGTGAAGTTGCCAGAGAGCAATGGCTCTCTTCACTCCG TGGAAGTTGTCCTCTCAGAAGCTGGGCTTTTTCCTGGTGACTTTTGGCTTCATTTGGGGTATGATGCTTCTGCACTTTACCATCCAGCAG CGAACTCAGCCTGAAAGCAGCTCCATGCTGCGCGAGCAGATCCTGGACCTCAGCAAAAGGTACATCAAGGCACTGGCAGAAGAAAACAGG AATGTGGTGGATGGGCCATACGCTGGAGTCATGACAGCTTATGTCATCAGCAGTGAGGACTCTGAAGGATCCACTGACGTTGATGAGCCC TTAGAAGTCTTCATCTCAGCACCGAGAAGTGAGCCTGTGATCAATAATGACAACCCTTTAGAATCAAATGATGAAAAGGAGGGCCAAGAA GCCACTTGCTCACGACCCCAGATTGTACCAGAGCCCATGGATTTCAGAAAATTATCTACATTCAGAGAAAGTTTTAAGAAAAGAGTGATA GGACAAGACCACGACTTTTCAGAATCCAGTGAGGAGGAGGCGCCCGCAGAAGCCTCGAGCGGGGCACTGAGAAGCAAGCATGGTGAGAAG GCTCCTATGACTTCTAGAAGTACATCTACTTGGAGAATACCCAGCAGGAAGAGACGTTTCAGCAGTAGTGACTTTTCAGACCTGAGTAAT GGAGAAGAGCTTCAGGAAACCTGCAGCTCATCCCTAAGAAGAGGGTCAGGTAAAGAAGATTAGGATGCCAAGACTTGGCCTGCAGAATGT CAGGAATGTGAATTAAAAGCTGCTGTTTCCAGACGCTTTTTATTCTGAGCACCTTCACTACCTTGTATCCAGTTCATCTGGGAACTCCTT TTTGCATTTTAGAAAATGGAAAGAGGCAGGAAATTATGATAAACTCATGTTTAACAGAAAGAGTTTCACTGACTAAATGTATGTAATTAT ATTTTGTTGTTGTAGAAGAAATAAATAGCAAATTTGTGGTATTCTTTTTTTTAAACCTGCTCTCATTCCTATTAACACTAAGATCTTAGA TTTTTATAGTGATAAATGGGTTGACATCATTGTCATTTGTAATTGTAAAGCCTCAAAAGACAACTGTTCCTACTATGTAATTATAGACAG >53360_53360_13_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000409645_SP100_chr2_231327150_ENST00000409341_length(amino acids)=236AA_BP=80 MALFTPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQPESSSMLREQILDLSKRYIKALAEENRNVVDGPYAGVMTAYVISSEDSEGS TDVDEPLEVFISAPRSEPVINNDNPLESNDEKEGQEATCSRPQIVPEPMDFRKLSTFRESFKKRVIGQDHDFSESSEEEAPAEASSGALR -------------------------------------------------------------- >53360_53360_14_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000409645_SP100_chr2_231327150_ENST00000409824_length(transcript)=1340nt_BP=493nt AGCCCAGGTAGCCGCGCCGCAGGCTCGGGGCGTCGCGACCTCGCGCCTCGGGCCGCGTGGGCACGGCGGCCGGCGGGTGCTCCCGGCTGC TGCTGCTGCCCAACAAGGAGCAGAATGGAAGTGAGGAAAGGCAACCAGCTGACACAGGAGCCAGAGTGAGACCAGCAGACTCTCACACTC AACCTACACCATGAATTTGTGTCTATCTTCTACGCGTTAAGAGCCAAGGACAGGTGAAGTTGCCAGAGAGCAATGGCTCTCTTCACTCCG TGGAAGTTGTCCTCTCAGAAGCTGGGCTTTTTCCTGGTGACTTTTGGCTTCATTTGGGGTATGATGCTTCTGCACTTTACCATCCAGCAG CGAACTCAGCCTGAAAGCAGCTCCATGCTGCGCGAGCAGATCCTGGACCTCAGCAAAAGGTACATCAAGGCACTGGCAGAAGAAAACAGG AATGTGGTGGATGGGCCATACGCTGGAGTCATGACAGCTTATGTCATCAGCAGTGAGGACTCTGAAGGATCCACTGACGTTGATGAGCCC TTAGAAGTCTTCATCTCAGCACCGAGAAGTGAGCCTGTGATCAATAATGACAACCCTTTAGAATCAAATGATGAAAAGGAGGGCCAAGAA GCCACTTGCTCACGACCCCAGATTGTACCAGAGCCCATGGATTTCAGAAAATTATCTACATTCAGAGAAAGTTTTAAGAAAAGAGTGATA GGACAAGACCACGACTTTTCAGAATCCAGTGAGGAGGAGGCGCCCGCAGAAGCCTCGAGCGGGGCACTGAGAAGCAAGCATGGTGAGAAG GCTCCTATGACTTCTAGAAGTACATCTACTTGGAGAATACCCAGCAGGAAGAGACGTTTCAGCAGTAGTGACTTTTCAGACCTGAGTAAT GGAGAAGAGCTTCAGGAAACCTGCAGCTCATCCCTAAGAAGAGGGTCAGGTAAAGAAGATTAGGATGCCAAGACTTGGCCTGCAGAATGT CAGGAATGTGAATTAAAAGCTGCTGTTTCCAGACGCTTTTTATTCTGAGCACCTTCACTACCTTGTATCCAGTTCATCTGGGAACTCCTT TTTGCATTTTAGAAAATGGAAAGAGGCAGGAAATTATGATAAACTCATGTTTAACAGAAAGAGTTTCACTGACTAAATGTATGTAATTAT ATTTTGTTGTTGTAGAAGAAATAAATAGCAAATTTGTGGTATTCTTTTTTTTAAACCTGCTCTCATTCCTATTAACACTAAGATCTTAGA >53360_53360_14_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000409645_SP100_chr2_231327150_ENST00000409824_length(amino acids)=236AA_BP=80 MALFTPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQPESSSMLREQILDLSKRYIKALAEENRNVVDGPYAGVMTAYVISSEDSEGS TDVDEPLEVFISAPRSEPVINNDNPLESNDEKEGQEATCSRPQIVPEPMDFRKLSTFRESFKKRVIGQDHDFSESSEEEAPAEASSGALR -------------------------------------------------------------- >53360_53360_15_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000409645_SP100_chr2_231327150_ENST00000409897_length(transcript)=1381nt_BP=493nt AGCCCAGGTAGCCGCGCCGCAGGCTCGGGGCGTCGCGACCTCGCGCCTCGGGCCGCGTGGGCACGGCGGCCGGCGGGTGCTCCCGGCTGC TGCTGCTGCCCAACAAGGAGCAGAATGGAAGTGAGGAAAGGCAACCAGCTGACACAGGAGCCAGAGTGAGACCAGCAGACTCTCACACTC AACCTACACCATGAATTTGTGTCTATCTTCTACGCGTTAAGAGCCAAGGACAGGTGAAGTTGCCAGAGAGCAATGGCTCTCTTCACTCCG TGGAAGTTGTCCTCTCAGAAGCTGGGCTTTTTCCTGGTGACTTTTGGCTTCATTTGGGGTATGATGCTTCTGCACTTTACCATCCAGCAG CGAACTCAGCCTGAAAGCAGCTCCATGCTGCGCGAGCAGATCCTGGACCTCAGCAAAAGGTACATCAAGGCACTGGCAGAAGAAAACAGG AATGTGGTGGATGGGCCATACGCTGGAGTCATGACAGCTTATGTCATCAGCAGTGAGGACTCTGAAGGATCCACTGACGTTGATGAGCCC TTAGAAGTCTTCATCTCAGCACCGAGAAGTGAGCCTGTGATCAATAATGACAACCCTTTAGAATCAAATGATGAAAAGGAGGGCCAAGAA GCCACTTGCTCACGACCCCAGATTGTACCAGAGCCCATGGATTTCAGAAAATTATCTACATTCAGAGAAAGTTTTAAGAAAAGAGTGATA GGACAAGACCACGACTTTTCAGAATCCAGTGAGGAGGAGGCGCCCGCAGAAGCCTCGAGCGGGGCACTGAGAAGCAAGCATGGTGAGAAG GCTCCTATGACTTCTAGAAGTACATCTACTTGGAGAATACCCAGCAGGAAGAGACGTTTCAGCAGTAGTGACTTTTCAGACCTGAGTAAT GGAGAAGAGCTTCAGGAAACCTGCAGCTCATCCCTAAGAAGAGGGTCAGGTAAAGAAGATTAGGATGCCAAGACTTGGCCTGCAGAATGT CAGGAATGTGAATTAAAAGCTGCTGTTTCCAGACGCTTTTTATTCTGAGCACCTTCACTACCTTGTATCCAGTTCATCTGGGAACTCCTT TTTGCATTTTAGAAAATGGAAAGAGGCAGGAAATTATGATAAACTCATGTTTAACAGAAAGAGTTTCACTGACTAAATGTATGTAATTAT ATTTTGTTGTTGTAGAAGAAATAAATAGCAAATTTGTGGTATTCTTTTTTTTAAACCTGCTCTCATTCCTATTAACACTAAGATCTTAGA TTTTTATAGTGATAAATGGGTTGACATCATTGTCATTTGTAATTGTAAAGCCTCAAAAGACAACTGTTCCTACTATGTAATTATAGACAG >53360_53360_15_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000409645_SP100_chr2_231327150_ENST00000409897_length(amino acids)=236AA_BP=80 MALFTPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQPESSSMLREQILDLSKRYIKALAEENRNVVDGPYAGVMTAYVISSEDSEGS TDVDEPLEVFISAPRSEPVINNDNPLESNDEKEGQEATCSRPQIVPEPMDFRKLSTFRESFKKRVIGQDHDFSESSEEEAPAEASSGALR -------------------------------------------------------------- >53360_53360_16_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000409645_SP100_chr2_231327150_ENST00000427101_length(transcript)=1363nt_BP=493nt AGCCCAGGTAGCCGCGCCGCAGGCTCGGGGCGTCGCGACCTCGCGCCTCGGGCCGCGTGGGCACGGCGGCCGGCGGGTGCTCCCGGCTGC TGCTGCTGCCCAACAAGGAGCAGAATGGAAGTGAGGAAAGGCAACCAGCTGACACAGGAGCCAGAGTGAGACCAGCAGACTCTCACACTC AACCTACACCATGAATTTGTGTCTATCTTCTACGCGTTAAGAGCCAAGGACAGGTGAAGTTGCCAGAGAGCAATGGCTCTCTTCACTCCG TGGAAGTTGTCCTCTCAGAAGCTGGGCTTTTTCCTGGTGACTTTTGGCTTCATTTGGGGTATGATGCTTCTGCACTTTACCATCCAGCAG CGAACTCAGCCTGAAAGCAGCTCCATGCTGCGCGAGCAGATCCTGGACCTCAGCAAAAGGTACATCAAGGCACTGGCAGAAGAAAACAGG AATGTGGTGGATGGGCCATACGCTGGAGTCATGACAGCTTATGTCATCAGCAGTGAGGACTCTGAAGGATCCACTGACGTTGATGAGCCC TTAGAAGTCTTCATCTCAGCACCGAGAAGTGAGCCTGTGATCAATAATGACAACCCTTTAGAATCAAATGATGAAAAGGAGGGCCAAGAA GCCACTTGCTCACGACCCCAGATTGTACCAGAGCCCATGGATTTCAGAAAATTATCTACATTCAGAGAAAGTTTTAAGAAAAGAGTGATA GGACAAGACCACGACTTTTCAGAATCCAGTGAGGAGGAGGCGCCCGCAGAAGCCTCGAGCGGGGCACTGAGAAGCAAGCATGCTCCTATG ACTTCTAGAAGTACATCTACTTGGAGAATACCCAGCAGGAAGAGACGTTTCAGCAGTAGTGACTTTTCAGACCTGAGTAATGGAGAAGAG CTTCAGGAAACCTGCAGCTCATCCCTAAGAAGAGGGTCAGGTAAAGAAGATTAGGATGCCAAGACTTGGCCTGCAGAATGTCAGGAATGT GAATTAAAAGCTGCTGTTTCCAGACGCTTTTTATTCTGAGCACCTTCACTACCTTGTATCCAGTTCATCTGGGAACTCCTTTTTGCATTT TAGAAAATGGAAAGAGGCAGGAAATTATGATAAACTCATGTTTAACAGAAAGAGTTTCACTGACTAAATGTATGTAATTATATTTTGTTG TTGTAGAAGAAATAAATAGCAAATTTGTGGTATTCTTTTTTTTAAACCTGCTCTCATTCCTATTAACACTAAGATCTTAGATTTTTATAG TGATAAATGGGTTGACATCATTGTCATTTGTAATTGTAAAGCCTCAAAAGACAACTGTTCCTACTATGTAATTATAGACAGAAATAAAAA >53360_53360_16_MGAT5-SP100_MGAT5_chr2_135012215_ENST00000409645_SP100_chr2_231327150_ENST00000427101_length(amino acids)=233AA_BP=80 MALFTPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQPESSSMLREQILDLSKRYIKALAEENRNVVDGPYAGVMTAYVISSEDSEGS TDVDEPLEVFISAPRSEPVINNDNPLESNDEKEGQEATCSRPQIVPEPMDFRKLSTFRESFKKRVIGQDHDFSESSEEEAPAEASSGALR -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for MGAT5-SP100 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for MGAT5-SP100 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for MGAT5-SP100 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
