
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:MGP-ESR1 (FusionGDB2 ID:53416) |
Fusion Gene Summary for MGP-ESR1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: MGP-ESR1 | Fusion gene ID: 53416 | Hgene | Tgene | Gene symbol | MGP | ESR1 | Gene ID | 4256 | 2099 |
| Gene name | matrix Gla protein | estrogen receptor 1 | |
| Synonyms | GIG36|MGLAP|NTI | ER|ESR|ESRA|ESTRR|Era|NR3A1 | |
| Cytomap | 12p12.3 | 6q25.1-q25.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | matrix Gla proteincell growth-inhibiting gene 36 protein | estrogen receptorE2 receptor alphaER-alphaestradiol receptorestrogen nuclear receptor alphaestrogen receptor alpha E1-E2-1-2estrogen receptor alpha E1-N2-E2-1-2nuclear receptor subfamily 3 group A member 1oestrogen receptor alpha | |
| Modification date | 20200313 | 20200329 | |
| UniProtAcc | P08493 | P03372 | |
| Ensembl transtripts involved in fusion gene | ENST00000228938, ENST00000539261, | ENST00000206249, ENST00000406599, ENST00000427531, ENST00000443427, ENST00000456483, ENST00000482101, ENST00000338799, ENST00000440973, | |
| Fusion gene scores | * DoF score | 7 X 8 X 2=112 | 10 X 10 X 6=600 |
| # samples | 8 | 12 | |
| ** MAII score | log2(8/112*10)=-0.485426827170242 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(12/600*10)=-2.32192809488736 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: MGP [Title/Abstract] AND ESR1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | MGP(15037147)-ESR1(152265308), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | ESR1 | GO:0006366 | transcription by RNA polymerase II | 15831516 |
| Tgene | ESR1 | GO:0010629 | negative regulation of gene expression | 21695196 |
| Tgene | ESR1 | GO:0030520 | intracellular estrogen receptor signaling pathway | 9841876 |
| Tgene | ESR1 | GO:0032355 | response to estradiol | 15304487 |
| Tgene | ESR1 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling | 7651415|16043358 |
| Tgene | ESR1 | GO:0043433 | negative regulation of DNA-binding transcription factor activity | 10816575 |
| Tgene | ESR1 | GO:0043627 | response to estrogen | 11581164 |
| Tgene | ESR1 | GO:0045893 | positive regulation of transcription, DNA-templated | 9841876|20074560 |
| Tgene | ESR1 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly | 9841876 |
| Tgene | ESR1 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 11544182|12047722|15345745|15831516|18563714 |
| Tgene | ESR1 | GO:0051091 | positive regulation of DNA-binding transcription factor activity | 9328340|10681512 |
| Tgene | ESR1 | GO:0071392 | cellular response to estradiol stimulus | 15831516 |
 Fusion gene breakpoints across MGP (5'-gene) Fusion gene breakpoints across MGP (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
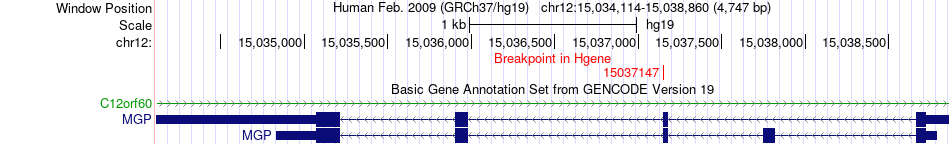 |
 Fusion gene breakpoints across ESR1 (3'-gene) Fusion gene breakpoints across ESR1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
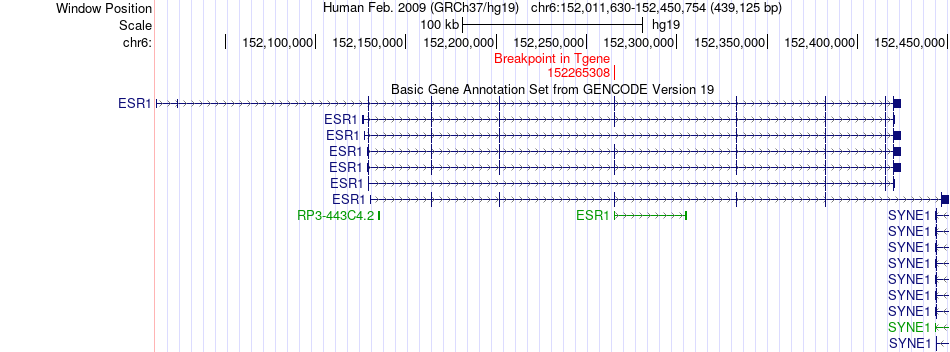 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-AR-A0U3-01A | MGP | chr12 | 15037147 | - | ESR1 | chr6 | 152265308 | + |
Top |
Fusion Gene ORF analysis for MGP-ESR1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000228938 | ENST00000206249 | MGP | chr12 | 15037147 | - | ESR1 | chr6 | 152265308 | + |
| 5CDS-intron | ENST00000228938 | ENST00000406599 | MGP | chr12 | 15037147 | - | ESR1 | chr6 | 152265308 | + |
| 5CDS-intron | ENST00000228938 | ENST00000427531 | MGP | chr12 | 15037147 | - | ESR1 | chr6 | 152265308 | + |
| 5CDS-intron | ENST00000228938 | ENST00000443427 | MGP | chr12 | 15037147 | - | ESR1 | chr6 | 152265308 | + |
| 5CDS-intron | ENST00000228938 | ENST00000456483 | MGP | chr12 | 15037147 | - | ESR1 | chr6 | 152265308 | + |
| 5CDS-intron | ENST00000228938 | ENST00000482101 | MGP | chr12 | 15037147 | - | ESR1 | chr6 | 152265308 | + |
| 5CDS-intron | ENST00000539261 | ENST00000206249 | MGP | chr12 | 15037147 | - | ESR1 | chr6 | 152265308 | + |
| 5CDS-intron | ENST00000539261 | ENST00000406599 | MGP | chr12 | 15037147 | - | ESR1 | chr6 | 152265308 | + |
| 5CDS-intron | ENST00000539261 | ENST00000427531 | MGP | chr12 | 15037147 | - | ESR1 | chr6 | 152265308 | + |
| 5CDS-intron | ENST00000539261 | ENST00000443427 | MGP | chr12 | 15037147 | - | ESR1 | chr6 | 152265308 | + |
| 5CDS-intron | ENST00000539261 | ENST00000456483 | MGP | chr12 | 15037147 | - | ESR1 | chr6 | 152265308 | + |
| 5CDS-intron | ENST00000539261 | ENST00000482101 | MGP | chr12 | 15037147 | - | ESR1 | chr6 | 152265308 | + |
| In-frame | ENST00000228938 | ENST00000338799 | MGP | chr12 | 15037147 | - | ESR1 | chr6 | 152265308 | + |
| In-frame | ENST00000228938 | ENST00000440973 | MGP | chr12 | 15037147 | - | ESR1 | chr6 | 152265308 | + |
| In-frame | ENST00000539261 | ENST00000338799 | MGP | chr12 | 15037147 | - | ESR1 | chr6 | 152265308 | + |
| In-frame | ENST00000539261 | ENST00000440973 | MGP | chr12 | 15037147 | - | ESR1 | chr6 | 152265308 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000539261 | MGP | chr12 | 15037147 | - | ENST00000440973 | ESR1 | chr6 | 152265308 | + | 5565 | 229 | 120 | 1256 | 378 |
| ENST00000539261 | MGP | chr12 | 15037147 | - | ENST00000338799 | ESR1 | chr6 | 152265308 | + | 1344 | 229 | 120 | 1256 | 378 |
| ENST00000228938 | MGP | chr12 | 15037147 | - | ENST00000440973 | ESR1 | chr6 | 152265308 | + | 5571 | 235 | 51 | 1262 | 403 |
| ENST00000228938 | MGP | chr12 | 15037147 | - | ENST00000338799 | ESR1 | chr6 | 152265308 | + | 1350 | 235 | 51 | 1262 | 403 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000539261 | ENST00000440973 | MGP | chr12 | 15037147 | - | ESR1 | chr6 | 152265308 | + | 0.000268249 | 0.9997317 |
| ENST00000539261 | ENST00000338799 | MGP | chr12 | 15037147 | - | ESR1 | chr6 | 152265308 | + | 0.001521871 | 0.9984781 |
| ENST00000228938 | ENST00000440973 | MGP | chr12 | 15037147 | - | ESR1 | chr6 | 152265308 | + | 0.000232732 | 0.99976724 |
| ENST00000228938 | ENST00000338799 | MGP | chr12 | 15037147 | - | ESR1 | chr6 | 152265308 | + | 0.001832375 | 0.99816763 |
Top |
Fusion Genomic Features for MGP-ESR1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
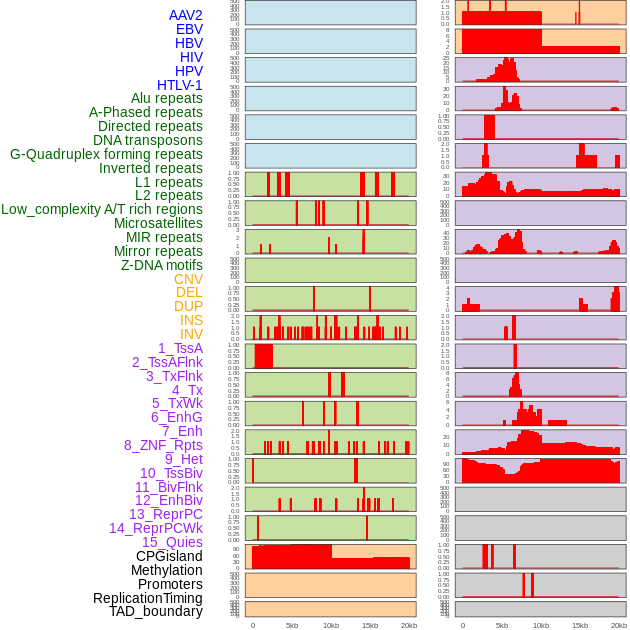 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
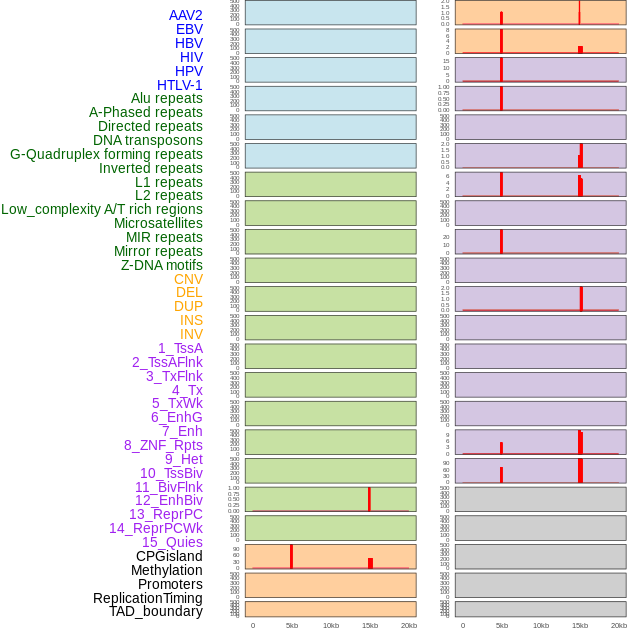 |
Top |
Fusion Protein Features for MGP-ESR1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:15037147/chr6:152265308) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MGP | ESR1 |
| FUNCTION: Associates with the organic matrix of bone and cartilage. Thought to act as an inhibitor of bone formation. | FUNCTION: Nuclear hormone receptor. The steroid hormones and their receptors are involved in the regulation of eukaryotic gene expression and affect cellular proliferation and differentiation in target tissues. Ligand-dependent nuclear transactivation involves either direct homodimer binding to a palindromic estrogen response element (ERE) sequence or association with other DNA-binding transcription factors, such as AP-1/c-Jun, c-Fos, ATF-2, Sp1 and Sp3, to mediate ERE-independent signaling. Ligand binding induces a conformational change allowing subsequent or combinatorial association with multiprotein coactivator complexes through LXXLL motifs of their respective components. Mutual transrepression occurs between the estrogen receptor (ER) and NF-kappa-B in a cell-type specific manner. Decreases NF-kappa-B DNA-binding activity and inhibits NF-kappa-B-mediated transcription from the IL6 promoter and displace RELA/p65 and associated coregulators from the promoter. Recruited to the NF-kappa-B response element of the CCL2 and IL8 promoters and can displace CREBBP. Present with NF-kappa-B components RELA/p65 and NFKB1/p50 on ERE sequences. Can also act synergistically with NF-kappa-B to activate transcription involving respective recruitment adjacent response elements; the function involves CREBBP. Can activate the transcriptional activity of TFF1. Also mediates membrane-initiated estrogen signaling involving various kinase cascades. Isoform 3 is involved in activation of NOS3 and endothelial nitric oxide production. Isoforms lacking one or several functional domains are thought to modulate transcriptional activity by competitive ligand or DNA binding and/or heterodimerization with the full-length receptor. Essential for MTA1-mediated transcriptional regulation of BRCA1 and BCAS3. Isoform 3 can bind to ERE and inhibit isoform 1. {ECO:0000269|PubMed:10681512, ECO:0000269|PubMed:10816575, ECO:0000269|PubMed:11477071, ECO:0000269|PubMed:11682626, ECO:0000269|PubMed:14764652, ECO:0000269|PubMed:15078875, ECO:0000269|PubMed:15891768, ECO:0000269|PubMed:16043358, ECO:0000269|PubMed:16617102, ECO:0000269|PubMed:16684779, ECO:0000269|PubMed:17922032, ECO:0000269|PubMed:17932106, ECO:0000269|PubMed:18247370, ECO:0000269|PubMed:19350539, ECO:0000269|PubMed:20074560, ECO:0000269|PubMed:20705611, ECO:0000269|PubMed:21330404, ECO:0000269|PubMed:22083956, ECO:0000269|PubMed:7651415, ECO:0000269|PubMed:9328340}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000456483 | 0 | 8 | 185_250 | 0 | 484.0 | DNA binding | Nuclear receptor | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000206249 | 2 | 8 | 311_547 | 253 | 596.0 | Domain | NR LBD | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000338799 | 3 | 9 | 311_547 | 253 | 596.0 | Domain | NR LBD | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000440973 | 4 | 10 | 311_547 | 253 | 596.0 | Domain | NR LBD | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000443427 | 3 | 9 | 311_547 | 253 | 596.0 | Domain | NR LBD | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000456483 | 0 | 8 | 311_547 | 0 | 484.0 | Domain | NR LBD | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000206249 | 2 | 8 | 251_310 | 253 | 596.0 | Region | Note=Hinge | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000206249 | 2 | 8 | 264_595 | 253 | 596.0 | Region | Note=Self-association | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000206249 | 2 | 8 | 311_595 | 253 | 596.0 | Region | Note=Transactivation AF-2 | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000338799 | 3 | 9 | 251_310 | 253 | 596.0 | Region | Note=Hinge | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000338799 | 3 | 9 | 264_595 | 253 | 596.0 | Region | Note=Self-association | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000338799 | 3 | 9 | 311_595 | 253 | 596.0 | Region | Note=Transactivation AF-2 | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000440973 | 4 | 10 | 251_310 | 253 | 596.0 | Region | Note=Hinge | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000440973 | 4 | 10 | 264_595 | 253 | 596.0 | Region | Note=Self-association | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000440973 | 4 | 10 | 311_595 | 253 | 596.0 | Region | Note=Transactivation AF-2 | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000443427 | 3 | 9 | 251_310 | 253 | 596.0 | Region | Note=Hinge | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000443427 | 3 | 9 | 264_595 | 253 | 596.0 | Region | Note=Self-association | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000443427 | 3 | 9 | 311_595 | 253 | 596.0 | Region | Note=Transactivation AF-2 | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000456483 | 0 | 8 | 251_310 | 0 | 484.0 | Region | Note=Hinge | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000456483 | 0 | 8 | 264_595 | 0 | 484.0 | Region | Note=Self-association | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000456483 | 0 | 8 | 311_595 | 0 | 484.0 | Region | Note=Transactivation AF-2 | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000456483 | 0 | 8 | 185_205 | 0 | 484.0 | Zinc finger | NR C4-type | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000456483 | 0 | 8 | 221_245 | 0 | 484.0 | Zinc finger | NR C4-type |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MGP | chr12:15037147 | chr6:152265308 | ENST00000228938 | - | 3 | 5 | 51_97 | 56 | 129.0 | Domain | Gla |
| Hgene | MGP | chr12:15037147 | chr6:152265308 | ENST00000539261 | - | 2 | 4 | 51_97 | 31 | 104.0 | Domain | Gla |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000206249 | 2 | 8 | 185_250 | 253 | 596.0 | DNA binding | Nuclear receptor | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000338799 | 3 | 9 | 185_250 | 253 | 596.0 | DNA binding | Nuclear receptor | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000440973 | 4 | 10 | 185_250 | 253 | 596.0 | DNA binding | Nuclear receptor | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000443427 | 3 | 9 | 185_250 | 253 | 596.0 | DNA binding | Nuclear receptor | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000206249 | 2 | 8 | 185_205 | 253 | 596.0 | Zinc finger | NR C4-type | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000206249 | 2 | 8 | 221_245 | 253 | 596.0 | Zinc finger | NR C4-type | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000338799 | 3 | 9 | 185_205 | 253 | 596.0 | Zinc finger | NR C4-type | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000338799 | 3 | 9 | 221_245 | 253 | 596.0 | Zinc finger | NR C4-type | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000440973 | 4 | 10 | 185_205 | 253 | 596.0 | Zinc finger | NR C4-type | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000440973 | 4 | 10 | 221_245 | 253 | 596.0 | Zinc finger | NR C4-type | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000443427 | 3 | 9 | 185_205 | 253 | 596.0 | Zinc finger | NR C4-type | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000443427 | 3 | 9 | 221_245 | 253 | 596.0 | Zinc finger | NR C4-type |
Top |
Fusion Gene Sequence for MGP-ESR1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >53416_53416_1_MGP-ESR1_MGP_chr12_15037147_ENST00000228938_ESR1_chr6_152265308_ENST00000338799_length(transcript)=1350nt_BP=235nt CCCGTAGGAGCCTCTCTCCCTACTGCTGCTACACAAGACCCTGAGACTGACCTGCAGGACGAAACCATGAAGAGCCTGATCCTTCTTGCC ATCCTGGCCGCCTTAGCGGTAGTAACTTTGTGTTATGGAGAGTGGCAGAAAGAAGAAAACTTCGGCTTTGATATCGTTTCAGTTCTCTCT CTGAACTGGCATCGTGCCCAGGAATCACATGAAAGCATGGAATCTTATGAACTTAGGATACGAAAAGACCGAAGAGGAGGGAGAATGTTG AAACACAAGCGCCAGAGAGATGATGGGGAGGGCAGGGGTGAAGTGGGGTCTGCTGGAGACATGAGAGCTGCCAACCTTTGGCCAAGCCCG CTCATGATCAAACGCTCTAAGAAGAACAGCCTGGCCTTGTCCCTGACGGCCGACCAGATGGTCAGTGCCTTGTTGGATGCTGAGCCCCCG ATACTCTATTCCGAGTATGATCCTACCAGACCCTTCAGTGAAGCTTCGATGATGGGCTTACTGACCAACCTGGCAGACAGGGAGCTGGTT CACATGATCAACTGGGCGAAGAGGGTGCCAGGCTTTGTGGATTTGACCCTCCATGATCAGGTCCACCTTCTAGAATGTGCCTGGCTAGAG ATCCTGATGATTGGTCTCGTCTGGCGCTCCATGGAGCACCCAGGGAAGCTACTGTTTGCTCCTAACTTGCTCTTGGACAGGAACCAGGGA AAATGTGTAGAGGGCATGGTGGAGATCTTCGACATGCTGCTGGCTACATCATCTCGGTTCCGCATGATGAATCTGCAGGGAGAGGAGTTT GTGTGCCTCAAATCTATTATTTTGCTTAATTCTGGAGTGTACACATTTCTGTCCAGCACCCTGAAGTCTCTGGAAGAGAAGGACCATATC CACCGAGTCCTGGACAAGATCACAGACACTTTGATCCACCTGATGGCCAAGGCAGGCCTGACCCTGCAGCAGCAGCACCAGCGGCTGGCC CAGCTCCTCCTCATCCTCTCCCACATCAGGCACATGAGTAACAAAGGCATGGAGCATCTGTACAGCATGAAGTGCAAGAACGTGGTGCCC CTCTATGACCTGCTGCTGGAGATGCTGGACGCCCACCGCCTACATGCGCCCACTAGCCGTGGAGGGGCATCCGTGGAGGAGACGGACCAA AGCCACTTGGCCACTGCGGGCTCTACTTCATCGCATTCCTTGCAAAAGTATTACATCACGGGGGAGGCAGAGGGTTTCCCTGCCACGGTC TGAGAGCTCCCTGGCTCCCACACGGTTCAGATAATCCCTGCTGCATTTTACCCTCATCATGCACCACTTTAGCCAAATTCTGTCTCCTGC >53416_53416_1_MGP-ESR1_MGP_chr12_15037147_ENST00000228938_ESR1_chr6_152265308_ENST00000338799_length(amino acids)=403AA_BP=62 MQDETMKSLILLAILAALAVVTLCYGEWQKEENFGFDIVSVLSLNWHRAQESHESMESYELRIRKDRRGGRMLKHKRQRDDGEGRGEVGS AGDMRAANLWPSPLMIKRSKKNSLALSLTADQMVSALLDAEPPILYSEYDPTRPFSEASMMGLLTNLADRELVHMINWAKRVPGFVDLTL HDQVHLLECAWLEILMIGLVWRSMEHPGKLLFAPNLLLDRNQGKCVEGMVEIFDMLLATSSRFRMMNLQGEEFVCLKSIILLNSGVYTFL SSTLKSLEEKDHIHRVLDKITDTLIHLMAKAGLTLQQQHQRLAQLLLILSHIRHMSNKGMEHLYSMKCKNVVPLYDLLLEMLDAHRLHAP -------------------------------------------------------------- >53416_53416_2_MGP-ESR1_MGP_chr12_15037147_ENST00000228938_ESR1_chr6_152265308_ENST00000440973_length(transcript)=5571nt_BP=235nt CCCGTAGGAGCCTCTCTCCCTACTGCTGCTACACAAGACCCTGAGACTGACCTGCAGGACGAAACCATGAAGAGCCTGATCCTTCTTGCC ATCCTGGCCGCCTTAGCGGTAGTAACTTTGTGTTATGGAGAGTGGCAGAAAGAAGAAAACTTCGGCTTTGATATCGTTTCAGTTCTCTCT CTGAACTGGCATCGTGCCCAGGAATCACATGAAAGCATGGAATCTTATGAACTTAGGATACGAAAAGACCGAAGAGGAGGGAGAATGTTG AAACACAAGCGCCAGAGAGATGATGGGGAGGGCAGGGGTGAAGTGGGGTCTGCTGGAGACATGAGAGCTGCCAACCTTTGGCCAAGCCCG CTCATGATCAAACGCTCTAAGAAGAACAGCCTGGCCTTGTCCCTGACGGCCGACCAGATGGTCAGTGCCTTGTTGGATGCTGAGCCCCCG ATACTCTATTCCGAGTATGATCCTACCAGACCCTTCAGTGAAGCTTCGATGATGGGCTTACTGACCAACCTGGCAGACAGGGAGCTGGTT CACATGATCAACTGGGCGAAGAGGGTGCCAGGCTTTGTGGATTTGACCCTCCATGATCAGGTCCACCTTCTAGAATGTGCCTGGCTAGAG ATCCTGATGATTGGTCTCGTCTGGCGCTCCATGGAGCACCCAGGGAAGCTACTGTTTGCTCCTAACTTGCTCTTGGACAGGAACCAGGGA AAATGTGTAGAGGGCATGGTGGAGATCTTCGACATGCTGCTGGCTACATCATCTCGGTTCCGCATGATGAATCTGCAGGGAGAGGAGTTT GTGTGCCTCAAATCTATTATTTTGCTTAATTCTGGAGTGTACACATTTCTGTCCAGCACCCTGAAGTCTCTGGAAGAGAAGGACCATATC CACCGAGTCCTGGACAAGATCACAGACACTTTGATCCACCTGATGGCCAAGGCAGGCCTGACCCTGCAGCAGCAGCACCAGCGGCTGGCC CAGCTCCTCCTCATCCTCTCCCACATCAGGCACATGAGTAACAAAGGCATGGAGCATCTGTACAGCATGAAGTGCAAGAACGTGGTGCCC CTCTATGACCTGCTGCTGGAGATGCTGGACGCCCACCGCCTACATGCGCCCACTAGCCGTGGAGGGGCATCCGTGGAGGAGACGGACCAA AGCCACTTGGCCACTGCGGGCTCTACTTCATCGCATTCCTTGCAAAAGTATTACATCACGGGGGAGGCAGAGGGTTTCCCTGCCACGGTC TGAGAGCTCCCTGGCTCCCACACGGTTCAGATAATCCCTGCTGCATTTTACCCTCATCATGCACCACTTTAGCCAAATTCTGTCTCCTGC ATACACTCCGGCATGCATCCAACACCAATGGCTTTCTAGATGAGTGGCCATTCATTTGCTTGCTCAGTTCTTAGTGGCACATCTTCTGTC TTCTGTTGGGAACAGCCAAAGGGATTCCAAGGCTAAATCTTTGTAACAGCTCTCTTTCCCCCTTGCTATGTTACTAAGCGTGAGGATTCC CGTAGCTCTTCACAGCTGAACTCAGTCTATGGGTTGGGGCTCAGATAACTCTGTGCATTTAAGCTACTTGTAGAGACCCAGGCCTGGAGA GTAGACATTTTGCCTCTGATAAGCACTTTTTAAATGGCTCTAAGAATAAGCCACAGCAAAGAATTTAAAGTGGCTCCTTTAATTGGTGAC TTGGAGAAAGCTAGGTCAAGGGTTTATTATAGCACCCTCTTGTATTCCTATGGCAATGCATCCTTTTATGAAAGTGGTACACCTTAAAGC TTTTATATGACTGTAGCAGAGTATCTGGTGATTGTCAATTCATTCCCCCTATAGGAATACAAGGGGCACACAGGGAAGGCAGATCCCCTA GTTGGCAAGACTATTTTAACTTGATACACTGCAGATTCAGATGTGCTGAAAGCTCTGCCTCTGGCTTTCCGGTCATGGGTTCCAGTTAAT TCATGCCTCCCATGGACCTATGGAGAGCAGCAAGTTGATCTTAGTTAAGTCTCCCTATATGAGGGATAAGTTCCTGATTTTTGTTTTTAT TTTTGTGTTACAAAAGAAAGCCCTCCCTCCCTGAACTTGCAGTAAGGTCAGCTTCAGGACCTGTTCCAGTGGGCACTGTACTTGGATCTT CCCGGCGTGTGTGTGCCTTACACAGGGGTGAACTGTTCACTGTGGTGATGCATGATGAGGGTAAATGGTAGTTGAAAGGAGCAGGGGCCC TGGTGTTGCATTTAGCCCTGGGGCATGGAGCTGAACAGTACTTGTGCAGGATTGTTGTGGCTACTAGAGAACAAGAGGGAAAGTAGGGCA GAAACTGGATACAGTTCTGAGGCACAGCCAGACTTGCTCAGGGTGGCCCTGCCACAGGCTGCAGCTACCTAGGAACATTCCTTGCAGACC CCGCATTGCCCTTTGGGGGTGCCCTGGGATCCCTGGGGTAGTCCAGCTCTTCTTCATTTCCCAGCGTGGCCCTGGTTGGAAGAAGCAGCT GTCACAGCTGCTGTAGACAGCTGTGTTCCTACAATTGGCCCAGCACCCTGGGGCACGGGAGAAGGGTGGGGACCGTTGCTGTCACTACTC AGGCTGACTGGGGCCTGGTCAGATTACGTATGCCCTTGGTGGTTTAGAGATAATCCAAAATCAGGGTTTGGTTTGGGGAAGAAAATCCTC CCCCTTCCTCCCCCGCCCCGTTCCCTACCGCCTCCACTCCTGCCAGCTCATTTCCTTCAATTTCCTTTGACCTATAGGCTAAAAAAGAAA GGCTCATTCCAGCCACAGGGCAGCCTTCCCTGGGCCTTTGCTTCTCTAGCACAATTATGGGTTACTTCCTTTTTCTTAACAAAAAAGAAT GTTTGATTTCCTCTGGGTGACCTTATTGTCTGTAATTGAAACCCTATTGAGAGGTGATGTCTGTGTTAGCCAATGACCCAGGTGAGCTGC TCGGGCTTCTCTTGGTATGTCTTGTTTGGAAAAGTGGATTTCATTCATTTCTGATTGTCCAGTTAAGTGATCACCAAAGGACTGAGAATC TGGGAGGGCAAAAAAAAAAAAAAAGTTTTTATGTGCACTTAAATTTGGGGACAATTTTATGTATCTGTGTTAAGGATATGTTTAAGAACA TAATTCTTTTGTTGCTGTTTGTTTAAGAAGCACCTTAGTTTGTTTAAGAAGCACCTTATATAGTATAATATATATTTTTTTGAAATTACA TTGCTTGTTTATCAGACAATTGAATGTAGTAATTCTGTTCTGGATTTAATTTGACTGGGTTAACATGCAAAAACCAAGGAAAAATATTTA GTTTTTTTTTTTTTTTTTGTATACTTTTCAAGCTACCTTGTCATGTATACAGTCATTTATGCCTAAAGCCTGGTGATTATTCATTTAAAT GAAGATCACATTTCATATCAACTTTTGTATCCACAGTAGACAAAATAGCACTAATCCAGATGCCTATTGTTGGATACTGAATGACAGACA ATCTTATGTAGCAAAGATTATGCCTGAAAAGGAAAATTATTCAGGGCAGCTAATTTTGCTTTTACCAAAATATCAGTAGTAATATTTTTG GACAGTAGCTAATGGGTCAGTGGGTTCTTTTTAATGTTTATACTTAGATTTTCTTTTAAAAAAATTAAAATAAAACAAAAAAAAATTTCT AGGACTAGACGATGTAATACCAGCTAAAGCCAAACAATTATACAGTGGAAGGTTTTACATTATTCATCCAATGTGTTTCTATTCATGTTA AGATACTACTACATTTGAAGTGGGCAGAGAACATCAGATGATTGAAATGTTCGCCCAGGGGTCTCCAGCAACTTTGGAAATCTCTTTGTA TTTTTACTTGAAGTGCCACTAATGGACAGCAGATATTTTCTGGCTGATGTTGGTATTGGGTGTAGGAACATGATTTAAAAAAAAACTCTT GCCTCTGCTTTCCCCCACTCTGAGGCAAGTTAAAATGTAAAAGATGTGATTTATCTGGGGGGCTCAGGTATGGTGGGGAAGTGGATTCAG GAATCTGGGGAATGGCAAATATATTAAGAAGAGTATTGAAAGTATTTGGAGGAAAATGGTTAATTCTGGGTGTGCACCAGGGTTCAGTAG AGTCCACTTCTGCCCTGGAGACCACAAATCAACTAGCTCCATTTACAGCCATTTCTAAAATGGCAGCTTCAGTTCTAGAGAAGAAAGAAC AACATCAGCAGTAAAGTCCATGGAATAGCTAGTGGTCTGTGTTTCTTTTCGCCATTGCCTAGCTTGCCGTAATGATTCTATAATGCCATC ATGCAGCAATTATGAGAGGCTAGGTCATCCAAAGAGAAGACCCTATCAATGTAGGTTGCAAAATCTAACCCCTAAGGAAGTGCAGTCTTT GATTTGATTTCCCTAGTAACCTTGCAGATATGTTTAACCAAGCCATAGCCCATGCCTTTTGAGGGCTGAACAAATAAGGGACTTACTGAT AATTTACTTTTGATCACATTAAGGTGTTCTCACCTTGAAATCTTATACACTGAAATGGCCATTGATTTAGGCCACTGGCTTAGAGTACTC CTTCCCCTGCATGACACTGATTACAAATACTTTCCTATTCATACTTTCCAATTATGAGATGGACTGTGGGTACTGGGAGTGATCACTAAC ACCATAGTAATGTCTAATATTCACAGGCAGATCTGCTTGGGGAAGCTAGTTATGTGAAAGGCAAATAGAGTCATACAGTAGCTCAAAAGG CAACCATAATTCTCTTTGGTGCAGGTCTTGGGAGCGTGATCTAGATTACACTGCACCATTCCCAAGTTAATCCCCTGAAAACTTACTCTC AACTGGAGCAAATGAACTTTGGTCCCAAATATCCATCTTTTCAGTAGCGTTAATTATGCTCTGTTTCCAACTGCATTTCCTTTCCAATTG AATTAAAGTGTGGCCTCGTTTTTAGTCATTTAAAATTGTTTTCTAAGTAATTGCTGCCTCTATTATGGCACTTCAATTTTGCACTGTCTT TTGAGATTCAAGAAAAATTTCTATTCTTTTTTTTGCATCCAATTGTGCCTGAACTTTTAAAATATGTAAATGCTGCCATGTTCCAAACCC ATCGTCAGTGTGTGTGTTTAGAGCTGTGCACCCTAGAAACAACATATTGTCCCATGAGCAGGTGCCTGAGACACAGACCCCTTTGCATTC ACAGAGAGGTCATTGGTTATAGAGACTTGAATTAATAAGTGACATTATGCCAGTTTCTGTTCTCTCACAGGTGATAAACAATGCTTTTTG TGCACTACATACTCTTCAGTGTAGAGCTCTTGTTTTATGGGAAAAGGCTCAAATGCCAAATTGTGTTTGATGGATTAATATGCCCTTTTG CCGATGCATACTATTACTGATGTGACTCGGTTTTGTCGCAGCTTTGCTTTGTTTAATGAAACACACTTGTAAACCTCTTTTGCACTTTGA >53416_53416_2_MGP-ESR1_MGP_chr12_15037147_ENST00000228938_ESR1_chr6_152265308_ENST00000440973_length(amino acids)=403AA_BP=62 MQDETMKSLILLAILAALAVVTLCYGEWQKEENFGFDIVSVLSLNWHRAQESHESMESYELRIRKDRRGGRMLKHKRQRDDGEGRGEVGS AGDMRAANLWPSPLMIKRSKKNSLALSLTADQMVSALLDAEPPILYSEYDPTRPFSEASMMGLLTNLADRELVHMINWAKRVPGFVDLTL HDQVHLLECAWLEILMIGLVWRSMEHPGKLLFAPNLLLDRNQGKCVEGMVEIFDMLLATSSRFRMMNLQGEEFVCLKSIILLNSGVYTFL SSTLKSLEEKDHIHRVLDKITDTLIHLMAKAGLTLQQQHQRLAQLLLILSHIRHMSNKGMEHLYSMKCKNVVPLYDLLLEMLDAHRLHAP -------------------------------------------------------------- >53416_53416_3_MGP-ESR1_MGP_chr12_15037147_ENST00000539261_ESR1_chr6_152265308_ENST00000338799_length(transcript)=1344nt_BP=229nt TCTCCCACTGGTTCCTCCCCTCTCAACTGCTCTGGTTCTTATAAAAACCTCACAGCCTTCCACTAACATCCCGTAGGAGCCTCTCTCCCT ACTGCTGCTACACAAGACCCTGAGACTGACCTGCAGGACGAAACCATGAAGAGCCTGATCCTTCTTGCCATCCTGGCCGCCTTAGCGGTA GTAACTTTGTGTTATGAATCACATGAAAGCATGGAATCTTATGAACTTAGGATACGAAAAGACCGAAGAGGAGGGAGAATGTTGAAACAC AAGCGCCAGAGAGATGATGGGGAGGGCAGGGGTGAAGTGGGGTCTGCTGGAGACATGAGAGCTGCCAACCTTTGGCCAAGCCCGCTCATG ATCAAACGCTCTAAGAAGAACAGCCTGGCCTTGTCCCTGACGGCCGACCAGATGGTCAGTGCCTTGTTGGATGCTGAGCCCCCGATACTC TATTCCGAGTATGATCCTACCAGACCCTTCAGTGAAGCTTCGATGATGGGCTTACTGACCAACCTGGCAGACAGGGAGCTGGTTCACATG ATCAACTGGGCGAAGAGGGTGCCAGGCTTTGTGGATTTGACCCTCCATGATCAGGTCCACCTTCTAGAATGTGCCTGGCTAGAGATCCTG ATGATTGGTCTCGTCTGGCGCTCCATGGAGCACCCAGGGAAGCTACTGTTTGCTCCTAACTTGCTCTTGGACAGGAACCAGGGAAAATGT GTAGAGGGCATGGTGGAGATCTTCGACATGCTGCTGGCTACATCATCTCGGTTCCGCATGATGAATCTGCAGGGAGAGGAGTTTGTGTGC CTCAAATCTATTATTTTGCTTAATTCTGGAGTGTACACATTTCTGTCCAGCACCCTGAAGTCTCTGGAAGAGAAGGACCATATCCACCGA GTCCTGGACAAGATCACAGACACTTTGATCCACCTGATGGCCAAGGCAGGCCTGACCCTGCAGCAGCAGCACCAGCGGCTGGCCCAGCTC CTCCTCATCCTCTCCCACATCAGGCACATGAGTAACAAAGGCATGGAGCATCTGTACAGCATGAAGTGCAAGAACGTGGTGCCCCTCTAT GACCTGCTGCTGGAGATGCTGGACGCCCACCGCCTACATGCGCCCACTAGCCGTGGAGGGGCATCCGTGGAGGAGACGGACCAAAGCCAC TTGGCCACTGCGGGCTCTACTTCATCGCATTCCTTGCAAAAGTATTACATCACGGGGGAGGCAGAGGGTTTCCCTGCCACGGTCTGAGAG >53416_53416_3_MGP-ESR1_MGP_chr12_15037147_ENST00000539261_ESR1_chr6_152265308_ENST00000338799_length(amino acids)=378AA_BP=37 MQDETMKSLILLAILAALAVVTLCYESHESMESYELRIRKDRRGGRMLKHKRQRDDGEGRGEVGSAGDMRAANLWPSPLMIKRSKKNSLA LSLTADQMVSALLDAEPPILYSEYDPTRPFSEASMMGLLTNLADRELVHMINWAKRVPGFVDLTLHDQVHLLECAWLEILMIGLVWRSME HPGKLLFAPNLLLDRNQGKCVEGMVEIFDMLLATSSRFRMMNLQGEEFVCLKSIILLNSGVYTFLSSTLKSLEEKDHIHRVLDKITDTLI HLMAKAGLTLQQQHQRLAQLLLILSHIRHMSNKGMEHLYSMKCKNVVPLYDLLLEMLDAHRLHAPTSRGGASVEETDQSHLATAGSTSSH -------------------------------------------------------------- >53416_53416_4_MGP-ESR1_MGP_chr12_15037147_ENST00000539261_ESR1_chr6_152265308_ENST00000440973_length(transcript)=5565nt_BP=229nt TCTCCCACTGGTTCCTCCCCTCTCAACTGCTCTGGTTCTTATAAAAACCTCACAGCCTTCCACTAACATCCCGTAGGAGCCTCTCTCCCT ACTGCTGCTACACAAGACCCTGAGACTGACCTGCAGGACGAAACCATGAAGAGCCTGATCCTTCTTGCCATCCTGGCCGCCTTAGCGGTA GTAACTTTGTGTTATGAATCACATGAAAGCATGGAATCTTATGAACTTAGGATACGAAAAGACCGAAGAGGAGGGAGAATGTTGAAACAC AAGCGCCAGAGAGATGATGGGGAGGGCAGGGGTGAAGTGGGGTCTGCTGGAGACATGAGAGCTGCCAACCTTTGGCCAAGCCCGCTCATG ATCAAACGCTCTAAGAAGAACAGCCTGGCCTTGTCCCTGACGGCCGACCAGATGGTCAGTGCCTTGTTGGATGCTGAGCCCCCGATACTC TATTCCGAGTATGATCCTACCAGACCCTTCAGTGAAGCTTCGATGATGGGCTTACTGACCAACCTGGCAGACAGGGAGCTGGTTCACATG ATCAACTGGGCGAAGAGGGTGCCAGGCTTTGTGGATTTGACCCTCCATGATCAGGTCCACCTTCTAGAATGTGCCTGGCTAGAGATCCTG ATGATTGGTCTCGTCTGGCGCTCCATGGAGCACCCAGGGAAGCTACTGTTTGCTCCTAACTTGCTCTTGGACAGGAACCAGGGAAAATGT GTAGAGGGCATGGTGGAGATCTTCGACATGCTGCTGGCTACATCATCTCGGTTCCGCATGATGAATCTGCAGGGAGAGGAGTTTGTGTGC CTCAAATCTATTATTTTGCTTAATTCTGGAGTGTACACATTTCTGTCCAGCACCCTGAAGTCTCTGGAAGAGAAGGACCATATCCACCGA GTCCTGGACAAGATCACAGACACTTTGATCCACCTGATGGCCAAGGCAGGCCTGACCCTGCAGCAGCAGCACCAGCGGCTGGCCCAGCTC CTCCTCATCCTCTCCCACATCAGGCACATGAGTAACAAAGGCATGGAGCATCTGTACAGCATGAAGTGCAAGAACGTGGTGCCCCTCTAT GACCTGCTGCTGGAGATGCTGGACGCCCACCGCCTACATGCGCCCACTAGCCGTGGAGGGGCATCCGTGGAGGAGACGGACCAAAGCCAC TTGGCCACTGCGGGCTCTACTTCATCGCATTCCTTGCAAAAGTATTACATCACGGGGGAGGCAGAGGGTTTCCCTGCCACGGTCTGAGAG CTCCCTGGCTCCCACACGGTTCAGATAATCCCTGCTGCATTTTACCCTCATCATGCACCACTTTAGCCAAATTCTGTCTCCTGCATACAC TCCGGCATGCATCCAACACCAATGGCTTTCTAGATGAGTGGCCATTCATTTGCTTGCTCAGTTCTTAGTGGCACATCTTCTGTCTTCTGT TGGGAACAGCCAAAGGGATTCCAAGGCTAAATCTTTGTAACAGCTCTCTTTCCCCCTTGCTATGTTACTAAGCGTGAGGATTCCCGTAGC TCTTCACAGCTGAACTCAGTCTATGGGTTGGGGCTCAGATAACTCTGTGCATTTAAGCTACTTGTAGAGACCCAGGCCTGGAGAGTAGAC ATTTTGCCTCTGATAAGCACTTTTTAAATGGCTCTAAGAATAAGCCACAGCAAAGAATTTAAAGTGGCTCCTTTAATTGGTGACTTGGAG AAAGCTAGGTCAAGGGTTTATTATAGCACCCTCTTGTATTCCTATGGCAATGCATCCTTTTATGAAAGTGGTACACCTTAAAGCTTTTAT ATGACTGTAGCAGAGTATCTGGTGATTGTCAATTCATTCCCCCTATAGGAATACAAGGGGCACACAGGGAAGGCAGATCCCCTAGTTGGC AAGACTATTTTAACTTGATACACTGCAGATTCAGATGTGCTGAAAGCTCTGCCTCTGGCTTTCCGGTCATGGGTTCCAGTTAATTCATGC CTCCCATGGACCTATGGAGAGCAGCAAGTTGATCTTAGTTAAGTCTCCCTATATGAGGGATAAGTTCCTGATTTTTGTTTTTATTTTTGT GTTACAAAAGAAAGCCCTCCCTCCCTGAACTTGCAGTAAGGTCAGCTTCAGGACCTGTTCCAGTGGGCACTGTACTTGGATCTTCCCGGC GTGTGTGTGCCTTACACAGGGGTGAACTGTTCACTGTGGTGATGCATGATGAGGGTAAATGGTAGTTGAAAGGAGCAGGGGCCCTGGTGT TGCATTTAGCCCTGGGGCATGGAGCTGAACAGTACTTGTGCAGGATTGTTGTGGCTACTAGAGAACAAGAGGGAAAGTAGGGCAGAAACT GGATACAGTTCTGAGGCACAGCCAGACTTGCTCAGGGTGGCCCTGCCACAGGCTGCAGCTACCTAGGAACATTCCTTGCAGACCCCGCAT TGCCCTTTGGGGGTGCCCTGGGATCCCTGGGGTAGTCCAGCTCTTCTTCATTTCCCAGCGTGGCCCTGGTTGGAAGAAGCAGCTGTCACA GCTGCTGTAGACAGCTGTGTTCCTACAATTGGCCCAGCACCCTGGGGCACGGGAGAAGGGTGGGGACCGTTGCTGTCACTACTCAGGCTG ACTGGGGCCTGGTCAGATTACGTATGCCCTTGGTGGTTTAGAGATAATCCAAAATCAGGGTTTGGTTTGGGGAAGAAAATCCTCCCCCTT CCTCCCCCGCCCCGTTCCCTACCGCCTCCACTCCTGCCAGCTCATTTCCTTCAATTTCCTTTGACCTATAGGCTAAAAAAGAAAGGCTCA TTCCAGCCACAGGGCAGCCTTCCCTGGGCCTTTGCTTCTCTAGCACAATTATGGGTTACTTCCTTTTTCTTAACAAAAAAGAATGTTTGA TTTCCTCTGGGTGACCTTATTGTCTGTAATTGAAACCCTATTGAGAGGTGATGTCTGTGTTAGCCAATGACCCAGGTGAGCTGCTCGGGC TTCTCTTGGTATGTCTTGTTTGGAAAAGTGGATTTCATTCATTTCTGATTGTCCAGTTAAGTGATCACCAAAGGACTGAGAATCTGGGAG GGCAAAAAAAAAAAAAAAGTTTTTATGTGCACTTAAATTTGGGGACAATTTTATGTATCTGTGTTAAGGATATGTTTAAGAACATAATTC TTTTGTTGCTGTTTGTTTAAGAAGCACCTTAGTTTGTTTAAGAAGCACCTTATATAGTATAATATATATTTTTTTGAAATTACATTGCTT GTTTATCAGACAATTGAATGTAGTAATTCTGTTCTGGATTTAATTTGACTGGGTTAACATGCAAAAACCAAGGAAAAATATTTAGTTTTT TTTTTTTTTTTTGTATACTTTTCAAGCTACCTTGTCATGTATACAGTCATTTATGCCTAAAGCCTGGTGATTATTCATTTAAATGAAGAT CACATTTCATATCAACTTTTGTATCCACAGTAGACAAAATAGCACTAATCCAGATGCCTATTGTTGGATACTGAATGACAGACAATCTTA TGTAGCAAAGATTATGCCTGAAAAGGAAAATTATTCAGGGCAGCTAATTTTGCTTTTACCAAAATATCAGTAGTAATATTTTTGGACAGT AGCTAATGGGTCAGTGGGTTCTTTTTAATGTTTATACTTAGATTTTCTTTTAAAAAAATTAAAATAAAACAAAAAAAAATTTCTAGGACT AGACGATGTAATACCAGCTAAAGCCAAACAATTATACAGTGGAAGGTTTTACATTATTCATCCAATGTGTTTCTATTCATGTTAAGATAC TACTACATTTGAAGTGGGCAGAGAACATCAGATGATTGAAATGTTCGCCCAGGGGTCTCCAGCAACTTTGGAAATCTCTTTGTATTTTTA CTTGAAGTGCCACTAATGGACAGCAGATATTTTCTGGCTGATGTTGGTATTGGGTGTAGGAACATGATTTAAAAAAAAACTCTTGCCTCT GCTTTCCCCCACTCTGAGGCAAGTTAAAATGTAAAAGATGTGATTTATCTGGGGGGCTCAGGTATGGTGGGGAAGTGGATTCAGGAATCT GGGGAATGGCAAATATATTAAGAAGAGTATTGAAAGTATTTGGAGGAAAATGGTTAATTCTGGGTGTGCACCAGGGTTCAGTAGAGTCCA CTTCTGCCCTGGAGACCACAAATCAACTAGCTCCATTTACAGCCATTTCTAAAATGGCAGCTTCAGTTCTAGAGAAGAAAGAACAACATC AGCAGTAAAGTCCATGGAATAGCTAGTGGTCTGTGTTTCTTTTCGCCATTGCCTAGCTTGCCGTAATGATTCTATAATGCCATCATGCAG CAATTATGAGAGGCTAGGTCATCCAAAGAGAAGACCCTATCAATGTAGGTTGCAAAATCTAACCCCTAAGGAAGTGCAGTCTTTGATTTG ATTTCCCTAGTAACCTTGCAGATATGTTTAACCAAGCCATAGCCCATGCCTTTTGAGGGCTGAACAAATAAGGGACTTACTGATAATTTA CTTTTGATCACATTAAGGTGTTCTCACCTTGAAATCTTATACACTGAAATGGCCATTGATTTAGGCCACTGGCTTAGAGTACTCCTTCCC CTGCATGACACTGATTACAAATACTTTCCTATTCATACTTTCCAATTATGAGATGGACTGTGGGTACTGGGAGTGATCACTAACACCATA GTAATGTCTAATATTCACAGGCAGATCTGCTTGGGGAAGCTAGTTATGTGAAAGGCAAATAGAGTCATACAGTAGCTCAAAAGGCAACCA TAATTCTCTTTGGTGCAGGTCTTGGGAGCGTGATCTAGATTACACTGCACCATTCCCAAGTTAATCCCCTGAAAACTTACTCTCAACTGG AGCAAATGAACTTTGGTCCCAAATATCCATCTTTTCAGTAGCGTTAATTATGCTCTGTTTCCAACTGCATTTCCTTTCCAATTGAATTAA AGTGTGGCCTCGTTTTTAGTCATTTAAAATTGTTTTCTAAGTAATTGCTGCCTCTATTATGGCACTTCAATTTTGCACTGTCTTTTGAGA TTCAAGAAAAATTTCTATTCTTTTTTTTGCATCCAATTGTGCCTGAACTTTTAAAATATGTAAATGCTGCCATGTTCCAAACCCATCGTC AGTGTGTGTGTTTAGAGCTGTGCACCCTAGAAACAACATATTGTCCCATGAGCAGGTGCCTGAGACACAGACCCCTTTGCATTCACAGAG AGGTCATTGGTTATAGAGACTTGAATTAATAAGTGACATTATGCCAGTTTCTGTTCTCTCACAGGTGATAAACAATGCTTTTTGTGCACT ACATACTCTTCAGTGTAGAGCTCTTGTTTTATGGGAAAAGGCTCAAATGCCAAATTGTGTTTGATGGATTAATATGCCCTTTTGCCGATG CATACTATTACTGATGTGACTCGGTTTTGTCGCAGCTTTGCTTTGTTTAATGAAACACACTTGTAAACCTCTTTTGCACTTTGAAAAAGA >53416_53416_4_MGP-ESR1_MGP_chr12_15037147_ENST00000539261_ESR1_chr6_152265308_ENST00000440973_length(amino acids)=378AA_BP=37 MQDETMKSLILLAILAALAVVTLCYESHESMESYELRIRKDRRGGRMLKHKRQRDDGEGRGEVGSAGDMRAANLWPSPLMIKRSKKNSLA LSLTADQMVSALLDAEPPILYSEYDPTRPFSEASMMGLLTNLADRELVHMINWAKRVPGFVDLTLHDQVHLLECAWLEILMIGLVWRSME HPGKLLFAPNLLLDRNQGKCVEGMVEIFDMLLATSSRFRMMNLQGEEFVCLKSIILLNSGVYTFLSSTLKSLEEKDHIHRVLDKITDTLI HLMAKAGLTLQQQHQRLAQLLLILSHIRHMSNKGMEHLYSMKCKNVVPLYDLLLEMLDAHRLHAPTSRGGASVEETDQSHLATAGSTSSH -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for MGP-ESR1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000206249 | 2 | 8 | 262_595 | 253.33333333333334 | 596.0 | AKAP13 | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000338799 | 3 | 9 | 262_595 | 253.33333333333334 | 596.0 | AKAP13 | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000440973 | 4 | 10 | 262_595 | 253.33333333333334 | 596.0 | AKAP13 | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000443427 | 3 | 9 | 262_595 | 253.33333333333334 | 596.0 | AKAP13 | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000456483 | 0 | 8 | 262_595 | 0 | 484.0 | AKAP13 | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000456483 | 0 | 8 | 35_174 | 0 | 484.0 | DDX5%3B self-association |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000206249 | 2 | 8 | 35_174 | 253.33333333333334 | 596.0 | DDX5%3B self-association | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000338799 | 3 | 9 | 35_174 | 253.33333333333334 | 596.0 | DDX5%3B self-association | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000440973 | 4 | 10 | 35_174 | 253.33333333333334 | 596.0 | DDX5%3B self-association | |
| Tgene | ESR1 | chr12:15037147 | chr6:152265308 | ENST00000443427 | 3 | 9 | 35_174 | 253.33333333333334 | 596.0 | DDX5%3B self-association |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for MGP-ESR1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for MGP-ESR1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
