
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:MICAL3-BID (FusionGDB2 ID:53514) |
Fusion Gene Summary for MICAL3-BID |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: MICAL3-BID | Fusion gene ID: 53514 | Hgene | Tgene | Gene symbol | MICAL3 | BID | Gene ID | 57553 | 637 |
| Gene name | microtubule associated monooxygenase, calponin and LIM domain containing 3 | BH3 interacting domain death agonist | |
| Synonyms | MICAL-3 | FP497 | |
| Cytomap | 22q11.21 | 22q11.21 | |
| Type of gene | protein-coding | protein-coding | |
| Description | [F-actin]-monooxygenase MICAL3[F-actin]-methionine sulfoxide oxidase MICAL3flavoprotein oxidoreductase MICAL3molecule interacting with CasL protein 3protein MICAL-3protein-methionine sulfoxide oxidase MICAL3 | BH3-interacting domain death agonistBH3 interacting domain death agonist Si6 isoformBID isoform ES(1b)BID isoform L(2)BID isoform Si6Human BID coding sequenceapoptic death agonistdesmocollin type 4p22 BID | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q7RTP6 | P55957 | |
| Ensembl transtripts involved in fusion gene | ENST00000207726, ENST00000383094, ENST00000400561, ENST00000414725, ENST00000429452, ENST00000441493, ENST00000444520, ENST00000585038, ENST00000580469, | ENST00000399767, ENST00000399774, ENST00000473439, ENST00000342111, ENST00000399765, ENST00000551952, ENST00000317361, | |
| Fusion gene scores | * DoF score | 17 X 13 X 10=2210 | 8 X 6 X 7=336 |
| # samples | 17 | 9 | |
| ** MAII score | log2(17/2210*10)=-3.70043971814109 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/336*10)=-1.90046432644909 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: MICAL3 [Title/Abstract] AND BID [Title/Abstract] AND fusion [Title/Abstract] | ||
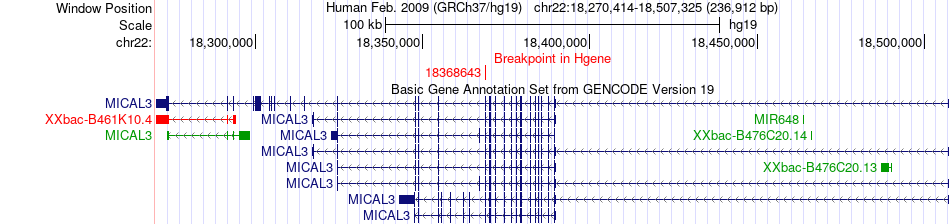
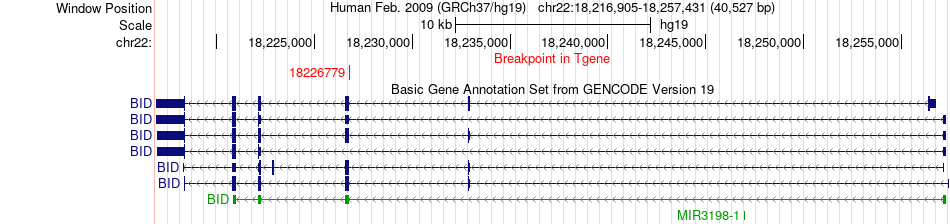
| Most frequent breakpoint | BID(18257146)-MICAL3(18274067), # samples:1 BID(18257147)-MICAL3(18274067), # samples:1 MICAL3(18368643)-BID(18226779), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | MICAL3-BID seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. MICAL3-BID seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | MICAL3 | GO:0007010 | cytoskeleton organization | 24440334 |
| Hgene | MICAL3 | GO:0030042 | actin filament depolymerization | 24440334 |
| Tgene | BID | GO:0001836 | release of cytochrome c from mitochondria | 17052454 |
| Tgene | BID | GO:0031334 | positive regulation of protein complex assembly | 19074440|21041309 |
| Tgene | BID | GO:0090150 | establishment of protein localization to membrane | 21041309 |
 Fusion gene breakpoints across MICAL3 (5'-gene) Fusion gene breakpoints across MICAL3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across BID (3'-gene) Fusion gene breakpoints across BID (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LIHC | TCGA-BC-4073 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
Top |
Fusion Gene ORF analysis for MICAL3-BID |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000207726 | ENST00000399767 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-5UTR | ENST00000207726 | ENST00000399774 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-5UTR | ENST00000207726 | ENST00000473439 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-5UTR | ENST00000383094 | ENST00000399767 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-5UTR | ENST00000383094 | ENST00000399774 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-5UTR | ENST00000383094 | ENST00000473439 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-5UTR | ENST00000400561 | ENST00000399767 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-5UTR | ENST00000400561 | ENST00000399774 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-5UTR | ENST00000400561 | ENST00000473439 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-5UTR | ENST00000414725 | ENST00000399767 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-5UTR | ENST00000414725 | ENST00000399774 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-5UTR | ENST00000414725 | ENST00000473439 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-5UTR | ENST00000429452 | ENST00000399767 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-5UTR | ENST00000429452 | ENST00000399774 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-5UTR | ENST00000429452 | ENST00000473439 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-5UTR | ENST00000441493 | ENST00000399767 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-5UTR | ENST00000441493 | ENST00000399774 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-5UTR | ENST00000441493 | ENST00000473439 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-5UTR | ENST00000444520 | ENST00000399767 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-5UTR | ENST00000444520 | ENST00000399774 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-5UTR | ENST00000444520 | ENST00000473439 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-5UTR | ENST00000585038 | ENST00000399767 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-5UTR | ENST00000585038 | ENST00000399774 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-5UTR | ENST00000585038 | ENST00000473439 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-intron | ENST00000207726 | ENST00000342111 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-intron | ENST00000207726 | ENST00000399765 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-intron | ENST00000207726 | ENST00000551952 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-intron | ENST00000383094 | ENST00000342111 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-intron | ENST00000383094 | ENST00000399765 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-intron | ENST00000383094 | ENST00000551952 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-intron | ENST00000400561 | ENST00000342111 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-intron | ENST00000400561 | ENST00000399765 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-intron | ENST00000400561 | ENST00000551952 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-intron | ENST00000414725 | ENST00000342111 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-intron | ENST00000414725 | ENST00000399765 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-intron | ENST00000414725 | ENST00000551952 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-intron | ENST00000429452 | ENST00000342111 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-intron | ENST00000429452 | ENST00000399765 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-intron | ENST00000429452 | ENST00000551952 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-intron | ENST00000441493 | ENST00000342111 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-intron | ENST00000441493 | ENST00000399765 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-intron | ENST00000441493 | ENST00000551952 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-intron | ENST00000444520 | ENST00000342111 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-intron | ENST00000444520 | ENST00000399765 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-intron | ENST00000444520 | ENST00000551952 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-intron | ENST00000585038 | ENST00000342111 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-intron | ENST00000585038 | ENST00000399765 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| 5CDS-intron | ENST00000585038 | ENST00000551952 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| Frame-shift | ENST00000207726 | ENST00000317361 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| Frame-shift | ENST00000429452 | ENST00000317361 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| Frame-shift | ENST00000441493 | ENST00000317361 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| Frame-shift | ENST00000444520 | ENST00000317361 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| In-frame | ENST00000383094 | ENST00000317361 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| In-frame | ENST00000400561 | ENST00000317361 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| In-frame | ENST00000414725 | ENST00000317361 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| In-frame | ENST00000585038 | ENST00000317361 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| intron-3CDS | ENST00000580469 | ENST00000317361 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| intron-5UTR | ENST00000580469 | ENST00000399767 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| intron-5UTR | ENST00000580469 | ENST00000399774 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| intron-5UTR | ENST00000580469 | ENST00000473439 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| intron-intron | ENST00000580469 | ENST00000342111 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| intron-intron | ENST00000580469 | ENST00000399765 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
| intron-intron | ENST00000580469 | ENST00000551952 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000400561 | MICAL3 | chr22 | 18368643 | - | ENST00000317361 | BID | chr22 | 18226779 | - | 4280 | 2264 | 23 | 2839 | 938 |
| ENST00000414725 | MICAL3 | chr22 | 18368643 | - | ENST00000317361 | BID | chr22 | 18226779 | - | 4257 | 2241 | 0 | 2816 | 938 |
| ENST00000383094 | MICAL3 | chr22 | 18368643 | - | ENST00000317361 | BID | chr22 | 18226779 | - | 4272 | 2256 | 15 | 2831 | 938 |
| ENST00000585038 | MICAL3 | chr22 | 18368643 | - | ENST00000317361 | BID | chr22 | 18226779 | - | 4319 | 2303 | 62 | 2878 | 938 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000400561 | ENST00000317361 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - | 0.000898868 | 0.99910116 |
| ENST00000414725 | ENST00000317361 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - | 0.000859743 | 0.99914026 |
| ENST00000383094 | ENST00000317361 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - | 0.00089737 | 0.9991027 |
| ENST00000585038 | ENST00000317361 | MICAL3 | chr22 | 18368643 | - | BID | chr22 | 18226779 | - | 0.000849174 | 0.9991509 |
Top |
Fusion Genomic Features for MICAL3-BID |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for MICAL3-BID |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr22:18257146/chr22:18274067) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MICAL3 | BID |
| FUNCTION: Monooxygenase that promotes depolymerization of F-actin by mediating oxidation of specific methionine residues on actin to form methionine-sulfoxide, resulting in actin filament disassembly and preventing repolymerization. In the absence of actin, it also functions as a NADPH oxidase producing H(2)O(2). Seems to act as Rab effector protein and plays a role in vesicle trafficking. Involved in exocytic vesicles tethering and fusion: the monooxygenase activity is required for this process and implicates RAB8A associated with exocytotic vesicles. Required for cytokinesis. Contributes to stabilization and/or maturation of the intercellular bridge independently of its monooxygenase activity. Promotes recruitment of Rab8 and ERC1 to the intercellular bridge, and together these proteins are proposed to function in timely abscission. {ECO:0000269|PubMed:21596566, ECO:0000269|PubMed:24440334}. | FUNCTION: The major proteolytic product p15 BID allows the release of cytochrome c (By similarity). Isoform 1, isoform 2 and isoform 4 induce ICE-like proteases and apoptosis. Isoform 3 does not induce apoptosis. Counters the protective effect of Bcl-2. {ECO:0000250|UniProtKB:P70444, ECO:0000269|PubMed:14583606}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000207726 | - | 16 | 21 | 688_691 | 747 | 977.0 | Compositional bias | Note=Poly-Glu |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000383094 | - | 15 | 19 | 688_691 | 747 | 949.0 | Compositional bias | Note=Poly-Glu |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000400561 | - | 15 | 20 | 688_691 | 747 | 967.0 | Compositional bias | Note=Poly-Glu |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000414725 | - | 15 | 20 | 688_691 | 747 | 977.0 | Compositional bias | Note=Poly-Glu |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000429452 | - | 16 | 23 | 688_691 | 747 | 1074.0 | Compositional bias | Note=Poly-Glu |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000441493 | - | 16 | 32 | 688_691 | 747 | 2003.0 | Compositional bias | Note=Poly-Glu |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000444520 | - | 16 | 21 | 688_691 | 747 | 967.0 | Compositional bias | Note=Poly-Glu |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000585038 | - | 15 | 22 | 688_691 | 747 | 1074.0 | Compositional bias | Note=Poly-Glu |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000207726 | - | 16 | 21 | 518_624 | 747 | 977.0 | Domain | Calponin-homology (CH) |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000383094 | - | 15 | 19 | 518_624 | 747 | 949.0 | Domain | Calponin-homology (CH) |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000400561 | - | 15 | 20 | 518_624 | 747 | 967.0 | Domain | Calponin-homology (CH) |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000414725 | - | 15 | 20 | 518_624 | 747 | 977.0 | Domain | Calponin-homology (CH) |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000429452 | - | 16 | 23 | 518_624 | 747 | 1074.0 | Domain | Calponin-homology (CH) |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000441493 | - | 16 | 32 | 518_624 | 747 | 2003.0 | Domain | Calponin-homology (CH) |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000444520 | - | 16 | 21 | 518_624 | 747 | 967.0 | Domain | Calponin-homology (CH) |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000585038 | - | 15 | 22 | 518_624 | 747 | 1074.0 | Domain | Calponin-homology (CH) |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000207726 | - | 16 | 21 | 663_684 | 747 | 977.0 | Motif | Nuclear localization signal |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000383094 | - | 15 | 19 | 663_684 | 747 | 949.0 | Motif | Nuclear localization signal |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000400561 | - | 15 | 20 | 663_684 | 747 | 967.0 | Motif | Nuclear localization signal |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000414725 | - | 15 | 20 | 663_684 | 747 | 977.0 | Motif | Nuclear localization signal |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000429452 | - | 16 | 23 | 663_684 | 747 | 1074.0 | Motif | Nuclear localization signal |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000441493 | - | 16 | 32 | 663_684 | 747 | 2003.0 | Motif | Nuclear localization signal |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000444520 | - | 16 | 21 | 663_684 | 747 | 967.0 | Motif | Nuclear localization signal |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000585038 | - | 15 | 22 | 663_684 | 747 | 1074.0 | Motif | Nuclear localization signal |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000207726 | - | 16 | 21 | 116_118 | 747 | 977.0 | Nucleotide binding | FAD |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000207726 | - | 16 | 21 | 123_125 | 747 | 977.0 | Nucleotide binding | FAD |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000383094 | - | 15 | 19 | 116_118 | 747 | 949.0 | Nucleotide binding | FAD |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000383094 | - | 15 | 19 | 123_125 | 747 | 949.0 | Nucleotide binding | FAD |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000400561 | - | 15 | 20 | 116_118 | 747 | 967.0 | Nucleotide binding | FAD |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000400561 | - | 15 | 20 | 123_125 | 747 | 967.0 | Nucleotide binding | FAD |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000414725 | - | 15 | 20 | 116_118 | 747 | 977.0 | Nucleotide binding | FAD |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000414725 | - | 15 | 20 | 123_125 | 747 | 977.0 | Nucleotide binding | FAD |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000429452 | - | 16 | 23 | 116_118 | 747 | 1074.0 | Nucleotide binding | FAD |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000429452 | - | 16 | 23 | 123_125 | 747 | 1074.0 | Nucleotide binding | FAD |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000441493 | - | 16 | 32 | 116_118 | 747 | 2003.0 | Nucleotide binding | FAD |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000441493 | - | 16 | 32 | 123_125 | 747 | 2003.0 | Nucleotide binding | FAD |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000444520 | - | 16 | 21 | 116_118 | 747 | 967.0 | Nucleotide binding | FAD |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000444520 | - | 16 | 21 | 123_125 | 747 | 967.0 | Nucleotide binding | FAD |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000585038 | - | 15 | 22 | 116_118 | 747 | 1074.0 | Nucleotide binding | FAD |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000585038 | - | 15 | 22 | 123_125 | 747 | 1074.0 | Nucleotide binding | FAD |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000207726 | - | 16 | 21 | 2_494 | 747 | 977.0 | Region | Monooxygenase domain |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000383094 | - | 15 | 19 | 2_494 | 747 | 949.0 | Region | Monooxygenase domain |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000400561 | - | 15 | 20 | 2_494 | 747 | 967.0 | Region | Monooxygenase domain |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000414725 | - | 15 | 20 | 2_494 | 747 | 977.0 | Region | Monooxygenase domain |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000429452 | - | 16 | 23 | 2_494 | 747 | 1074.0 | Region | Monooxygenase domain |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000441493 | - | 16 | 32 | 2_494 | 747 | 2003.0 | Region | Monooxygenase domain |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000444520 | - | 16 | 21 | 2_494 | 747 | 967.0 | Region | Monooxygenase domain |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000585038 | - | 15 | 22 | 2_494 | 747 | 1074.0 | Region | Monooxygenase domain |
| Tgene | BID | chr22:18368643 | chr22:18226779 | ENST00000317361 | 1 | 6 | 86_100 | 50 | 242.0 | Motif | BH3 | |
| Tgene | BID | chr22:18368643 | chr22:18226779 | ENST00000342111 | 1 | 7 | 86_100 | 4 | 182.33333333333334 | Motif | BH3 | |
| Tgene | BID | chr22:18368643 | chr22:18226779 | ENST00000399765 | 0 | 4 | 86_100 | 0 | 100.0 | Motif | BH3 | |
| Tgene | BID | chr22:18368643 | chr22:18226779 | ENST00000399767 | 0 | 5 | 86_100 | 0 | 100.0 | Motif | BH3 | |
| Tgene | BID | chr22:18368643 | chr22:18226779 | ENST00000399774 | 1 | 6 | 86_100 | 4 | 196.0 | Motif | BH3 | |
| Tgene | BID | chr22:18368643 | chr22:18226779 | ENST00000551952 | 1 | 6 | 86_100 | 4 | 196.0 | Motif | BH3 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000207726 | - | 16 | 21 | 1821_1992 | 747 | 977.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000383094 | - | 15 | 19 | 1821_1992 | 747 | 949.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000400561 | - | 15 | 20 | 1821_1992 | 747 | 967.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000414725 | - | 15 | 20 | 1821_1992 | 747 | 977.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000429452 | - | 16 | 23 | 1821_1992 | 747 | 1074.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000441493 | - | 16 | 32 | 1821_1992 | 747 | 2003.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000444520 | - | 16 | 21 | 1821_1992 | 747 | 967.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000585038 | - | 15 | 22 | 1821_1992 | 747 | 1074.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000207726 | - | 16 | 21 | 1141_1505 | 747 | 977.0 | Compositional bias | Note=Pro-rich |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000207726 | - | 16 | 21 | 889_1138 | 747 | 977.0 | Compositional bias | Note=Glu-rich |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000383094 | - | 15 | 19 | 1141_1505 | 747 | 949.0 | Compositional bias | Note=Pro-rich |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000383094 | - | 15 | 19 | 889_1138 | 747 | 949.0 | Compositional bias | Note=Glu-rich |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000400561 | - | 15 | 20 | 1141_1505 | 747 | 967.0 | Compositional bias | Note=Pro-rich |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000400561 | - | 15 | 20 | 889_1138 | 747 | 967.0 | Compositional bias | Note=Glu-rich |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000414725 | - | 15 | 20 | 1141_1505 | 747 | 977.0 | Compositional bias | Note=Pro-rich |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000414725 | - | 15 | 20 | 889_1138 | 747 | 977.0 | Compositional bias | Note=Glu-rich |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000429452 | - | 16 | 23 | 1141_1505 | 747 | 1074.0 | Compositional bias | Note=Pro-rich |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000429452 | - | 16 | 23 | 889_1138 | 747 | 1074.0 | Compositional bias | Note=Glu-rich |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000441493 | - | 16 | 32 | 1141_1505 | 747 | 2003.0 | Compositional bias | Note=Pro-rich |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000441493 | - | 16 | 32 | 889_1138 | 747 | 2003.0 | Compositional bias | Note=Glu-rich |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000444520 | - | 16 | 21 | 1141_1505 | 747 | 967.0 | Compositional bias | Note=Pro-rich |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000444520 | - | 16 | 21 | 889_1138 | 747 | 967.0 | Compositional bias | Note=Glu-rich |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000585038 | - | 15 | 22 | 1141_1505 | 747 | 1074.0 | Compositional bias | Note=Pro-rich |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000585038 | - | 15 | 22 | 889_1138 | 747 | 1074.0 | Compositional bias | Note=Glu-rich |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000207726 | - | 16 | 21 | 1841_1990 | 747 | 977.0 | Domain | bMERB |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000207726 | - | 16 | 21 | 762_824 | 747 | 977.0 | Domain | LIM zinc-binding |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000383094 | - | 15 | 19 | 1841_1990 | 747 | 949.0 | Domain | bMERB |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000383094 | - | 15 | 19 | 762_824 | 747 | 949.0 | Domain | LIM zinc-binding |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000400561 | - | 15 | 20 | 1841_1990 | 747 | 967.0 | Domain | bMERB |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000400561 | - | 15 | 20 | 762_824 | 747 | 967.0 | Domain | LIM zinc-binding |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000414725 | - | 15 | 20 | 1841_1990 | 747 | 977.0 | Domain | bMERB |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000414725 | - | 15 | 20 | 762_824 | 747 | 977.0 | Domain | LIM zinc-binding |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000429452 | - | 16 | 23 | 1841_1990 | 747 | 1074.0 | Domain | bMERB |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000429452 | - | 16 | 23 | 762_824 | 747 | 1074.0 | Domain | LIM zinc-binding |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000441493 | - | 16 | 32 | 1841_1990 | 747 | 2003.0 | Domain | bMERB |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000441493 | - | 16 | 32 | 762_824 | 747 | 2003.0 | Domain | LIM zinc-binding |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000444520 | - | 16 | 21 | 1841_1990 | 747 | 967.0 | Domain | bMERB |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000444520 | - | 16 | 21 | 762_824 | 747 | 967.0 | Domain | LIM zinc-binding |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000585038 | - | 15 | 22 | 1841_1990 | 747 | 1074.0 | Domain | bMERB |
| Hgene | MICAL3 | chr22:18368643 | chr22:18226779 | ENST00000585038 | - | 15 | 22 | 762_824 | 747 | 1074.0 | Domain | LIM zinc-binding |
Top |
Fusion Gene Sequence for MICAL3-BID |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >53514_53514_1_MICAL3-BID_MICAL3_chr22_18368643_ENST00000383094_BID_chr22_18226779_ENST00000317361_length(transcript)=4272nt_BP=2256nt TGAGAGTGAGGCAGCATGGAGGAGAGGAAGCATGAGACCATGAACCCAGCTCATGTCCTCTTTGACCGGTTTGTCCAGGCCACCACCTGC AAGGGAACCCTCAAGGCTTTCCAGGAGCTCTGTGACCACCTGGAACTAAAGCCAAAGGACTACCGCTCCTTCTATCACAAGCTCAAGTCC AAGCTTAACTACTGGAAAGCCAAAGCCCTCTGGGCAAAATTGGACAAACGGGGCAGTCACAAAGACTACAAAAAGGGAAAAGCGTGCACT AACACCAAGTGTCTCATCATTGGGGCTGGCCCCTGTGGTCTCCGTACAGCCATCGACTTATCCTTACTGGGGGCCAAGGTGGTTGTTATT GAGAAACGAGATGCCTTCTCCCGCAACAACGTCTTGCATCTCTGGCCATTCACCATACATGATCTACGAGGTCTGGGTGCCAAGAAGTTC TATGGCAAGTTCTGTGCTGGAGCCATCGACCATATCAGTATCCGTCAGCTCCAACTAATACTTTTGAAAGTAGCCTTGATCCTAGGCATT GAAATCCACGTCAATGTGGAATTCCAAGGACTTATACAGCCTCCTGAGGACCAAGAGAATGAACGGATAGGCTGGCGGGCACTGGTGCAC CCCAAGACTCATCCTGTGTCAGAGTATGAATTTGAAGTGATCATCGGTGGGGATGGTCGGAGGAACACCTTGGAAGGGTTTCGTCGGAAA GAATTCCGTGGCAAACTGGCCATCGCCATCACGGCAAATTTTATCAACCGAAATACAACAGCAGAAGCTAAAGTGGAAGAGATCAGTGGT GTGGCTTTTATATTCAACCAAAAATTTTTCCAGGAACTGAGGGAAGCCACAGGTATTGACTTGGAGAACATCGTTTACTACAAAGATGAC ACACACTATTTCGTTATGACAGCCAAAAAGCAGAGTTTGCTGGACAAAGGAGTGATACTACATGACTACGCCGACACAGAGCTCCTGCTT TCCCGAGAAAACGTGGACCAGGAGGCTCTGCTCAGCTATGCCAGGGAGGCGGCAGACTTCTCTACCCAGCAGCAGCTGCCGTCTCTGGAT TTTGCCATCAATCACTATGGGCAGCCCGATGTGGCCATGTTTGACTTCACTTGTATGTATGCCTCCGAGAACGCCGCCTTGGTGCGGGAG CAGAACGGACACCAGTTACTAGTGGCTCTGGTCGGGGACAGCCTCCTAGAGCCTTTCTGGCCAATGGGAACAGGAATAGCCCGGGGCTTT CTAGCTGCTATGGACTCTGCCTGGATGGTCCGAAGTTGGTCTCTAGGAACGAGCCCTTTGGAAGTGCTGGCAGAGAGGGAAAGTATTTAC AGGTTGCTGCCTCAGACCACCCCTGAGAATGTGAGTAAGAACTTCAGCCAGTACAGTATCGACCCTGTCACTCGGTATCCCAATATCAAC GTCAACTTCCTCCGGCCAAGCCAGGTGCGCCATTTATATGATACTGGCGAAACAAAAGATATTCACCTGGAAATGGAGAGCCTGGTGAAT TCCCGAACCACCCCCAAATTGACTCGCAATGAGTCTGTAGCTCGTTCAAGCAAACTGCTGGGTTGGTGCCAGAGGCAGACAGATGGCTAT GCAGGGGTAAACGTGACAGATCTCACCATGTCCTGGAAAAGTGGCTTGGCCCTTTGTGCAATTATCCATAGATACCGCCCTGACCTGATA GATTTTGATTCTTTGGATGAGCAAAATGTGGAGAAGAATAACCAACTGGCCTTTGACATTGCTGAGAAGGAATTGGGCATTTCTCCCATC ATGACAGGCAAAGAAATGGCCTCCGTGGGGGAGCCTGATAAGCTGTCCATGGTGATGTACCTGACTCAGTTCTACGAGATGTTTAAGGAC TCCCTCCCCTCTAGCGACACCTTGGACCTAAATGCCGAGGAGAAAGCAGTCCTGATAGCCAGCACCAGATCCCCTATCTCCTTCCTAAGC AAACTTGGCCAGACCATCTCTCGGAAGCGTTCTCCCAAGGATAAAAAGGAAAAGGACTTGGATGGTGCTGGGAAGAGGAGAAAGACCAGT CAATCAGAGGAGGAGGAAGCTCCTCGGGGCCACAGAGGAGAAAGACCGACCCTGGTGAGCACTCTGACAGACAGGAGGATGGACGTTGCC GTTGGGAACCAGAACAAAGTGAAGTACATGGCGACCCAGCTGCTGGCCAAATTTGAAGAGAATGCGCCCGCACAGTCCATCGGCATACGG AGACAGGTCAACAACGGTTCCAGCCTCAGGGATGAGTGCATCACAAACCTACTGGTGTTTGGCTTCCTCCAAAGCTGTTCTGACAACAGC TTCCGCAGAGAGCTGGACGCACTGGGCCACGAGCTGCCAGTGCTGGCTCCCCAGTGGGAGGGCTACGATGAGCTGCAGACTGATGGCAAC CGCAGCAGCCACTCCCGCTTGGGAAGAATAGAGGCAGATTCTGAAAGTCAAGAAGACATCATCCGGAATATTGCCAGGCACCTCGCCCAG GTCGGGGACAGCATGGACCGTAGCATCCCTCCGGGCCTGGTGAACGGCCTGGCCCTGCAGCTCAGGAACACCAGCCGGTCGGAGGAGGAC CGGAACAGGGACCTGGCCACTGCCCTGGAGCAGCTGCTGCAGGCCTACCCTAGAGACATGGAGAAGGAGAAGACCATGCTGGTGCTGGCC CTGCTGCTGGCCAAGAAGGTGGCCAGTCACACGCCGTCCTTGCTCCGTGATGTCTTTCACACAACAGTGAATTTTATTAACCAGAACCTA CGCACCTACGTGAGGAGCTTAGCCAGAAATGGGATGGACTGAACGGACAGTTCCAGAAGTGTGACTGGCTAAAGCTCGATGTGGTCACAG CTGTATAGCTGCTTCCAGTGTAGACGGAGCCCTGGCATGTCAACAGCGTTCCTAGAGAAGACAGGCTGGAAGATAGCTGTGACTTCTATT TTAAAGACAATGTTAAACTTATAACCCACTTTAAAATATCTACATTAATATACTTGAATGAAAATGTCCATTTACACGTATTTGAATGGC CTTCATATCATCCACACATGAATCTGCACATCTGTAAATCTACACACGGTGCCTTTATTTCCACTGTGCAGGTTCCCACTTAAAAATTAA ATTGGAAAGCAGGTTTCAAGGAAGTAGAAACAAAATACAATTTTTTTGGTAAAAAAAAATTACTGTTTATTAAAGTACAACCATAGAGGA TGGTCTTACAGCAGGCAGTATCCTGTTTGAGGAAAGCAAGAATCAGAGAAGGAACATACCCCTTACAAATGAAAAATTCCACTCAAAATA GGGACTATCTATCTTAATACTAAGGAACCAACAATCTTCCTGTTTAAAAAACCACATGGCACAGAGATTCTGAACTAAAGTGCTGCACTC AAATGATGGGAAGTCCGGCCCCAGTACACAGGGGCTTGACTTTTTCAACTTCGTTTCCTTTGTTGGAGTCAAAAAGAACCACTTGTGGTT CTAAAAGGTGTGAAGGTGATTTAAGGGCCCAGGTCAGCCACTGTTTGTTTACAAAATCAGGTAACTAACTGCATACACTTTTTCTCTTTC CATGACATCAAGACTTTGCTAAAGACATGAAGCCACGGGTGCCAGAAGCTACTGCGATGCCCCGGGAGTTAGCCCCCTGGTAATAGCTGT AAACTTCCAATTTCTAGCCATACGCTCAGCTCATCCATGCCTCAGAAGTGCATCTGGAGAGAACAGGTTTCTAAGCATAAAAGATGAAAG AGCAGTTGGACTTTTTAAAAATTCAGCAAAGTGGTTCCCTCTCTTAGGGACAGTCAAAACCAAGTCACTTAGGTAGTACCAAAATAAATA AGGAAAAGCTTAGCTTTAGAAACAGTGCAACACTGGTCTGCTGTTCCAGTGGTAAGCTATGTCCCAGGAATCAGTTTAAAAGCACGACAG TGGATGCTGGGTCCATATCACACACATTGCTGTGAACAGGAAACTCCTGTGACCACAACATGAGGCCACTGGAGACGCATATGAGTAAGG GCACTGACGGACTCATGATTTCTTCTTACCAGATGCTTTCCTGTTCTTTAAGAGTTTAAAATCATCAGAAAGGAAAAACAAACTCTATAT TGTTCAGCATGCAATACATACCACGCTAGGGCTGGCTCAATTGAAAGTGGGCAAAAGCTTACAAATACTAAAAAGAAGTGCTGCCGCGCA >53514_53514_1_MICAL3-BID_MICAL3_chr22_18368643_ENST00000383094_BID_chr22_18226779_ENST00000317361_length(amino acids)=938AA_BP=0 MEERKHETMNPAHVLFDRFVQATTCKGTLKAFQELCDHLELKPKDYRSFYHKLKSKLNYWKAKALWAKLDKRGSHKDYKKGKACTNTKCL IIGAGPCGLRTAIDLSLLGAKVVVIEKRDAFSRNNVLHLWPFTIHDLRGLGAKKFYGKFCAGAIDHISIRQLQLILLKVALILGIEIHVN VEFQGLIQPPEDQENERIGWRALVHPKTHPVSEYEFEVIIGGDGRRNTLEGFRRKEFRGKLAIAITANFINRNTTAEAKVEEISGVAFIF NQKFFQELREATGIDLENIVYYKDDTHYFVMTAKKQSLLDKGVILHDYADTELLLSRENVDQEALLSYAREAADFSTQQQLPSLDFAINH YGQPDVAMFDFTCMYASENAALVREQNGHQLLVALVGDSLLEPFWPMGTGIARGFLAAMDSAWMVRSWSLGTSPLEVLAERESIYRLLPQ TTPENVSKNFSQYSIDPVTRYPNINVNFLRPSQVRHLYDTGETKDIHLEMESLVNSRTTPKLTRNESVARSSKLLGWCQRQTDGYAGVNV TDLTMSWKSGLALCAIIHRYRPDLIDFDSLDEQNVEKNNQLAFDIAEKELGISPIMTGKEMASVGEPDKLSMVMYLTQFYEMFKDSLPSS DTLDLNAEEKAVLIASTRSPISFLSKLGQTISRKRSPKDKKEKDLDGAGKRRKTSQSEEEEAPRGHRGERPTLVSTLTDRRMDVAVGNQN KVKYMATQLLAKFEENAPAQSIGIRRQVNNGSSLRDECITNLLVFGFLQSCSDNSFRRELDALGHELPVLAPQWEGYDELQTDGNRSSHS RLGRIEADSESQEDIIRNIARHLAQVGDSMDRSIPPGLVNGLALQLRNTSRSEEDRNRDLATALEQLLQAYPRDMEKEKTMLVLALLLAK -------------------------------------------------------------- >53514_53514_2_MICAL3-BID_MICAL3_chr22_18368643_ENST00000400561_BID_chr22_18226779_ENST00000317361_length(transcript)=4280nt_BP=2264nt GGGAGTGCTGAGAGTGAGGCAGCATGGAGGAGAGGAAGCATGAGACCATGAACCCAGCTCATGTCCTCTTTGACCGGTTTGTCCAGGCCA CCACCTGCAAGGGAACCCTCAAGGCTTTCCAGGAGCTCTGTGACCACCTGGAACTAAAGCCAAAGGACTACCGCTCCTTCTATCACAAGC TCAAGTCCAAGCTTAACTACTGGAAAGCCAAAGCCCTCTGGGCAAAATTGGACAAACGGGGCAGTCACAAAGACTACAAAAAGGGAAAAG CGTGCACTAACACCAAGTGTCTCATCATTGGGGCTGGCCCCTGTGGTCTCCGTACAGCCATCGACTTATCCTTACTGGGGGCCAAGGTGG TTGTTATTGAGAAACGAGATGCCTTCTCCCGCAACAACGTCTTGCATCTCTGGCCATTCACCATACATGATCTACGAGGTCTGGGTGCCA AGAAGTTCTATGGCAAGTTCTGTGCTGGAGCCATCGACCATATCAGTATCCGTCAGCTCCAACTAATACTTTTGAAAGTAGCCTTGATCC TAGGCATTGAAATCCACGTCAATGTGGAATTCCAAGGACTTATACAGCCTCCTGAGGACCAAGAGAATGAACGGATAGGCTGGCGGGCAC TGGTGCACCCCAAGACTCATCCTGTGTCAGAGTATGAATTTGAAGTGATCATCGGTGGGGATGGTCGGAGGAACACCTTGGAAGGGTTTC GTCGGAAAGAATTCCGTGGCAAACTGGCCATCGCCATCACGGCAAATTTTATCAACCGAAATACAACAGCAGAAGCTAAAGTGGAAGAGA TCAGTGGTGTGGCTTTTATATTCAACCAAAAATTTTTCCAGGAACTGAGGGAAGCCACAGGTATTGACTTGGAGAACATCGTTTACTACA AAGATGACACACACTATTTCGTTATGACAGCCAAAAAGCAGAGTTTGCTGGACAAAGGAGTGATACTACATGACTACGCCGACACAGAGC TCCTGCTTTCCCGAGAAAACGTGGACCAGGAGGCTCTGCTCAGCTATGCCAGGGAGGCGGCAGACTTCTCTACCCAGCAGCAGCTGCCGT CTCTGGATTTTGCCATCAATCACTATGGGCAGCCCGATGTGGCCATGTTTGACTTCACTTGTATGTATGCCTCCGAGAACGCCGCCTTGG TGCGGGAGCAGAACGGACACCAGTTACTAGTGGCTCTGGTCGGGGACAGCCTCCTAGAGCCTTTCTGGCCAATGGGAACAGGAATAGCCC GGGGCTTTCTAGCTGCTATGGACTCTGCCTGGATGGTCCGAAGTTGGTCTCTAGGAACGAGCCCTTTGGAAGTGCTGGCAGAGAGGGAAA GTATTTACAGGTTGCTGCCTCAGACCACCCCTGAGAATGTGAGTAAGAACTTCAGCCAGTACAGTATCGACCCTGTCACTCGGTATCCCA ATATCAACGTCAACTTCCTCCGGCCAAGCCAGGTGCGCCATTTATATGATACTGGCGAAACAAAAGATATTCACCTGGAAATGGAGAGCC TGGTGAATTCCCGAACCACCCCCAAATTGACTCGCAATGAGTCTGTAGCTCGTTCAAGCAAACTGCTGGGTTGGTGCCAGAGGCAGACAG ATGGCTATGCAGGGGTAAACGTGACAGATCTCACCATGTCCTGGAAAAGTGGCTTGGCCCTTTGTGCAATTATCCATAGATACCGCCCTG ACCTGATAGATTTTGATTCTTTGGATGAGCAAAATGTGGAGAAGAATAACCAACTGGCCTTTGACATTGCTGAGAAGGAATTGGGCATTT CTCCCATCATGACAGGCAAAGAAATGGCCTCCGTGGGGGAGCCTGATAAGCTGTCCATGGTGATGTACCTGACTCAGTTCTACGAGATGT TTAAGGACTCCCTCCCCTCTAGCGACACCTTGGACCTAAATGCCGAGGAGAAAGCAGTCCTGATAGCCAGCACCAGATCCCCTATCTCCT TCCTAAGCAAACTTGGCCAGACCATCTCTCGGAAGCGTTCTCCCAAGGATAAAAAGGAAAAGGACTTGGATGGTGCTGGGAAGAGGAGAA AGACCAGTCAATCAGAGGAGGAGGAAGCTCCTCGGGGCCACAGAGGAGAAAGACCGACCCTGGTGAGCACTCTGACAGACAGGAGGATGG ACGTTGCCGTTGGGAACCAGAACAAAGTGAAGTACATGGCGACCCAGCTGCTGGCCAAATTTGAAGAGAATGCGCCCGCACAGTCCATCG GCATACGGAGACAGGTCAACAACGGTTCCAGCCTCAGGGATGAGTGCATCACAAACCTACTGGTGTTTGGCTTCCTCCAAAGCTGTTCTG ACAACAGCTTCCGCAGAGAGCTGGACGCACTGGGCCACGAGCTGCCAGTGCTGGCTCCCCAGTGGGAGGGCTACGATGAGCTGCAGACTG ATGGCAACCGCAGCAGCCACTCCCGCTTGGGAAGAATAGAGGCAGATTCTGAAAGTCAAGAAGACATCATCCGGAATATTGCCAGGCACC TCGCCCAGGTCGGGGACAGCATGGACCGTAGCATCCCTCCGGGCCTGGTGAACGGCCTGGCCCTGCAGCTCAGGAACACCAGCCGGTCGG AGGAGGACCGGAACAGGGACCTGGCCACTGCCCTGGAGCAGCTGCTGCAGGCCTACCCTAGAGACATGGAGAAGGAGAAGACCATGCTGG TGCTGGCCCTGCTGCTGGCCAAGAAGGTGGCCAGTCACACGCCGTCCTTGCTCCGTGATGTCTTTCACACAACAGTGAATTTTATTAACC AGAACCTACGCACCTACGTGAGGAGCTTAGCCAGAAATGGGATGGACTGAACGGACAGTTCCAGAAGTGTGACTGGCTAAAGCTCGATGT GGTCACAGCTGTATAGCTGCTTCCAGTGTAGACGGAGCCCTGGCATGTCAACAGCGTTCCTAGAGAAGACAGGCTGGAAGATAGCTGTGA CTTCTATTTTAAAGACAATGTTAAACTTATAACCCACTTTAAAATATCTACATTAATATACTTGAATGAAAATGTCCATTTACACGTATT TGAATGGCCTTCATATCATCCACACATGAATCTGCACATCTGTAAATCTACACACGGTGCCTTTATTTCCACTGTGCAGGTTCCCACTTA AAAATTAAATTGGAAAGCAGGTTTCAAGGAAGTAGAAACAAAATACAATTTTTTTGGTAAAAAAAAATTACTGTTTATTAAAGTACAACC ATAGAGGATGGTCTTACAGCAGGCAGTATCCTGTTTGAGGAAAGCAAGAATCAGAGAAGGAACATACCCCTTACAAATGAAAAATTCCAC TCAAAATAGGGACTATCTATCTTAATACTAAGGAACCAACAATCTTCCTGTTTAAAAAACCACATGGCACAGAGATTCTGAACTAAAGTG CTGCACTCAAATGATGGGAAGTCCGGCCCCAGTACACAGGGGCTTGACTTTTTCAACTTCGTTTCCTTTGTTGGAGTCAAAAAGAACCAC TTGTGGTTCTAAAAGGTGTGAAGGTGATTTAAGGGCCCAGGTCAGCCACTGTTTGTTTACAAAATCAGGTAACTAACTGCATACACTTTT TCTCTTTCCATGACATCAAGACTTTGCTAAAGACATGAAGCCACGGGTGCCAGAAGCTACTGCGATGCCCCGGGAGTTAGCCCCCTGGTA ATAGCTGTAAACTTCCAATTTCTAGCCATACGCTCAGCTCATCCATGCCTCAGAAGTGCATCTGGAGAGAACAGGTTTCTAAGCATAAAA GATGAAAGAGCAGTTGGACTTTTTAAAAATTCAGCAAAGTGGTTCCCTCTCTTAGGGACAGTCAAAACCAAGTCACTTAGGTAGTACCAA AATAAATAAGGAAAAGCTTAGCTTTAGAAACAGTGCAACACTGGTCTGCTGTTCCAGTGGTAAGCTATGTCCCAGGAATCAGTTTAAAAG CACGACAGTGGATGCTGGGTCCATATCACACACATTGCTGTGAACAGGAAACTCCTGTGACCACAACATGAGGCCACTGGAGACGCATAT GAGTAAGGGCACTGACGGACTCATGATTTCTTCTTACCAGATGCTTTCCTGTTCTTTAAGAGTTTAAAATCATCAGAAAGGAAAAACAAA CTCTATATTGTTCAGCATGCAATACATACCACGCTAGGGCTGGCTCAATTGAAAGTGGGCAAAAGCTTACAAATACTAAAAAGAAGTGCT >53514_53514_2_MICAL3-BID_MICAL3_chr22_18368643_ENST00000400561_BID_chr22_18226779_ENST00000317361_length(amino acids)=938AA_BP=0 MEERKHETMNPAHVLFDRFVQATTCKGTLKAFQELCDHLELKPKDYRSFYHKLKSKLNYWKAKALWAKLDKRGSHKDYKKGKACTNTKCL IIGAGPCGLRTAIDLSLLGAKVVVIEKRDAFSRNNVLHLWPFTIHDLRGLGAKKFYGKFCAGAIDHISIRQLQLILLKVALILGIEIHVN VEFQGLIQPPEDQENERIGWRALVHPKTHPVSEYEFEVIIGGDGRRNTLEGFRRKEFRGKLAIAITANFINRNTTAEAKVEEISGVAFIF NQKFFQELREATGIDLENIVYYKDDTHYFVMTAKKQSLLDKGVILHDYADTELLLSRENVDQEALLSYAREAADFSTQQQLPSLDFAINH YGQPDVAMFDFTCMYASENAALVREQNGHQLLVALVGDSLLEPFWPMGTGIARGFLAAMDSAWMVRSWSLGTSPLEVLAERESIYRLLPQ TTPENVSKNFSQYSIDPVTRYPNINVNFLRPSQVRHLYDTGETKDIHLEMESLVNSRTTPKLTRNESVARSSKLLGWCQRQTDGYAGVNV TDLTMSWKSGLALCAIIHRYRPDLIDFDSLDEQNVEKNNQLAFDIAEKELGISPIMTGKEMASVGEPDKLSMVMYLTQFYEMFKDSLPSS DTLDLNAEEKAVLIASTRSPISFLSKLGQTISRKRSPKDKKEKDLDGAGKRRKTSQSEEEEAPRGHRGERPTLVSTLTDRRMDVAVGNQN KVKYMATQLLAKFEENAPAQSIGIRRQVNNGSSLRDECITNLLVFGFLQSCSDNSFRRELDALGHELPVLAPQWEGYDELQTDGNRSSHS RLGRIEADSESQEDIIRNIARHLAQVGDSMDRSIPPGLVNGLALQLRNTSRSEEDRNRDLATALEQLLQAYPRDMEKEKTMLVLALLLAK -------------------------------------------------------------- >53514_53514_3_MICAL3-BID_MICAL3_chr22_18368643_ENST00000414725_BID_chr22_18226779_ENST00000317361_length(transcript)=4257nt_BP=2241nt ATGGAGGAGAGGAAGCATGAGACCATGAACCCAGCTCATGTCCTCTTTGACCGGTTTGTCCAGGCCACCACCTGCAAGGGAACCCTCAAG GCTTTCCAGGAGCTCTGTGACCACCTGGAACTAAAGCCAAAGGACTACCGCTCCTTCTATCACAAGCTCAAGTCCAAGCTTAACTACTGG AAAGCCAAAGCCCTCTGGGCAAAATTGGACAAACGGGGCAGTCACAAAGACTACAAAAAGGGAAAAGCGTGCACTAACACCAAGTGTCTC ATCATTGGGGCTGGCCCCTGTGGTCTCCGTACAGCCATCGACTTATCCTTACTGGGGGCCAAGGTGGTTGTTATTGAGAAACGAGATGCC TTCTCCCGCAACAACGTCTTGCATCTCTGGCCATTCACCATACATGATCTACGAGGTCTGGGTGCCAAGAAGTTCTATGGCAAGTTCTGT GCTGGAGCCATCGACCATATCAGTATCCGTCAGCTCCAACTAATACTTTTGAAAGTAGCCTTGATCCTAGGCATTGAAATCCACGTCAAT GTGGAATTCCAAGGACTTATACAGCCTCCTGAGGACCAAGAGAATGAACGGATAGGCTGGCGGGCACTGGTGCACCCCAAGACTCATCCT GTGTCAGAGTATGAATTTGAAGTGATCATCGGTGGGGATGGTCGGAGGAACACCTTGGAAGGGTTTCGTCGGAAAGAATTCCGTGGCAAA CTGGCCATCGCCATCACGGCAAATTTTATCAACCGAAATACAACAGCAGAAGCTAAAGTGGAAGAGATCAGTGGTGTGGCTTTTATATTC AACCAAAAATTTTTCCAGGAACTGAGGGAAGCCACAGGTATTGACTTGGAGAACATCGTTTACTACAAAGATGACACACACTATTTCGTT ATGACAGCCAAAAAGCAGAGTTTGCTGGACAAAGGAGTGATACTACATGACTACGCCGACACAGAGCTCCTGCTTTCCCGAGAAAACGTG GACCAGGAGGCTCTGCTCAGCTATGCCAGGGAGGCGGCAGACTTCTCTACCCAGCAGCAGCTGCCGTCTCTGGATTTTGCCATCAATCAC TATGGGCAGCCCGATGTGGCCATGTTTGACTTCACTTGTATGTATGCCTCCGAGAACGCCGCCTTGGTGCGGGAGCAGAACGGACACCAG TTACTAGTGGCTCTGGTCGGGGACAGCCTCCTAGAGCCTTTCTGGCCAATGGGAACAGGAATAGCCCGGGGCTTTCTAGCTGCTATGGAC TCTGCCTGGATGGTCCGAAGTTGGTCTCTAGGAACGAGCCCTTTGGAAGTGCTGGCAGAGAGGGAAAGTATTTACAGGTTGCTGCCTCAG ACCACCCCTGAGAATGTGAGTAAGAACTTCAGCCAGTACAGTATCGACCCTGTCACTCGGTATCCCAATATCAACGTCAACTTCCTCCGG CCAAGCCAGGTGCGCCATTTATATGATACTGGCGAAACAAAAGATATTCACCTGGAAATGGAGAGCCTGGTGAATTCCCGAACCACCCCC AAATTGACTCGCAATGAGTCTGTAGCTCGTTCAAGCAAACTGCTGGGTTGGTGCCAGAGGCAGACAGATGGCTATGCAGGGGTAAACGTG ACAGATCTCACCATGTCCTGGAAAAGTGGCTTGGCCCTTTGTGCAATTATCCATAGATACCGCCCTGACCTGATAGATTTTGATTCTTTG GATGAGCAAAATGTGGAGAAGAATAACCAACTGGCCTTTGACATTGCTGAGAAGGAATTGGGCATTTCTCCCATCATGACAGGCAAAGAA ATGGCCTCCGTGGGGGAGCCTGATAAGCTGTCCATGGTGATGTACCTGACTCAGTTCTACGAGATGTTTAAGGACTCCCTCCCCTCTAGC GACACCTTGGACCTAAATGCCGAGGAGAAAGCAGTCCTGATAGCCAGCACCAGATCCCCTATCTCCTTCCTAAGCAAACTTGGCCAGACC ATCTCTCGGAAGCGTTCTCCCAAGGATAAAAAGGAAAAGGACTTGGATGGTGCTGGGAAGAGGAGAAAGACCAGTCAATCAGAGGAGGAG GAAGCTCCTCGGGGCCACAGAGGAGAAAGACCGACCCTGGTGAGCACTCTGACAGACAGGAGGATGGACGTTGCCGTTGGGAACCAGAAC AAAGTGAAGTACATGGCGACCCAGCTGCTGGCCAAATTTGAAGAGAATGCGCCCGCACAGTCCATCGGCATACGGAGACAGGTCAACAAC GGTTCCAGCCTCAGGGATGAGTGCATCACAAACCTACTGGTGTTTGGCTTCCTCCAAAGCTGTTCTGACAACAGCTTCCGCAGAGAGCTG GACGCACTGGGCCACGAGCTGCCAGTGCTGGCTCCCCAGTGGGAGGGCTACGATGAGCTGCAGACTGATGGCAACCGCAGCAGCCACTCC CGCTTGGGAAGAATAGAGGCAGATTCTGAAAGTCAAGAAGACATCATCCGGAATATTGCCAGGCACCTCGCCCAGGTCGGGGACAGCATG GACCGTAGCATCCCTCCGGGCCTGGTGAACGGCCTGGCCCTGCAGCTCAGGAACACCAGCCGGTCGGAGGAGGACCGGAACAGGGACCTG GCCACTGCCCTGGAGCAGCTGCTGCAGGCCTACCCTAGAGACATGGAGAAGGAGAAGACCATGCTGGTGCTGGCCCTGCTGCTGGCCAAG AAGGTGGCCAGTCACACGCCGTCCTTGCTCCGTGATGTCTTTCACACAACAGTGAATTTTATTAACCAGAACCTACGCACCTACGTGAGG AGCTTAGCCAGAAATGGGATGGACTGAACGGACAGTTCCAGAAGTGTGACTGGCTAAAGCTCGATGTGGTCACAGCTGTATAGCTGCTTC CAGTGTAGACGGAGCCCTGGCATGTCAACAGCGTTCCTAGAGAAGACAGGCTGGAAGATAGCTGTGACTTCTATTTTAAAGACAATGTTA AACTTATAACCCACTTTAAAATATCTACATTAATATACTTGAATGAAAATGTCCATTTACACGTATTTGAATGGCCTTCATATCATCCAC ACATGAATCTGCACATCTGTAAATCTACACACGGTGCCTTTATTTCCACTGTGCAGGTTCCCACTTAAAAATTAAATTGGAAAGCAGGTT TCAAGGAAGTAGAAACAAAATACAATTTTTTTGGTAAAAAAAAATTACTGTTTATTAAAGTACAACCATAGAGGATGGTCTTACAGCAGG CAGTATCCTGTTTGAGGAAAGCAAGAATCAGAGAAGGAACATACCCCTTACAAATGAAAAATTCCACTCAAAATAGGGACTATCTATCTT AATACTAAGGAACCAACAATCTTCCTGTTTAAAAAACCACATGGCACAGAGATTCTGAACTAAAGTGCTGCACTCAAATGATGGGAAGTC CGGCCCCAGTACACAGGGGCTTGACTTTTTCAACTTCGTTTCCTTTGTTGGAGTCAAAAAGAACCACTTGTGGTTCTAAAAGGTGTGAAG GTGATTTAAGGGCCCAGGTCAGCCACTGTTTGTTTACAAAATCAGGTAACTAACTGCATACACTTTTTCTCTTTCCATGACATCAAGACT TTGCTAAAGACATGAAGCCACGGGTGCCAGAAGCTACTGCGATGCCCCGGGAGTTAGCCCCCTGGTAATAGCTGTAAACTTCCAATTTCT AGCCATACGCTCAGCTCATCCATGCCTCAGAAGTGCATCTGGAGAGAACAGGTTTCTAAGCATAAAAGATGAAAGAGCAGTTGGACTTTT TAAAAATTCAGCAAAGTGGTTCCCTCTCTTAGGGACAGTCAAAACCAAGTCACTTAGGTAGTACCAAAATAAATAAGGAAAAGCTTAGCT TTAGAAACAGTGCAACACTGGTCTGCTGTTCCAGTGGTAAGCTATGTCCCAGGAATCAGTTTAAAAGCACGACAGTGGATGCTGGGTCCA TATCACACACATTGCTGTGAACAGGAAACTCCTGTGACCACAACATGAGGCCACTGGAGACGCATATGAGTAAGGGCACTGACGGACTCA TGATTTCTTCTTACCAGATGCTTTCCTGTTCTTTAAGAGTTTAAAATCATCAGAAAGGAAAAACAAACTCTATATTGTTCAGCATGCAAT ACATACCACGCTAGGGCTGGCTCAATTGAAAGTGGGCAAAAGCTTACAAATACTAAAAAGAAGTGCTGCCGCGCAGTGTGGAGGCCACTG >53514_53514_3_MICAL3-BID_MICAL3_chr22_18368643_ENST00000414725_BID_chr22_18226779_ENST00000317361_length(amino acids)=938AA_BP=0 MEERKHETMNPAHVLFDRFVQATTCKGTLKAFQELCDHLELKPKDYRSFYHKLKSKLNYWKAKALWAKLDKRGSHKDYKKGKACTNTKCL IIGAGPCGLRTAIDLSLLGAKVVVIEKRDAFSRNNVLHLWPFTIHDLRGLGAKKFYGKFCAGAIDHISIRQLQLILLKVALILGIEIHVN VEFQGLIQPPEDQENERIGWRALVHPKTHPVSEYEFEVIIGGDGRRNTLEGFRRKEFRGKLAIAITANFINRNTTAEAKVEEISGVAFIF NQKFFQELREATGIDLENIVYYKDDTHYFVMTAKKQSLLDKGVILHDYADTELLLSRENVDQEALLSYAREAADFSTQQQLPSLDFAINH YGQPDVAMFDFTCMYASENAALVREQNGHQLLVALVGDSLLEPFWPMGTGIARGFLAAMDSAWMVRSWSLGTSPLEVLAERESIYRLLPQ TTPENVSKNFSQYSIDPVTRYPNINVNFLRPSQVRHLYDTGETKDIHLEMESLVNSRTTPKLTRNESVARSSKLLGWCQRQTDGYAGVNV TDLTMSWKSGLALCAIIHRYRPDLIDFDSLDEQNVEKNNQLAFDIAEKELGISPIMTGKEMASVGEPDKLSMVMYLTQFYEMFKDSLPSS DTLDLNAEEKAVLIASTRSPISFLSKLGQTISRKRSPKDKKEKDLDGAGKRRKTSQSEEEEAPRGHRGERPTLVSTLTDRRMDVAVGNQN KVKYMATQLLAKFEENAPAQSIGIRRQVNNGSSLRDECITNLLVFGFLQSCSDNSFRRELDALGHELPVLAPQWEGYDELQTDGNRSSHS RLGRIEADSESQEDIIRNIARHLAQVGDSMDRSIPPGLVNGLALQLRNTSRSEEDRNRDLATALEQLLQAYPRDMEKEKTMLVLALLLAK -------------------------------------------------------------- >53514_53514_4_MICAL3-BID_MICAL3_chr22_18368643_ENST00000585038_BID_chr22_18226779_ENST00000317361_length(transcript)=4319nt_BP=2303nt GTGTCAGGTGGACCCACAGCCATCCCACCTCCCCCTCTGGGGAGTGCTGAGAGTGAGGCAGCATGGAGGAGAGGAAGCATGAGACCATGA ACCCAGCTCATGTCCTCTTTGACCGGTTTGTCCAGGCCACCACCTGCAAGGGAACCCTCAAGGCTTTCCAGGAGCTCTGTGACCACCTGG AACTAAAGCCAAAGGACTACCGCTCCTTCTATCACAAGCTCAAGTCCAAGCTTAACTACTGGAAAGCCAAAGCCCTCTGGGCAAAATTGG ACAAACGGGGCAGTCACAAAGACTACAAAAAGGGAAAAGCGTGCACTAACACCAAGTGTCTCATCATTGGGGCTGGCCCCTGTGGTCTCC GTACAGCCATCGACTTATCCTTACTGGGGGCCAAGGTGGTTGTTATTGAGAAACGAGATGCCTTCTCCCGCAACAACGTCTTGCATCTCT GGCCATTCACCATACATGATCTACGAGGTCTGGGTGCCAAGAAGTTCTATGGCAAGTTCTGTGCTGGAGCCATCGACCATATCAGTATCC GTCAGCTCCAACTAATACTTTTGAAAGTAGCCTTGATCCTAGGCATTGAAATCCACGTCAATGTGGAATTCCAAGGACTTATACAGCCTC CTGAGGACCAAGAGAATGAACGGATAGGCTGGCGGGCACTGGTGCACCCCAAGACTCATCCTGTGTCAGAGTATGAATTTGAAGTGATCA TCGGTGGGGATGGTCGGAGGAACACCTTGGAAGGGTTTCGTCGGAAAGAATTCCGTGGCAAACTGGCCATCGCCATCACGGCAAATTTTA TCAACCGAAATACAACAGCAGAAGCTAAAGTGGAAGAGATCAGTGGTGTGGCTTTTATATTCAACCAAAAATTTTTCCAGGAACTGAGGG AAGCCACAGGTATTGACTTGGAGAACATCGTTTACTACAAAGATGACACACACTATTTCGTTATGACAGCCAAAAAGCAGAGTTTGCTGG ACAAAGGAGTGATACTACATGACTACGCCGACACAGAGCTCCTGCTTTCCCGAGAAAACGTGGACCAGGAGGCTCTGCTCAGCTATGCCA GGGAGGCGGCAGACTTCTCTACCCAGCAGCAGCTGCCGTCTCTGGATTTTGCCATCAATCACTATGGGCAGCCCGATGTGGCCATGTTTG ACTTCACTTGTATGTATGCCTCCGAGAACGCCGCCTTGGTGCGGGAGCAGAACGGACACCAGTTACTAGTGGCTCTGGTCGGGGACAGCC TCCTAGAGCCTTTCTGGCCAATGGGAACAGGAATAGCCCGGGGCTTTCTAGCTGCTATGGACTCTGCCTGGATGGTCCGAAGTTGGTCTC TAGGAACGAGCCCTTTGGAAGTGCTGGCAGAGAGGGAAAGTATTTACAGGTTGCTGCCTCAGACCACCCCTGAGAATGTGAGTAAGAACT TCAGCCAGTACAGTATCGACCCTGTCACTCGGTATCCCAATATCAACGTCAACTTCCTCCGGCCAAGCCAGGTGCGCCATTTATATGATA CTGGCGAAACAAAAGATATTCACCTGGAAATGGAGAGCCTGGTGAATTCCCGAACCACCCCCAAATTGACTCGCAATGAGTCTGTAGCTC GTTCAAGCAAACTGCTGGGTTGGTGCCAGAGGCAGACAGATGGCTATGCAGGGGTAAACGTGACAGATCTCACCATGTCCTGGAAAAGTG GCTTGGCCCTTTGTGCAATTATCCATAGATACCGCCCTGACCTGATAGATTTTGATTCTTTGGATGAGCAAAATGTGGAGAAGAATAACC AACTGGCCTTTGACATTGCTGAGAAGGAATTGGGCATTTCTCCCATCATGACAGGCAAAGAAATGGCCTCCGTGGGGGAGCCTGATAAGC TGTCCATGGTGATGTACCTGACTCAGTTCTACGAGATGTTTAAGGACTCCCTCCCCTCTAGCGACACCTTGGACCTAAATGCCGAGGAGA AAGCAGTCCTGATAGCCAGCACCAGATCCCCTATCTCCTTCCTAAGCAAACTTGGCCAGACCATCTCTCGGAAGCGTTCTCCCAAGGATA AAAAGGAAAAGGACTTGGATGGTGCTGGGAAGAGGAGAAAGACCAGTCAATCAGAGGAGGAGGAAGCTCCTCGGGGCCACAGAGGAGAAA GACCGACCCTGGTGAGCACTCTGACAGACAGGAGGATGGACGTTGCCGTTGGGAACCAGAACAAAGTGAAGTACATGGCGACCCAGCTGC TGGCCAAATTTGAAGAGAATGCGCCCGCACAGTCCATCGGCATACGGAGACAGGTCAACAACGGTTCCAGCCTCAGGGATGAGTGCATCA CAAACCTACTGGTGTTTGGCTTCCTCCAAAGCTGTTCTGACAACAGCTTCCGCAGAGAGCTGGACGCACTGGGCCACGAGCTGCCAGTGC TGGCTCCCCAGTGGGAGGGCTACGATGAGCTGCAGACTGATGGCAACCGCAGCAGCCACTCCCGCTTGGGAAGAATAGAGGCAGATTCTG AAAGTCAAGAAGACATCATCCGGAATATTGCCAGGCACCTCGCCCAGGTCGGGGACAGCATGGACCGTAGCATCCCTCCGGGCCTGGTGA ACGGCCTGGCCCTGCAGCTCAGGAACACCAGCCGGTCGGAGGAGGACCGGAACAGGGACCTGGCCACTGCCCTGGAGCAGCTGCTGCAGG CCTACCCTAGAGACATGGAGAAGGAGAAGACCATGCTGGTGCTGGCCCTGCTGCTGGCCAAGAAGGTGGCCAGTCACACGCCGTCCTTGC TCCGTGATGTCTTTCACACAACAGTGAATTTTATTAACCAGAACCTACGCACCTACGTGAGGAGCTTAGCCAGAAATGGGATGGACTGAA CGGACAGTTCCAGAAGTGTGACTGGCTAAAGCTCGATGTGGTCACAGCTGTATAGCTGCTTCCAGTGTAGACGGAGCCCTGGCATGTCAA CAGCGTTCCTAGAGAAGACAGGCTGGAAGATAGCTGTGACTTCTATTTTAAAGACAATGTTAAACTTATAACCCACTTTAAAATATCTAC ATTAATATACTTGAATGAAAATGTCCATTTACACGTATTTGAATGGCCTTCATATCATCCACACATGAATCTGCACATCTGTAAATCTAC ACACGGTGCCTTTATTTCCACTGTGCAGGTTCCCACTTAAAAATTAAATTGGAAAGCAGGTTTCAAGGAAGTAGAAACAAAATACAATTT TTTTGGTAAAAAAAAATTACTGTTTATTAAAGTACAACCATAGAGGATGGTCTTACAGCAGGCAGTATCCTGTTTGAGGAAAGCAAGAAT CAGAGAAGGAACATACCCCTTACAAATGAAAAATTCCACTCAAAATAGGGACTATCTATCTTAATACTAAGGAACCAACAATCTTCCTGT TTAAAAAACCACATGGCACAGAGATTCTGAACTAAAGTGCTGCACTCAAATGATGGGAAGTCCGGCCCCAGTACACAGGGGCTTGACTTT TTCAACTTCGTTTCCTTTGTTGGAGTCAAAAAGAACCACTTGTGGTTCTAAAAGGTGTGAAGGTGATTTAAGGGCCCAGGTCAGCCACTG TTTGTTTACAAAATCAGGTAACTAACTGCATACACTTTTTCTCTTTCCATGACATCAAGACTTTGCTAAAGACATGAAGCCACGGGTGCC AGAAGCTACTGCGATGCCCCGGGAGTTAGCCCCCTGGTAATAGCTGTAAACTTCCAATTTCTAGCCATACGCTCAGCTCATCCATGCCTC AGAAGTGCATCTGGAGAGAACAGGTTTCTAAGCATAAAAGATGAAAGAGCAGTTGGACTTTTTAAAAATTCAGCAAAGTGGTTCCCTCTC TTAGGGACAGTCAAAACCAAGTCACTTAGGTAGTACCAAAATAAATAAGGAAAAGCTTAGCTTTAGAAACAGTGCAACACTGGTCTGCTG TTCCAGTGGTAAGCTATGTCCCAGGAATCAGTTTAAAAGCACGACAGTGGATGCTGGGTCCATATCACACACATTGCTGTGAACAGGAAA CTCCTGTGACCACAACATGAGGCCACTGGAGACGCATATGAGTAAGGGCACTGACGGACTCATGATTTCTTCTTACCAGATGCTTTCCTG TTCTTTAAGAGTTTAAAATCATCAGAAAGGAAAAACAAACTCTATATTGTTCAGCATGCAATACATACCACGCTAGGGCTGGCTCAATTG >53514_53514_4_MICAL3-BID_MICAL3_chr22_18368643_ENST00000585038_BID_chr22_18226779_ENST00000317361_length(amino acids)=938AA_BP=0 MEERKHETMNPAHVLFDRFVQATTCKGTLKAFQELCDHLELKPKDYRSFYHKLKSKLNYWKAKALWAKLDKRGSHKDYKKGKACTNTKCL IIGAGPCGLRTAIDLSLLGAKVVVIEKRDAFSRNNVLHLWPFTIHDLRGLGAKKFYGKFCAGAIDHISIRQLQLILLKVALILGIEIHVN VEFQGLIQPPEDQENERIGWRALVHPKTHPVSEYEFEVIIGGDGRRNTLEGFRRKEFRGKLAIAITANFINRNTTAEAKVEEISGVAFIF NQKFFQELREATGIDLENIVYYKDDTHYFVMTAKKQSLLDKGVILHDYADTELLLSRENVDQEALLSYAREAADFSTQQQLPSLDFAINH YGQPDVAMFDFTCMYASENAALVREQNGHQLLVALVGDSLLEPFWPMGTGIARGFLAAMDSAWMVRSWSLGTSPLEVLAERESIYRLLPQ TTPENVSKNFSQYSIDPVTRYPNINVNFLRPSQVRHLYDTGETKDIHLEMESLVNSRTTPKLTRNESVARSSKLLGWCQRQTDGYAGVNV TDLTMSWKSGLALCAIIHRYRPDLIDFDSLDEQNVEKNNQLAFDIAEKELGISPIMTGKEMASVGEPDKLSMVMYLTQFYEMFKDSLPSS DTLDLNAEEKAVLIASTRSPISFLSKLGQTISRKRSPKDKKEKDLDGAGKRRKTSQSEEEEAPRGHRGERPTLVSTLTDRRMDVAVGNQN KVKYMATQLLAKFEENAPAQSIGIRRQVNNGSSLRDECITNLLVFGFLQSCSDNSFRRELDALGHELPVLAPQWEGYDELQTDGNRSSHS RLGRIEADSESQEDIIRNIARHLAQVGDSMDRSIPPGLVNGLALQLRNTSRSEEDRNRDLATALEQLLQAYPRDMEKEKTMLVLALLLAK -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for MICAL3-BID |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for MICAL3-BID |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for MICAL3-BID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
