
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:MICU2-ATP8A2 (FusionGDB2 ID:53580) |
Fusion Gene Summary for MICU2-ATP8A2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: MICU2-ATP8A2 | Fusion gene ID: 53580 | Hgene | Tgene | Gene symbol | MICU2 | ATP8A2 | Gene ID | 221154 | 51761 |
| Gene name | mitochondrial calcium uptake 2 | ATPase phospholipid transporting 8A2 | |
| Synonyms | 1110008L20Rik|EFHA1 | ATP|ATPIB|CAMRQ4|IB|ML-1 | |
| Cytomap | 13q12.11 | 13q12.13 | |
| Type of gene | protein-coding | protein-coding | |
| Description | calcium uptake protein 2, mitochondrialEF hand domain family A1EF hand domain family, member A1EF-hand domain-containing family member A1Smhs2 homolog | phospholipid-transporting ATPase IBATPase, aminophospholipid transporter, class I, type 8A, member 2ATPase, aminophospholipid transporter-like, class I, type 8A, member 2P4-ATPase flippase complex alpha subunit ATP8A2probable phospholipid-transporting | |
| Modification date | 20200315 | 20200329 | |
| UniProtAcc | Q8IYU8 | Q9NTI2 | |
| Ensembl transtripts involved in fusion gene | ENST00000382374, ENST00000479790, | ENST00000491840, ENST00000255283, ENST00000381655, | |
| Fusion gene scores | * DoF score | 5 X 3 X 3=45 | 13 X 13 X 7=1183 |
| # samples | 5 | 15 | |
| ** MAII score | log2(5/45*10)=0.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(15/1183*10)=-2.97941566784391 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: MICU2 [Title/Abstract] AND ATP8A2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | MICU2(22178078)-ATP8A2(26402256), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | MICU2 | GO:0006851 | mitochondrial calcium ion transmembrane transport | 24560927 |
| Hgene | MICU2 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration | 24560927 |
 Fusion gene breakpoints across MICU2 (5'-gene) Fusion gene breakpoints across MICU2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
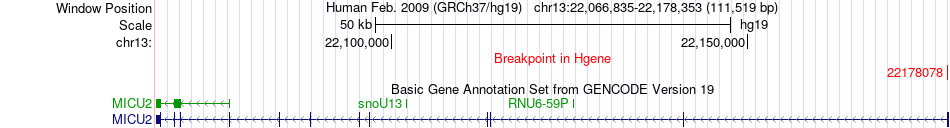 |
 Fusion gene breakpoints across ATP8A2 (3'-gene) Fusion gene breakpoints across ATP8A2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
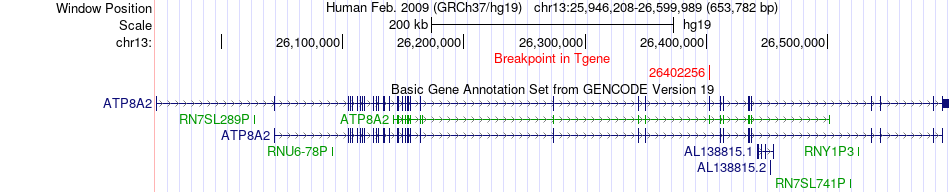 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-E9-A22D-01A | MICU2 | chr13 | 22178078 | - | ATP8A2 | chr13 | 26402256 | + |
Top |
Fusion Gene ORF analysis for MICU2-ATP8A2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000382374 | ENST00000491840 | MICU2 | chr13 | 22178078 | - | ATP8A2 | chr13 | 26402256 | + |
| 5CDS-intron | ENST00000382374 | ENST00000255283 | MICU2 | chr13 | 22178078 | - | ATP8A2 | chr13 | 26402256 | + |
| In-frame | ENST00000382374 | ENST00000381655 | MICU2 | chr13 | 22178078 | - | ATP8A2 | chr13 | 26402256 | + |
| intron-3CDS | ENST00000479790 | ENST00000381655 | MICU2 | chr13 | 22178078 | - | ATP8A2 | chr13 | 26402256 | + |
| intron-3UTR | ENST00000479790 | ENST00000491840 | MICU2 | chr13 | 22178078 | - | ATP8A2 | chr13 | 26402256 | + |
| intron-intron | ENST00000479790 | ENST00000255283 | MICU2 | chr13 | 22178078 | - | ATP8A2 | chr13 | 26402256 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000382374 | MICU2 | chr13 | 22178078 | - | ENST00000381655 | ATP8A2 | chr13 | 26402256 | + | 7030 | 276 | 66 | 1163 | 365 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000382374 | ENST00000381655 | MICU2 | chr13 | 22178078 | - | ATP8A2 | chr13 | 26402256 | + | 0.014314462 | 0.9856856 |
Top |
Fusion Genomic Features for MICU2-ATP8A2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
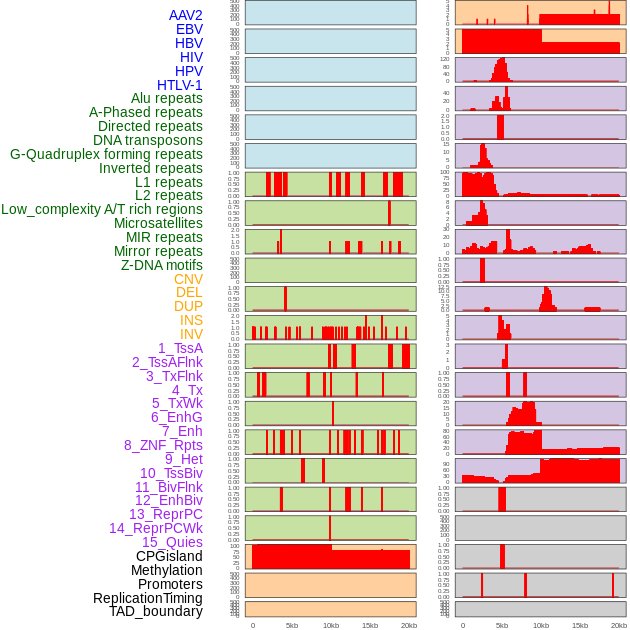 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
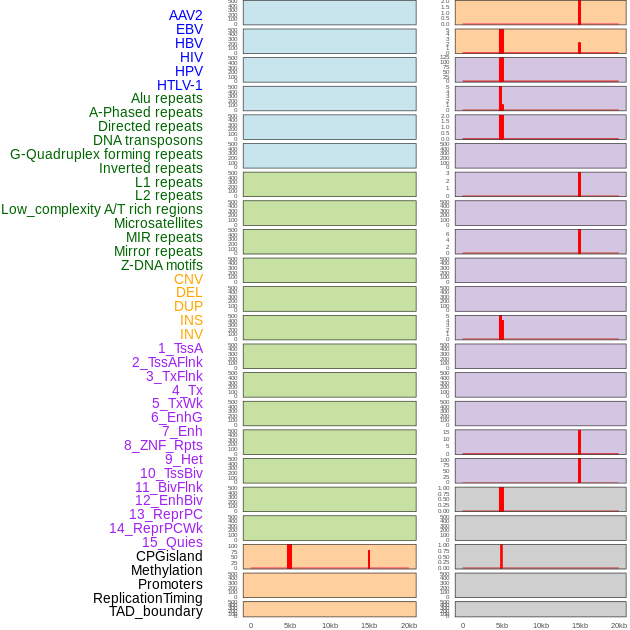 |
Top |
Fusion Protein Features for MICU2-ATP8A2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr13:22178078/chr13:26402256) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MICU2 | ATP8A2 |
| FUNCTION: Key regulator of mitochondrial calcium uniporter (MCU) required to limit calcium uptake by MCU when cytoplasmic calcium is low (PubMed:24503055, PubMed:24560927, PubMed:26903221). MICU1 and MICU2 form a disulfide-linked heterodimer that stimulate and inhibit MCU activity, depending on the concentration of calcium (PubMed:24560927). MICU2 acts as a gatekeeper of MCU that senses calcium level via its EF-hand domains: prevents channel opening at resting calcium, avoiding energy dissipation and cell-death triggering (PubMed:24560927). {ECO:0000269|PubMed:24503055, ECO:0000269|PubMed:24560927, ECO:0000269|PubMed:26387864, ECO:0000269|PubMed:26903221}. | FUNCTION: Catalytic component of a P4-ATPase flippase complex which catalyzes the hydrolysis of ATP coupled to the transport of aminophospholipids from the outer to the inner leaflet of various membranes and ensures the maintenance of asymmetric distribution of phospholipids. Phospholipid translocation seems also to be implicated in vesicle formation and in uptake of lipid signaling molecules. Reconstituted to liposomes, the ATP8A2:TMEM30A flippase complex predominantly transports phosphatidylserine (PS) and to a lesser extent phosphatidylethanolamine (PE). Phospholipid translocation is not associated with a countertransport of an inorganic ion or other charged substrate from the cytoplasmic side toward the exoplasm in connection with the phosphorylation from ATP. ATP8A2:TMEM30A may be involved in regulation of neurite outgrowth. Proposed to function in the generation and maintenance of phospholipid asymmetry in photoreceptor disk membranes and neuronal axon membranes. May be involved in vesicle trafficking in neuronal cells. Required for normal visual and auditory function; involved in photoreceptor and inner ear spiral ganglion cell survival. {ECO:0000250|UniProtKB:C7EXK4}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MICU2 | chr13:22178078 | chr13:26402256 | ENST00000382374 | - | 1 | 12 | 2_50 | 70 | 435.0 | Compositional bias | Note=Ala-rich |
| Tgene | ATP8A2 | chr13:22178078 | chr13:26402256 | ENST00000381655 | 26 | 37 | 1019_1028 | 893 | 1189.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP8A2 | chr13:22178078 | chr13:26402256 | ENST00000381655 | 26 | 37 | 1050_1063 | 893 | 1189.0 | Topological domain | Extracellular | |
| Tgene | ATP8A2 | chr13:22178078 | chr13:26402256 | ENST00000381655 | 26 | 37 | 1085_1188 | 893 | 1189.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP8A2 | chr13:22178078 | chr13:26402256 | ENST00000381655 | 26 | 37 | 909_910 | 893 | 1189.0 | Topological domain | Extracellular | |
| Tgene | ATP8A2 | chr13:22178078 | chr13:26402256 | ENST00000381655 | 26 | 37 | 932_959 | 893 | 1189.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP8A2 | chr13:22178078 | chr13:26402256 | ENST00000381655 | 26 | 37 | 981_997 | 893 | 1189.0 | Topological domain | Extracellular | |
| Tgene | ATP8A2 | chr13:22178078 | chr13:26402256 | ENST00000381655 | 26 | 37 | 1029_1049 | 893 | 1189.0 | Transmembrane | Helical | |
| Tgene | ATP8A2 | chr13:22178078 | chr13:26402256 | ENST00000381655 | 26 | 37 | 1064_1084 | 893 | 1189.0 | Transmembrane | Helical | |
| Tgene | ATP8A2 | chr13:22178078 | chr13:26402256 | ENST00000381655 | 26 | 37 | 911_931 | 893 | 1189.0 | Transmembrane | Helical | |
| Tgene | ATP8A2 | chr13:22178078 | chr13:26402256 | ENST00000381655 | 26 | 37 | 960_980 | 893 | 1189.0 | Transmembrane | Helical | |
| Tgene | ATP8A2 | chr13:22178078 | chr13:26402256 | ENST00000381655 | 26 | 37 | 998_1018 | 893 | 1189.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MICU2 | chr13:22178078 | chr13:26402256 | ENST00000382374 | - | 1 | 12 | 172_207 | 70 | 435.0 | Domain | EF-hand 1 |
| Hgene | MICU2 | chr13:22178078 | chr13:26402256 | ENST00000382374 | - | 1 | 12 | 227_262 | 70 | 435.0 | Domain | EF-hand 2 |
| Hgene | MICU2 | chr13:22178078 | chr13:26402256 | ENST00000382374 | - | 1 | 12 | 293_328 | 70 | 435.0 | Domain | EF-hand 3 |
| Hgene | MICU2 | chr13:22178078 | chr13:26402256 | ENST00000382374 | - | 1 | 12 | 362_397 | 70 | 435.0 | Domain | EF-hand 4 |
| Tgene | ATP8A2 | chr13:22178078 | chr13:26402256 | ENST00000381655 | 26 | 37 | 116_119 | 893 | 1189.0 | Topological domain | Extracellular | |
| Tgene | ATP8A2 | chr13:22178078 | chr13:26402256 | ENST00000381655 | 26 | 37 | 141_316 | 893 | 1189.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP8A2 | chr13:22178078 | chr13:26402256 | ENST00000381655 | 26 | 37 | 1_94 | 893 | 1189.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP8A2 | chr13:22178078 | chr13:26402256 | ENST00000381655 | 26 | 37 | 338_364 | 893 | 1189.0 | Topological domain | Extracellular | |
| Tgene | ATP8A2 | chr13:22178078 | chr13:26402256 | ENST00000381655 | 26 | 37 | 386_887 | 893 | 1189.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP8A2 | chr13:22178078 | chr13:26402256 | ENST00000381655 | 26 | 37 | 120_140 | 893 | 1189.0 | Transmembrane | Helical | |
| Tgene | ATP8A2 | chr13:22178078 | chr13:26402256 | ENST00000381655 | 26 | 37 | 317_337 | 893 | 1189.0 | Transmembrane | Helical | |
| Tgene | ATP8A2 | chr13:22178078 | chr13:26402256 | ENST00000381655 | 26 | 37 | 365_385 | 893 | 1189.0 | Transmembrane | Helical | |
| Tgene | ATP8A2 | chr13:22178078 | chr13:26402256 | ENST00000381655 | 26 | 37 | 888_908 | 893 | 1189.0 | Transmembrane | Helical | |
| Tgene | ATP8A2 | chr13:22178078 | chr13:26402256 | ENST00000381655 | 26 | 37 | 95_115 | 893 | 1189.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for MICU2-ATP8A2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >53580_53580_1_MICU2-ATP8A2_MICU2_chr13_22178078_ENST00000382374_ATP8A2_chr13_26402256_ENST00000381655_length(transcript)=7030nt_BP=276nt GAGAGTTCCCAAGCGGTAGGCGGCGGCGCCGGGAGAGAAGCGCCGCCTAGCTGCGCTTCCGCAAAGATGGCGGCGGCTGCGGGTAGCTGC GCGCGGGTGGCGGCCTGGGGCGGAAAACTGCGACGGGGGCTCGCTGTCAGCCGACAGGCTGTGCGGAGTCCCGGCCCCTTGGCAGCGGCA GTGGCCGGCGCGGCCCTGGCAGGAGCAGGAGCGGCCTGGCACCACAGCCGCGTCAGTGTTGCGGCGCGGGATGGCAGTTTTACAGTCTCC GCACAGCTTTGGTTCGCCTTTGTTAATGGATTTTCTGGGCAGATTTTATTTGAACGTTGGTGCATCGGCCTGTACAATGTGATTTTCACC GCTTTGCCGCCCTTCACTCTGGGAATCTTTGAGAGGTCTTGCACTCAGGAGAGCATGCTCAGGTTTCCCCAGCTCTACAAAATCACCCAG AATGGCGAAGGCTTCAACACAAAGGTTTTCTGGGGTCACTGCATCAACGCCTTGGTCCACTCCCTCATCCTCTTCTGGTTTCCCATGAAA GCTCTGGAGCATGATACTGTGTTGACAAGTGGTCATGCTACCGACTATTTATTTGTTGGAAATATTGTTTACACATATGTTGTTGTTACT GTTTGTCTGAAAGCTGGTTTGGAGACCACAGCTTGGACTAAATTCAGTCATCTGGCTGTCTGGGGAAGCATGCTGACCTGGCTGGTGTTT TTTGGCATCTACTCGACCATCTGGCCCACCATTCCCATTGCTCCAGATATGAGAGGACAGGCAACTATGGTCCTGAGCTCCGCACACTTC TGGTTGGGATTATTTCTGGTTCCTACTGCCTGTTTGATTGAAGATGTGGCATGGAGAGCAGCCAAGCACACCTGCAAAAAGACATTGCTG GAGGAGGTGCAGGAGCTGGAAACCAAGTCTCGAGTCCTGGGAAAAGCGGTGCTGCGGGATAGCAATGGAAAGAGGCTGAACGAGCGCGAC CGCCTGATCAAGAGGCTGGGCCGGAAGACGCCCCCGACGCTGTTCCGGGGCAGCTCCCTGCAGCAGGGCGTCCCGCATGGGTATGCTTTT TCTCAAGAAGAACACGGAGCTGTTAGTCAGGAAGAAGTCATCCGTGCTTATGACACCACCAAAAAGAAATCCAGGAAGAAATAAGACATG AATTTTCCTGACTGATCTTAGGAAAGAGATTCAGTTTGTTGCACCCAGTGTTAACACATCTTTGTCAGAGAAGACTGGCGTCAGCAGCCA AAACACCAGGAAACACATTTCTGTGGCCTTAGCCAAGCAGTTTGTTAGTTACATATTCCCTCGCAAACCTGGAGTGCAGACCACAGGGGA AGCTATCTTTGCCCTCCCAACTCGTCTGCAGTGCTTAGCCTAACTTTTGTTTATGTCGTTATGAAGCATTCAACTGTGCTCTGTGAGGTG TGAAATTAAAAACATTATGTTTCACCAATATTTAAACATCAGTACTAGTTGTCCTGGGAGAAAGGGAAAGGAGTTTTATGTTGCGTGAGA GGCCCATCCTGTGTAATTGGAGCAGGGCACACTTGCTTCCTGTTGAGTTAACTCAGAGGTTAAGTCCAACGGGCCACATGCAGACTTCAC TGTAGGCAGGTTGCTCTCCTGCTTTGATTCCTGTTTTGTGTGTAAAATTGGCATAAACTTCTTGATTGCAGTGAAATCACAAAATTCTCT ATCGGGGTGGTCAACCTGAGAACATTTATTTGAACCTCTTAGCCACATTTCCAGCAGGGCAAACCAACTGATGCCCTGGAAGAGTCTTCC GCACCCTCTCCAGGTGCACTCGGCCCAGTCCTGCCGCCGTGTCCAGGCGCCCCTCACCCCCACACCTGCTGCATGGCTGGCCACACCACT CAGTGCATGGCGGCCGATGGGCAGCCAACCCAAACCCGCGCCTTTCCTTGTTCCACTGCAGACTCAGATACAGATGCGAAAAATTCCTTC TTCCACCGCCCTTCTCGTTCTGTAAAGAAAGAAAAGAAACATAGCCTTTCTGCATATATTCTAAACGTCTCTCTGCCTCTGTCTGACATG GGGCCACCCCACAGGTCAGAGTGGTGGTAGAACCCCTTCAGGACTCCCAGCCGTGGTCAGGCTCTGAATACTCCCTTCCCAACATCCAGA CTGCTGGGCCTTTGGCATCCACTTACATTAGAACCCACGTTTGTTTCAGAGCACATTTTGGACTTTCACTGTTGGGAAATGAATGAATTT ATAACATGCCTGCACAGCGAAGGAACACACCTGTCGCTCTTAGCTCTAGAGTCAGAGGATGAGTAAACCCAGATGCAAGAGTATAGGACA TTGAGTGGGGAGAACAAGACGACCACAGAAGTCCTCAGAAGGAGAAGGAAGGACACGGAGACACTGAGAGGAGGACACAGAGGAATCGCC ACCAGATCTTTGCAGTAGAAACTCTGAAATAGGCCTTGAGACCCAGACAATTGTCATGATCAGCCCAGTGTGTATGAGAGCTTAAACAAG ACCTTCACACACAGTTCTAATTGAAAAGAATTCCAAACATTTCAGGGAAACACCTTTTTAAAAAATATACTGACATGGTTTCCTTTCTGT ATCCCATCAGTGTTAGACACAGGTACCTATCAGAATACGGCATATCCTGAATTATCTAAGACAAGGAAGTGAAGAGCGGAATCACTGACT TTCCCAACTGCAGAGTCCCACACTTAAATCTCAACAGAACCTAGATGTCTTCAGGCATGAATGTGAGCACCTGGAGCCCTTACTAAACAA CCTGCCTGGAGCCCTGTTCTCTGCTCTGGTTGCCAAACCAGATTTTTGGATGGCCTAAATACTTGTGGGCAACTGTTACTGGTGAAAAAT GGGAGTGGAGAGCATGTTTTGGATTTTCACTGTGGGGAAATGAATGAATTTATCACATGACTGTGCAGCGAAGGAGCACACCTGTCACTC TTAGCTCTAGAGTCGGAGGATGAGTCAACCCTGTAGGAATGATTGTGCTAAATTAGGAAATAACACTTTAGTGCACTTTGCTTCCTAACC CTTTTTTGTCAGAAATCTACAACCTACTGGGCATGGAAATGATTTAAAATATGATCTGCATTGACTAAGAAAACTCATCCTCACTTATGA GGTGACCAGGATGCAGAGAGGGCCTGAGGGGCAGCAGAACAATGCAATTCACAGGTGAGGCTCCAAAAAGGGCCATTTCTTTCTCCCCAA GGTGCATAGAACATTTTTAAATATCAGAGTACCCAGCAAGACGGAAGGACTGTCATGCAGGAACTCTAACACAAAATGTGCCTATGACTA ACACGACAAACCAAGGTGTATGGTTTCTTTGCACACCCAGAAAGGTGATGGCCAAGATACAGAGGAGAAACGAAAACTGGAATTGAATGT GCATTAGGCATCTTAACTATTCATATTTTGTCTCATTTGGGAACTAAGCCTATTTGGAAAGAGTGGATTAGAAATTGAATTCTTTGTCAA ACAGTTAGCTCTGTCTTGAACTTGAAGCCTTCCAAATCAGAATTATGTATTTAAGTGTGTGTTTGTGTTTGTTTGGGGTTTTTGTTTCTT ACATTAGAGTTCATATTTTCTGGGATTTAAGGATACAGGTGTATTTTCCTATTCTCTTGAGAAGTAGACTAAACAATATTTGCAGTGATG ACAGACGCACACAGAAGAAAACATTAGAAGAGATGCCTTTCTATCCTCTCATGTGGTTGAGCATTCTTACAGCCAAATGACTAAATTGGC TGTAAATTGGTTTCTAGGAGGAGCAGCTGTCACTACGTGTGATGTATAATATTTAGCCAGGGCCAACTCAGGGGCTGTGTCATTTGTCCT ATCTTTAAATTGAGGTCAGAGTGTCACACACAACCCCTCCCCTGGGGCAGGGTGGACTGGACTAGGTTGCATACGCTGGCTGTCAGCTCA GCAAAAATGCCACATGGGGCTTGTCACTCCGCCAGACACTCACTGACTCACGGGCTGGCAGGTCACGCTCGGTTAGCGCTCCAGGGCCTG ACTCTGGATTGAGCTAAACTGGCAGAATCCACTCAGGCCAACCCTTGGTCTACTGCTGTGTACAGACAGCCCAGTCCTGTGTTGAGTCTC TCTTCTCCTGGATGACCCTGCAAAAGGAAGAGACCAGTTGTACTGTGGGCTTAGTTCTGGGTTTCCACTGCACACTGACACGTGCCAGCA AGGGTAGCTGTGGAAAACACGTATCAGGAGAGCAGAGTAGTCTGGATGAATACTCTGTGCTGATCAGAACGGGCTCGGTCATAATTTAGG AGTGGAGCTGATGTGTCTGTCCTGCCTCAGTGCAGAATATTCCAGGCTTCTTGACCAGGGTTATGTTCACCCATCTCCCAACACACACAT GCACACATGAAAACAAAAACATGGTAACTCTTTTGGTTCGGCACACCATACAAAACGTGCTTTGGGGCGAGGAGTGCTGTTTCTTTTAAC ACTTGTAGAGGAATATCCTGAGAAGTGAACCTGGGCTTTGGGAGAGCTCAGCTGAGAATTCTGTTGACATTTTCTTTCCCTTGACAAGGC TTCAGGTTCGTCTCACTATAGGGAGCTGGCTGTGAAAATCAGTAATAACTTGTCAAGAAGCAGTGAAAAATGCGGGGGGGTGGAAAGAGC CTAGGTAACTAATGTCTTATAATGCATATGTATGTAAATATTACAGGATTTAAGGTTGAATTTTTTAAAAAGAAAGTTATAGTCTGTAAT TTCCATTTGTTATAATAATGACCTTTAATCTTGTCATTTGGAACCATAAAGCATTTTTATCAGGTACCTCTGTTCCAAGGGATTTATGTC TTAGACCATAGCTGAATTGAATGTTTGCAAAACACTGCTATAGGATAAGGTGGTCTTTAGTTTTGAACGTGTGAAAGGACTGCACACTTT TCAGCCAGGGTTTGAGTTACTGCCCAGGGTCATCGTCTCAAAGTAATTCAAGGAGTGATTTAACATCAGCATTTGAAATGTAGTCTTCAT CTCCTGGGATCCATAAAAAAATGTGAACAGGGAAATGGTGGCTAAGCAGAGCCTGAAATAATAACTTGGCAAAGAAATGAGTTTATCAGG TCGAGTCAAAACATGGCATCCCCTGTTACACTCAAGAAATGTTTTCTTCATGTAAATGTTTATACGGGCATATATAATCACAATGGGAAC AGTTAAAACCCCCTCCCTTCAAAAAAAGAAAATCTATATCAGTTGGGTTTGGTTTTGGTTCTTCATTGGCTCAAGGCAGTTAACTGTCTC AGTATAGCCTTCGGGGAGATTTAACCTCATTCTAGCCATTTTTCCATCCTGAAGGCCAAGAAGGACTATTAGAAGGGTTTTTGAGGGGTT TCGGAGGTGAGGGCCCAAGACCCCCATAATGACATCATTAGGCATTCTTGAAAGGTGTTAACAGACCAGCACGCTCGATGTGTTGTACCC TTCATTTATTTTTTTTTCTCAGCATCCCAAGTTGTCCATCAGTACATTCCCCTTCACCTTACCTGATCTTCATTTAATGCCCAGTCTGTA ACTAATGTTCTGTTTAAAGCTCTCTTAATTTGTTGGCTATGAGTGATTTGTGAATCTGGGATGTAACCCTGAGTGAGGAGAAAAGGGCAT TGAGCCAGCATAAATCTGAATGGGCAGAAGCCACAAACAAATGGGAGCAGAAAAGAAGGGCGAGCTCCCCCGCGCCGCACCCCGTGTATA ATCCAGTGTTAACCTCTGAGGAACACCTCGGCTCTCCCAACTGAGGAGAGACAAAAGAGGGATTCTTTTTACACCCAGGCTGGCGCATTT TCAAGTGACAATCTCGGGCTCTAGTTGCCTTTGAGATATCCACTCTGCTCTTTCTCCCAGGCCTCAGACCAAGAAAAACATGCTCTCAAG ATTAGCCCATAGGCAGTTCTTGTGACCTGGCTGAAGAAAGAAAGGAGACCTGTTTGTTTTAAAAGTCGGGCGCAAAGTGTCAGGTGGCTT TGATTTATGACAGAAAGAGGAAGAAGAAAGTTGGAGTAATAGCACTATCCAAATAACCCTGCCCAGGAAACTTGGGGGTCAAGAGAGCTT ATCAAGAGCCTTTTATAGGTAAGCTCTTCCGTGTGAAAGAGAAAAGGCCGGAAAGAGGGAGAGAGCCCAGAGAGCAGGCTGCGAACTCAC TTCCAATGGGATTGGAGTCTGTTGTATTCTTAAGAGATAGCCTGATATGTGGTTTATAGGTGAATCAATAAACACCAACAACAACAAAAA AAAAAAAAAAAAAAAAGGAAGAAAAGAAAAAAAGGAAACCAGCCCTGTCATGGAATTTCTCTCCTTCCCTGCACAGTAAAGACTTTTGGG TTTTCATGGATAAAATCAATGTCAGTACTGAAACTCCTACTCTCCCCTCCCGCCCCACTCTCCCCCGTTGCCCGAGATGGCCAAGTTCAG GCCTGTGCAATGCCGCTTCCCTCTGAGCCTCCCTCTCAAGGGCCACGCAGGCAGCTGCAGCAGGGCCAGCTGCAGGATGGGGCTGCCGGT CACTGAATTGTCGTTCAAATGCATCATCTTTGTGGCGTCTTTCTCATGCGAGCAAAGCCACGTGCTCTCCTGTCTGCTGTCACATCTGTG CCTGGATTGCTTAAATATTGTTTGTGATGGGGAGGTTTTAATCTGGTGATGCAGAGGGAAGCAGGGCTGTGGGGGCACGTTTAATTGGCT CCCAGCAGCGTGGGGGGTGCTTCTATGGTGTGTGGGGTTTTTTGTTGCCTCCCTCTAGAAGTGTTACCGTTTTCACGTCCTATTAATGTC CTCTGGTTGTTAAATTACAGCAGCACATTACAGTGCACTGGGTTCCCTCCTGGAGTGAATACAAACGGAGGGCATCTACTTGTATTTTTA GAAGTTTTGGGAGAATTTAGTGATTTGTGGCTTTGATCAATCCTGTTGACTGGTGTATGTCTGCGCAAACCTGTTTCAAATAAATCTTTT >53580_53580_1_MICU2-ATP8A2_MICU2_chr13_22178078_ENST00000382374_ATP8A2_chr13_26402256_ENST00000381655_length(amino acids)=365AA_BP=69 MAAAAGSCARVAAWGGKLRRGLAVSRQAVRSPGPLAAAVAGAALAGAGAAWHHSRVSVAARDGSFTVSAQLWFAFVNGFSGQILFERWCI GLYNVIFTALPPFTLGIFERSCTQESMLRFPQLYKITQNGEGFNTKVFWGHCINALVHSLILFWFPMKALEHDTVLTSGHATDYLFVGNI VYTYVVVTVCLKAGLETTAWTKFSHLAVWGSMLTWLVFFGIYSTIWPTIPIAPDMRGQATMVLSSAHFWLGLFLVPTACLIEDVAWRAAK HTCKKTLLEEVQELETKSRVLGKAVLRDSNGKRLNERDRLIKRLGRKTPPTLFRGSSLQQGVPHGYAFSQEEHGAVSQEEVIRAYDTTKK -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for MICU2-ATP8A2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for MICU2-ATP8A2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for MICU2-ATP8A2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
