
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:MITF-ARL6IP5 (FusionGDB2 ID:53980) |
Fusion Gene Summary for MITF-ARL6IP5 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: MITF-ARL6IP5 | Fusion gene ID: 53980 | Hgene | Tgene | Gene symbol | MITF | ARL6IP5 | Gene ID | 4286 | 10550 |
| Gene name | melanocyte inducing transcription factor | ADP ribosylation factor like GTPase 6 interacting protein 5 | |
| Synonyms | CMM8|COMMAD|MI|WS2|WS2A|bHLHe32 | DERP11|GTRAP3-18|HSPC127|JWA|PRAF3|Yip6b|addicsin|hp22|jmx | |
| Cytomap | 3p13 | 3p14.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | microphthalmia-associated transcription factorclass E basic helix-loop-helix protein 32melanogenesis associated transcription factormicrophtalmia-associated transcription factor | PRA1 family protein 3ADP-ribosylation factor GTPase 6 interacting protein 5ADP-ribosylation factor-like 6 interacting protein 5ADP-ribosylation factor-like protein 6-interacting protein 5ADP-ribosylation-like factor 6 interacting protein 5ARL-6-inter | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | O75030 | O75915 | |
| Ensembl transtripts involved in fusion gene | ENST00000352241, ENST00000448226, ENST00000314557, ENST00000314589, ENST00000328528, ENST00000394348, ENST00000394351, ENST00000394355, ENST00000472437, ENST00000531774, | ENST00000273258, ENST00000478935, | |
| Fusion gene scores | * DoF score | 11 X 6 X 6=396 | 8 X 7 X 3=168 |
| # samples | 9 | 8 | |
| ** MAII score | log2(9/396*10)=-2.13750352374993 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(8/168*10)=-1.0703893278914 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: MITF [Title/Abstract] AND ARL6IP5 [Title/Abstract] AND fusion [Title/Abstract] | ||
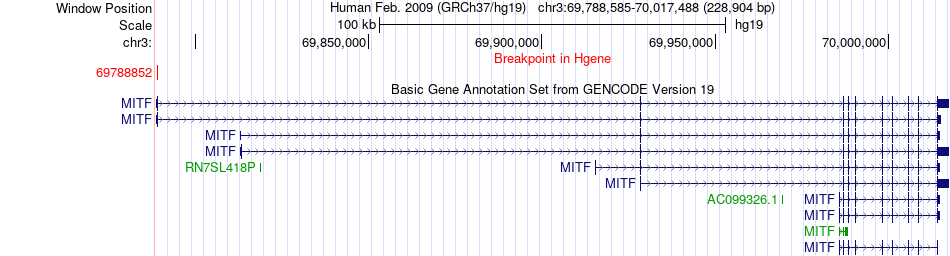
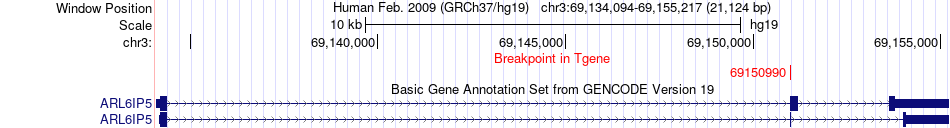
| Most frequent breakpoint | MITF(69788852)-ARL6IP5(69150990), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | MITF | GO:0010628 | positive regulation of gene expression | 22234890 |
| Hgene | MITF | GO:0045893 | positive regulation of transcription, DNA-templated | 9647758 |
| Hgene | MITF | GO:0045944 | positive regulation of transcription by RNA polymerase II | 20530484|21209915 |
| Hgene | MITF | GO:0065003 | protein-containing complex assembly | 20530484 |
| Hgene | MITF | GO:2000144 | positive regulation of DNA-templated transcription, initiation | 8995290|12204775 |
| Hgene | MITF | GO:2001141 | regulation of RNA biosynthetic process | 16411896 |
| Tgene | ARL6IP5 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress | 18387645 |
 Fusion gene breakpoints across MITF (5'-gene) Fusion gene breakpoints across MITF (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across ARL6IP5 (3'-gene) Fusion gene breakpoints across ARL6IP5 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-HC-8216-01A | MITF | chr3 | 69788852 | + | ARL6IP5 | chr3 | 69150990 | + |
| ChimerDB4 | PRAD | TCGA-HC-8216 | MITF | chr3 | 69788852 | + | ARL6IP5 | chr3 | 69150990 | + |
Top |
Fusion Gene ORF analysis for MITF-ARL6IP5 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000352241 | ENST00000273258 | MITF | chr3 | 69788852 | + | ARL6IP5 | chr3 | 69150990 | + |
| In-frame | ENST00000352241 | ENST00000478935 | MITF | chr3 | 69788852 | + | ARL6IP5 | chr3 | 69150990 | + |
| In-frame | ENST00000448226 | ENST00000273258 | MITF | chr3 | 69788852 | + | ARL6IP5 | chr3 | 69150990 | + |
| In-frame | ENST00000448226 | ENST00000478935 | MITF | chr3 | 69788852 | + | ARL6IP5 | chr3 | 69150990 | + |
| intron-3CDS | ENST00000314557 | ENST00000273258 | MITF | chr3 | 69788852 | + | ARL6IP5 | chr3 | 69150990 | + |
| intron-3CDS | ENST00000314557 | ENST00000478935 | MITF | chr3 | 69788852 | + | ARL6IP5 | chr3 | 69150990 | + |
| intron-3CDS | ENST00000314589 | ENST00000273258 | MITF | chr3 | 69788852 | + | ARL6IP5 | chr3 | 69150990 | + |
| intron-3CDS | ENST00000314589 | ENST00000478935 | MITF | chr3 | 69788852 | + | ARL6IP5 | chr3 | 69150990 | + |
| intron-3CDS | ENST00000328528 | ENST00000273258 | MITF | chr3 | 69788852 | + | ARL6IP5 | chr3 | 69150990 | + |
| intron-3CDS | ENST00000328528 | ENST00000478935 | MITF | chr3 | 69788852 | + | ARL6IP5 | chr3 | 69150990 | + |
| intron-3CDS | ENST00000394348 | ENST00000273258 | MITF | chr3 | 69788852 | + | ARL6IP5 | chr3 | 69150990 | + |
| intron-3CDS | ENST00000394348 | ENST00000478935 | MITF | chr3 | 69788852 | + | ARL6IP5 | chr3 | 69150990 | + |
| intron-3CDS | ENST00000394351 | ENST00000273258 | MITF | chr3 | 69788852 | + | ARL6IP5 | chr3 | 69150990 | + |
| intron-3CDS | ENST00000394351 | ENST00000478935 | MITF | chr3 | 69788852 | + | ARL6IP5 | chr3 | 69150990 | + |
| intron-3CDS | ENST00000394355 | ENST00000273258 | MITF | chr3 | 69788852 | + | ARL6IP5 | chr3 | 69150990 | + |
| intron-3CDS | ENST00000394355 | ENST00000478935 | MITF | chr3 | 69788852 | + | ARL6IP5 | chr3 | 69150990 | + |
| intron-3CDS | ENST00000472437 | ENST00000273258 | MITF | chr3 | 69788852 | + | ARL6IP5 | chr3 | 69150990 | + |
| intron-3CDS | ENST00000472437 | ENST00000478935 | MITF | chr3 | 69788852 | + | ARL6IP5 | chr3 | 69150990 | + |
| intron-3CDS | ENST00000531774 | ENST00000273258 | MITF | chr3 | 69788852 | + | ARL6IP5 | chr3 | 69150990 | + |
| intron-3CDS | ENST00000531774 | ENST00000478935 | MITF | chr3 | 69788852 | + | ARL6IP5 | chr3 | 69150990 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000352241 | MITF | chr3 | 69788852 | + | ENST00000273258 | ARL6IP5 | chr3 | 69150990 | + | 2088 | 267 | 58 | 657 | 199 |
| ENST00000352241 | MITF | chr3 | 69788852 | + | ENST00000478935 | ARL6IP5 | chr3 | 69150990 | + | 1513 | 267 | 58 | 357 | 99 |
| ENST00000448226 | MITF | chr3 | 69788852 | + | ENST00000273258 | ARL6IP5 | chr3 | 69150990 | + | 2052 | 231 | 22 | 621 | 199 |
| ENST00000448226 | MITF | chr3 | 69788852 | + | ENST00000478935 | ARL6IP5 | chr3 | 69150990 | + | 1477 | 231 | 22 | 321 | 99 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000352241 | ENST00000273258 | MITF | chr3 | 69788852 | + | ARL6IP5 | chr3 | 69150990 | + | 0.002338808 | 0.99766123 |
| ENST00000352241 | ENST00000478935 | MITF | chr3 | 69788852 | + | ARL6IP5 | chr3 | 69150990 | + | 0.21400735 | 0.7859927 |
| ENST00000448226 | ENST00000273258 | MITF | chr3 | 69788852 | + | ARL6IP5 | chr3 | 69150990 | + | 0.002349186 | 0.9976508 |
| ENST00000448226 | ENST00000478935 | MITF | chr3 | 69788852 | + | ARL6IP5 | chr3 | 69150990 | + | 0.27479237 | 0.7252076 |
Top |
Fusion Genomic Features for MITF-ARL6IP5 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
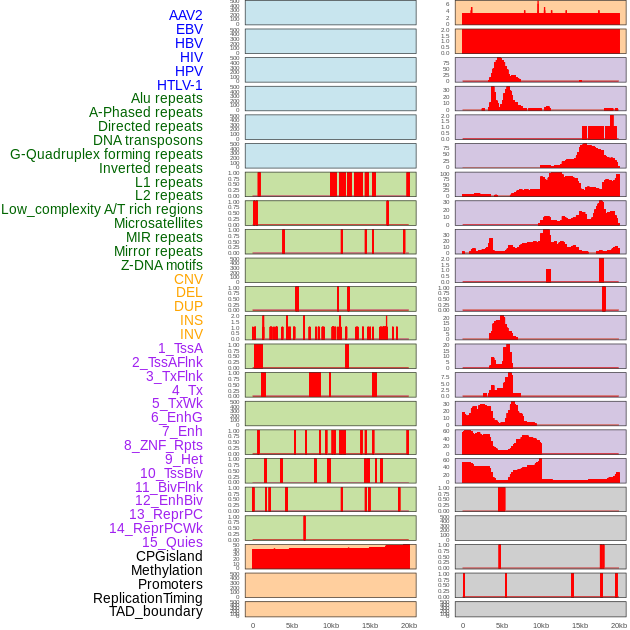
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
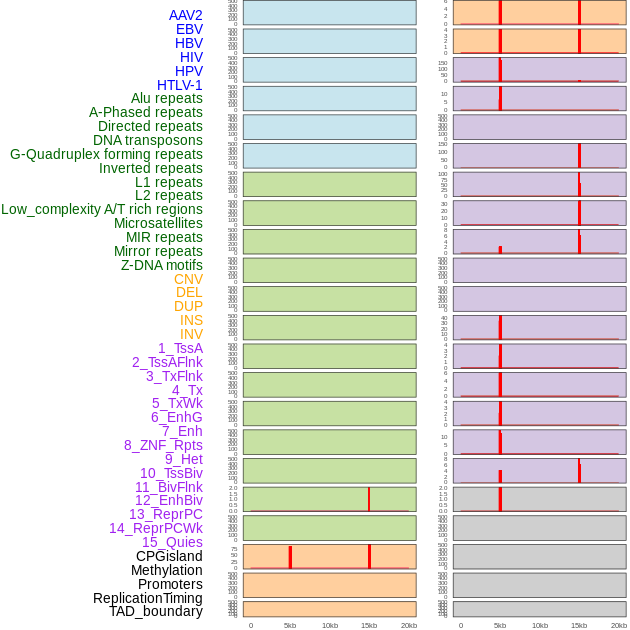
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for MITF-ARL6IP5 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:69788852/chr3:69150990) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MITF | ARL6IP5 |
| FUNCTION: Transcription factor that regulates the expression of genes with essential roles in cell differentiation, proliferation and survival. Binds to M-boxes (5'-TCATGTG-3') and symmetrical DNA sequences (E-boxes) (5'-CACGTG-3') found in the promoters of target genes, such as BCL2 and tyrosinase (TYR). Plays an important role in melanocyte development by regulating the expression of tyrosinase (TYR) and tyrosinase-related protein 1 (TYRP1). Plays a critical role in the differentiation of various cell types, such as neural crest-derived melanocytes, mast cells, osteoclasts and optic cup-derived retinal pigment epithelium. {ECO:0000269|PubMed:10587587, ECO:0000269|PubMed:22647378, ECO:0000269|PubMed:27889061, ECO:0000269|PubMed:9647758}. | FUNCTION: Regulates intracellular concentrations of taurine and glutamate. Negatively modulates SLC1A1/EAAC1 glutamate transport activity by decreasing its affinity for glutamate in a PKC activity-dependent manner. Plays a role in the retention of SLC1A1/EAAC1 in the endoplasmic reticulum. {ECO:0000250|UniProtKB:Q8R5J9, ECO:0000250|UniProtKB:Q9ES40}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | ARL6IP5 | chr3:69788852 | chr3:69150990 | ENST00000273258 | 0 | 3 | 103_117 | 58 | 189.0 | Region | Required for homodimer formation and heterodimer formation with ARL6IP1 | |
| Tgene | ARL6IP5 | chr3:69788852 | chr3:69150990 | ENST00000273258 | 0 | 3 | 136_188 | 58 | 189.0 | Region | Targeting to endoplasmic reticulum membrane | |
| Tgene | ARL6IP5 | chr3:69788852 | chr3:69150990 | ENST00000273258 | 0 | 3 | 78_93 | 58 | 189.0 | Topological domain | Cytoplasmic | |
| Tgene | ARL6IP5 | chr3:69788852 | chr3:69150990 | ENST00000273258 | 0 | 3 | 115_135 | 58 | 189.0 | Transmembrane | Helical | |
| Tgene | ARL6IP5 | chr3:69788852 | chr3:69150990 | ENST00000273258 | 0 | 3 | 57_77 | 58 | 189.0 | Transmembrane | Helical | |
| Tgene | ARL6IP5 | chr3:69788852 | chr3:69150990 | ENST00000273258 | 0 | 3 | 94_114 | 58 | 189.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000314557 | + | 1 | 9 | 355_402 | 0 | 414.0 | Coiled coil | Ontology_term=ECO:0000305 |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000314589 | + | 1 | 10 | 355_402 | 0 | 505.0 | Coiled coil | Ontology_term=ECO:0000305 |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000328528 | + | 1 | 10 | 355_402 | 0 | 520.0 | Coiled coil | Ontology_term=ECO:0000305 |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000352241 | + | 1 | 10 | 355_402 | 34 | 521.0 | Coiled coil | Ontology_term=ECO:0000305 |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000394351 | + | 1 | 9 | 355_402 | 0 | 420.0 | Coiled coil | Ontology_term=ECO:0000305 |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000394355 | + | 1 | 9 | 355_402 | 0 | 496.0 | Coiled coil | Ontology_term=ECO:0000305 |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000448226 | + | 1 | 10 | 355_402 | 34 | 527.0 | Coiled coil | Ontology_term=ECO:0000305 |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000472437 | + | 1 | 10 | 355_402 | 0 | 469.0 | Coiled coil | Ontology_term=ECO:0000305 |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000531774 | + | 1 | 9 | 355_402 | 0 | 358.0 | Coiled coil | Ontology_term=ECO:0000305 |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000314557 | + | 1 | 9 | 311_364 | 0 | 414.0 | Domain | bHLH |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000314589 | + | 1 | 10 | 311_364 | 0 | 505.0 | Domain | bHLH |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000328528 | + | 1 | 10 | 311_364 | 0 | 520.0 | Domain | bHLH |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000352241 | + | 1 | 10 | 311_364 | 34 | 521.0 | Domain | bHLH |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000394351 | + | 1 | 9 | 311_364 | 0 | 420.0 | Domain | bHLH |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000394355 | + | 1 | 9 | 311_364 | 0 | 496.0 | Domain | bHLH |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000448226 | + | 1 | 10 | 311_364 | 34 | 527.0 | Domain | bHLH |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000472437 | + | 1 | 10 | 311_364 | 0 | 469.0 | Domain | bHLH |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000531774 | + | 1 | 9 | 311_364 | 0 | 358.0 | Domain | bHLH |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000314557 | + | 1 | 9 | 224_295 | 0 | 414.0 | Region | Note=Transactivation |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000314557 | + | 1 | 9 | 374_395 | 0 | 414.0 | Region | Leucine-zipper |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000314557 | + | 1 | 9 | 401_431 | 0 | 414.0 | Region | Note=DNA binding regulation |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000314589 | + | 1 | 10 | 224_295 | 0 | 505.0 | Region | Note=Transactivation |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000314589 | + | 1 | 10 | 374_395 | 0 | 505.0 | Region | Leucine-zipper |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000314589 | + | 1 | 10 | 401_431 | 0 | 505.0 | Region | Note=DNA binding regulation |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000328528 | + | 1 | 10 | 224_295 | 0 | 520.0 | Region | Note=Transactivation |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000328528 | + | 1 | 10 | 374_395 | 0 | 520.0 | Region | Leucine-zipper |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000328528 | + | 1 | 10 | 401_431 | 0 | 520.0 | Region | Note=DNA binding regulation |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000352241 | + | 1 | 10 | 224_295 | 34 | 521.0 | Region | Note=Transactivation |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000352241 | + | 1 | 10 | 374_395 | 34 | 521.0 | Region | Leucine-zipper |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000352241 | + | 1 | 10 | 401_431 | 34 | 521.0 | Region | Note=DNA binding regulation |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000394351 | + | 1 | 9 | 224_295 | 0 | 420.0 | Region | Note=Transactivation |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000394351 | + | 1 | 9 | 374_395 | 0 | 420.0 | Region | Leucine-zipper |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000394351 | + | 1 | 9 | 401_431 | 0 | 420.0 | Region | Note=DNA binding regulation |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000394355 | + | 1 | 9 | 224_295 | 0 | 496.0 | Region | Note=Transactivation |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000394355 | + | 1 | 9 | 374_395 | 0 | 496.0 | Region | Leucine-zipper |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000394355 | + | 1 | 9 | 401_431 | 0 | 496.0 | Region | Note=DNA binding regulation |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000448226 | + | 1 | 10 | 224_295 | 34 | 527.0 | Region | Note=Transactivation |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000448226 | + | 1 | 10 | 374_395 | 34 | 527.0 | Region | Leucine-zipper |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000448226 | + | 1 | 10 | 401_431 | 34 | 527.0 | Region | Note=DNA binding regulation |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000472437 | + | 1 | 10 | 224_295 | 0 | 469.0 | Region | Note=Transactivation |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000472437 | + | 1 | 10 | 374_395 | 0 | 469.0 | Region | Leucine-zipper |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000472437 | + | 1 | 10 | 401_431 | 0 | 469.0 | Region | Note=DNA binding regulation |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000531774 | + | 1 | 9 | 224_295 | 0 | 358.0 | Region | Note=Transactivation |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000531774 | + | 1 | 9 | 374_395 | 0 | 358.0 | Region | Leucine-zipper |
| Hgene | MITF | chr3:69788852 | chr3:69150990 | ENST00000531774 | + | 1 | 9 | 401_431 | 0 | 358.0 | Region | Note=DNA binding regulation |
| Tgene | ARL6IP5 | chr3:69788852 | chr3:69150990 | ENST00000273258 | 0 | 3 | 1_35 | 58 | 189.0 | Topological domain | Cytoplasmic | |
| Tgene | ARL6IP5 | chr3:69788852 | chr3:69150990 | ENST00000273258 | 0 | 3 | 36_56 | 58 | 189.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for MITF-ARL6IP5 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >53980_53980_1_MITF-ARL6IP5_MITF_chr3_69788852_ENST00000352241_ARL6IP5_chr3_69150990_ENST00000273258_length(transcript)=2088nt_BP=267nt GTAAACTCCCCGCGCTGGGGCGGGCGGCCGCGAGCCGGCGAGCGGGCAGAGCTCGGCACTGCGCCGGGGCGCACGGCTCGGGGGACCCAG GCCCAGCTACCTTCCCTCCGCCCCCGGGCTCTGTTCTCACTTTCCAGCAGTGGAAGGACGGGAAGCGGGAGCCATGCAGTCCGAATCGGG GATCGTGCCGGATTTCGAAGTCGGGGAGGAGTTTCATGAAGAGCCCAAAACCTATTACGAACTCAAAAGTCAACCGCTGAAGAGCAGGTT TCTGAGTCCCTTCAACATGATCCTGGGAGGAATCGTGGTGGTGCTGGTGTTCACAGGGTTTGTGTGGGCAGCCCACAATAAAGACGTCCT TCGCCGGATGAAGAAGCGCTACCCCACGACGTTCGTTATGGTGGTCATGTTGGCGAGCTATTTCCTTATCTCCATGTTTGGAGGAGTCAT GGTCTTTGTGTTTGGCATTACTTTTCCTTTGCTGTTGATGTTTATCCATGCATCGTTGAGACTTCGGAACCTCAAGAACAAACTGGAGAA TAAAATGGAAGGAATAGGTTTGAAGAGGACACCGATGGGCATTGTCCTGGATGCCCTAGAACAGCAGGAAGAAGGCATCAACAGACTCAC TGACTATATCAGCAAAGTGAAGGAATAAACATAACTTACCTGAGCTAGGGTTGCAGCAGAAATTGAGTTGCAGCTTGCCCTTGTCCAGAC CTATGTTCTGCTTGCGTTTTTGAAACAGGAGGTGCACGTACCACCCAATTATCTATGGCAGCATGCATGTATAGGCCGAACTATTATCAG CTCTGATGTTTCAGAGAGAAGACCTCAGAAACCGAAAGAAAACCACCACCCTCCTATTGTGTCTGAAGTTTCACGTGTGTTTATGAAATC TAATGGGAAATGGATCACACGATTTCTTTAAGGGAATTAAAAAAAATAAAAGAATTACGGCTTTTACAGCAACAATACGATTATCTTATA GGAAAAAAAAAATCATTGTAAAGTATCAAGACAATACGAGTAAATGAAAAGGCTGTTAAAGTAGATGACATCATGTGTTAGCCTGTTCCT AATCCCCTAGAATTGTAATGTGTGGGATATAAATTAGTTTTTATTATTCTCTTAAAAATCAAAGATGATCTCTATCACTTTGCCACCTGT TTGATGTGCAGTGGAAACTGGTTAAGCCAGTTGTTCATACTTCCTTTACAAATATAAAGATAGCTGTTTAGGATATTTTGTTACATTTTT GTAAATTTTTGAAATGCTAGTAATGTGTTTTCACCAGCAAGTATTTGTTGCAAACTTAATGTCATTTTCCTTAAGATGGTTACAGCTATG TAACCTGTATTATTCTGGACGGACTTATTAAAATACAAACAGACAAAAAATAAAACAAAACTTGAGTTCTATTTACCTTGCACATTTTTT GTTGTTACAGTGAAAAAAATGGTCCAAGAAAATGTTTGCCATTTTTGCATTGTTTCGTTTTTAACTGGAACATTTAGAAAGAAGGAAATG AATGTGCATTTTATTAATTCCTTAGGGGCACAAGGAGGACAATAATAGCTGATCTTTTGAAATTTGAAAAACGTCTTTAGATGACCAAGC AAAAAGACTTTAAAAAATGGTAATGAAAATGGAATGCAGCTACTGCAGCTAATAAAAAATTTTAGATAGCAATTGTTACAACCATATGCC TTTATAGCTAGACATTAGAATTATGATAGCATGAGTTTATACATTCTATTATTTTTCCTCCCTTTCTCATGTTTTTATAAATAGGTAATA AAAAATGTTTTGCCTGCCAATTGAATGATTTCGTAGCTGAAGTAGAAACATTTAGGTTTCTGTAGCATTAAATTGTGAAGACAACTGGAG TGGTACTTACTGAAGAAACTCTCTGTATGTCCTAGAATAAGAAGCAATGATGTGCTGCTTCTGATTTTTCTTGCATTTTAAATTCTCAGC CAACCTACAGCCATGATCTTTAGCACAGTGATATCACCATGACTTCACAGACATGGTCTAGAATCTGTACCCTTACCCACATATGAAGAA >53980_53980_1_MITF-ARL6IP5_MITF_chr3_69788852_ENST00000352241_ARL6IP5_chr3_69150990_ENST00000273258_length(amino acids)=199AA_BP=69 MRRGARLGGPRPSYLPSAPGLCSHFPAVEGREAGAMQSESGIVPDFEVGEEFHEEPKTYYELKSQPLKSRFLSPFNMILGGIVVVLVFTG FVWAAHNKDVLRRMKKRYPTTFVMVVMLASYFLISMFGGVMVFVFGITFPLLLMFIHASLRLRNLKNKLENKMEGIGLKRTPMGIVLDAL -------------------------------------------------------------- >53980_53980_2_MITF-ARL6IP5_MITF_chr3_69788852_ENST00000352241_ARL6IP5_chr3_69150990_ENST00000478935_length(transcript)=1513nt_BP=267nt GTAAACTCCCCGCGCTGGGGCGGGCGGCCGCGAGCCGGCGAGCGGGCAGAGCTCGGCACTGCGCCGGGGCGCACGGCTCGGGGGACCCAG GCCCAGCTACCTTCCCTCCGCCCCCGGGCTCTGTTCTCACTTTCCAGCAGTGGAAGGACGGGAAGCGGGAGCCATGCAGTCCGAATCGGG GATCGTGCCGGATTTCGAAGTCGGGGAGGAGTTTCATGAAGAGCCCAAAACCTATTACGAACTCAAAAGTCAACCGCTGAAGAGCAGGTT TCTGAGTCCCTTCAACCCTCCTATTGTGTCTGAAGTTTCACGTGTGTTTATGAAATCTAATGGGAAATGGATCACACGATTTCTTTAAGG GAATTAAAAAAAATAAAAGAATTACGGCTTTTACAGCAACAATACGATTATCTTATAGGAAAAAAAAAATCATTGTAAAGTATCAAGACA ATACGAGTAAATGAAAAGGCTGTTAAAGTAGATGACATCATGTGTTAGCCTGTTCCTAATCCCCTAGAATTGTAATGTGTGGGATATAAA TTAGTTTTTATTATTCTCTTAAAAATCAAAGATGATCTCTATCACTTTGCCACCTGTTTGATGTGCAGTGGAAACTGGTTAAGCCAGTTG TTCATACTTCCTTTACAAATATAAAGATAGCTGTTTAGGATATTTTGTTACATTTTTGTAAATTTTTGAAATGCTAGTAATGTGTTTTCA CCAGCAAGTATTTGTTGCAAACTTAATGTCATTTTCCTTAAGATGGTTACAGCTATGTAACCTGTATTATTCTGGACGGACTTATTAAAA TACAAACAGACAAAAAATAAAACAAAACTTGAGTTCTATTTACCTTGCACATTTTTTGTTGTTACAGTGAAAAAAATGGTCCAAGAAAAT GTTTGCCATTTTTGCATTGTTTCGTTTTTAACTGGAACATTTAGAAAGAAGGAAATGAATGTGCATTTTATTAATTCCTTAGGGGCACAA GGAGGACAATAATAGCTGATCTTTTGAAATTTGAAAAACGTCTTTAGATGACCAAGCAAAAAGACTTTAAAAAATGGTAATGAAAATGGA ATGCAGCTACTGCAGCTAATAAAAAATTTTAGATAGCAATTGTTACAACCATATGCCTTTATAGCTAGACATTAGAATTATGATAGCATG AGTTTATACATTCTATTATTTTTCCTCCCTTTCTCATGTTTTTATAAATAGGTAATAAAAAATGTTTTGCCTGCCAATTGAATGATTTCG TAGCTGAAGTAGAAACATTTAGGTTTCTGTAGCATTAAATTGTGAAGACAACTGGAGTGGTACTTACTGAAGAAACTCTCTGTATGTCCT AGAATAAGAAGCAATGATGTGCTGCTTCTGATTTTTCTTGCATTTTAAATTCTCAGCCAACCTACAGCCATGATCTTTAGCACAGTGATA >53980_53980_2_MITF-ARL6IP5_MITF_chr3_69788852_ENST00000352241_ARL6IP5_chr3_69150990_ENST00000478935_length(amino acids)=99AA_BP=69 MRRGARLGGPRPSYLPSAPGLCSHFPAVEGREAGAMQSESGIVPDFEVGEEFHEEPKTYYELKSQPLKSRFLSPFNPPIVSEVSRVFMKS -------------------------------------------------------------- >53980_53980_3_MITF-ARL6IP5_MITF_chr3_69788852_ENST00000448226_ARL6IP5_chr3_69150990_ENST00000273258_length(transcript)=2052nt_BP=231nt GGCGAGCGGGCAGAGCTCGGCACTGCGCCGGGGCGCACGGCTCGGGGGACCCAGGCCCAGCTACCTTCCCTCCGCCCCCGGGCTCTGTTC TCACTTTCCAGCAGTGGAAGGACGGGAAGCGGGAGCCATGCAGTCCGAATCGGGGATCGTGCCGGATTTCGAAGTCGGGGAGGAGTTTCA TGAAGAGCCCAAAACCTATTACGAACTCAAAAGTCAACCGCTGAAGAGCAGGTTTCTGAGTCCCTTCAACATGATCCTGGGAGGAATCGT GGTGGTGCTGGTGTTCACAGGGTTTGTGTGGGCAGCCCACAATAAAGACGTCCTTCGCCGGATGAAGAAGCGCTACCCCACGACGTTCGT TATGGTGGTCATGTTGGCGAGCTATTTCCTTATCTCCATGTTTGGAGGAGTCATGGTCTTTGTGTTTGGCATTACTTTTCCTTTGCTGTT GATGTTTATCCATGCATCGTTGAGACTTCGGAACCTCAAGAACAAACTGGAGAATAAAATGGAAGGAATAGGTTTGAAGAGGACACCGAT GGGCATTGTCCTGGATGCCCTAGAACAGCAGGAAGAAGGCATCAACAGACTCACTGACTATATCAGCAAAGTGAAGGAATAAACATAACT TACCTGAGCTAGGGTTGCAGCAGAAATTGAGTTGCAGCTTGCCCTTGTCCAGACCTATGTTCTGCTTGCGTTTTTGAAACAGGAGGTGCA CGTACCACCCAATTATCTATGGCAGCATGCATGTATAGGCCGAACTATTATCAGCTCTGATGTTTCAGAGAGAAGACCTCAGAAACCGAA AGAAAACCACCACCCTCCTATTGTGTCTGAAGTTTCACGTGTGTTTATGAAATCTAATGGGAAATGGATCACACGATTTCTTTAAGGGAA TTAAAAAAAATAAAAGAATTACGGCTTTTACAGCAACAATACGATTATCTTATAGGAAAAAAAAAATCATTGTAAAGTATCAAGACAATA CGAGTAAATGAAAAGGCTGTTAAAGTAGATGACATCATGTGTTAGCCTGTTCCTAATCCCCTAGAATTGTAATGTGTGGGATATAAATTA GTTTTTATTATTCTCTTAAAAATCAAAGATGATCTCTATCACTTTGCCACCTGTTTGATGTGCAGTGGAAACTGGTTAAGCCAGTTGTTC ATACTTCCTTTACAAATATAAAGATAGCTGTTTAGGATATTTTGTTACATTTTTGTAAATTTTTGAAATGCTAGTAATGTGTTTTCACCA GCAAGTATTTGTTGCAAACTTAATGTCATTTTCCTTAAGATGGTTACAGCTATGTAACCTGTATTATTCTGGACGGACTTATTAAAATAC AAACAGACAAAAAATAAAACAAAACTTGAGTTCTATTTACCTTGCACATTTTTTGTTGTTACAGTGAAAAAAATGGTCCAAGAAAATGTT TGCCATTTTTGCATTGTTTCGTTTTTAACTGGAACATTTAGAAAGAAGGAAATGAATGTGCATTTTATTAATTCCTTAGGGGCACAAGGA GGACAATAATAGCTGATCTTTTGAAATTTGAAAAACGTCTTTAGATGACCAAGCAAAAAGACTTTAAAAAATGGTAATGAAAATGGAATG CAGCTACTGCAGCTAATAAAAAATTTTAGATAGCAATTGTTACAACCATATGCCTTTATAGCTAGACATTAGAATTATGATAGCATGAGT TTATACATTCTATTATTTTTCCTCCCTTTCTCATGTTTTTATAAATAGGTAATAAAAAATGTTTTGCCTGCCAATTGAATGATTTCGTAG CTGAAGTAGAAACATTTAGGTTTCTGTAGCATTAAATTGTGAAGACAACTGGAGTGGTACTTACTGAAGAAACTCTCTGTATGTCCTAGA ATAAGAAGCAATGATGTGCTGCTTCTGATTTTTCTTGCATTTTAAATTCTCAGCCAACCTACAGCCATGATCTTTAGCACAGTGATATCA >53980_53980_3_MITF-ARL6IP5_MITF_chr3_69788852_ENST00000448226_ARL6IP5_chr3_69150990_ENST00000273258_length(amino acids)=199AA_BP=69 MRRGARLGGPRPSYLPSAPGLCSHFPAVEGREAGAMQSESGIVPDFEVGEEFHEEPKTYYELKSQPLKSRFLSPFNMILGGIVVVLVFTG FVWAAHNKDVLRRMKKRYPTTFVMVVMLASYFLISMFGGVMVFVFGITFPLLLMFIHASLRLRNLKNKLENKMEGIGLKRTPMGIVLDAL -------------------------------------------------------------- >53980_53980_4_MITF-ARL6IP5_MITF_chr3_69788852_ENST00000448226_ARL6IP5_chr3_69150990_ENST00000478935_length(transcript)=1477nt_BP=231nt GGCGAGCGGGCAGAGCTCGGCACTGCGCCGGGGCGCACGGCTCGGGGGACCCAGGCCCAGCTACCTTCCCTCCGCCCCCGGGCTCTGTTC TCACTTTCCAGCAGTGGAAGGACGGGAAGCGGGAGCCATGCAGTCCGAATCGGGGATCGTGCCGGATTTCGAAGTCGGGGAGGAGTTTCA TGAAGAGCCCAAAACCTATTACGAACTCAAAAGTCAACCGCTGAAGAGCAGGTTTCTGAGTCCCTTCAACCCTCCTATTGTGTCTGAAGT TTCACGTGTGTTTATGAAATCTAATGGGAAATGGATCACACGATTTCTTTAAGGGAATTAAAAAAAATAAAAGAATTACGGCTTTTACAG CAACAATACGATTATCTTATAGGAAAAAAAAAATCATTGTAAAGTATCAAGACAATACGAGTAAATGAAAAGGCTGTTAAAGTAGATGAC ATCATGTGTTAGCCTGTTCCTAATCCCCTAGAATTGTAATGTGTGGGATATAAATTAGTTTTTATTATTCTCTTAAAAATCAAAGATGAT CTCTATCACTTTGCCACCTGTTTGATGTGCAGTGGAAACTGGTTAAGCCAGTTGTTCATACTTCCTTTACAAATATAAAGATAGCTGTTT AGGATATTTTGTTACATTTTTGTAAATTTTTGAAATGCTAGTAATGTGTTTTCACCAGCAAGTATTTGTTGCAAACTTAATGTCATTTTC CTTAAGATGGTTACAGCTATGTAACCTGTATTATTCTGGACGGACTTATTAAAATACAAACAGACAAAAAATAAAACAAAACTTGAGTTC TATTTACCTTGCACATTTTTTGTTGTTACAGTGAAAAAAATGGTCCAAGAAAATGTTTGCCATTTTTGCATTGTTTCGTTTTTAACTGGA ACATTTAGAAAGAAGGAAATGAATGTGCATTTTATTAATTCCTTAGGGGCACAAGGAGGACAATAATAGCTGATCTTTTGAAATTTGAAA AACGTCTTTAGATGACCAAGCAAAAAGACTTTAAAAAATGGTAATGAAAATGGAATGCAGCTACTGCAGCTAATAAAAAATTTTAGATAG CAATTGTTACAACCATATGCCTTTATAGCTAGACATTAGAATTATGATAGCATGAGTTTATACATTCTATTATTTTTCCTCCCTTTCTCA TGTTTTTATAAATAGGTAATAAAAAATGTTTTGCCTGCCAATTGAATGATTTCGTAGCTGAAGTAGAAACATTTAGGTTTCTGTAGCATT AAATTGTGAAGACAACTGGAGTGGTACTTACTGAAGAAACTCTCTGTATGTCCTAGAATAAGAAGCAATGATGTGCTGCTTCTGATTTTT CTTGCATTTTAAATTCTCAGCCAACCTACAGCCATGATCTTTAGCACAGTGATATCACCATGACTTCACAGACATGGTCTAGAATCTGTA >53980_53980_4_MITF-ARL6IP5_MITF_chr3_69788852_ENST00000448226_ARL6IP5_chr3_69150990_ENST00000478935_length(amino acids)=99AA_BP=69 MRRGARLGGPRPSYLPSAPGLCSHFPAVEGREAGAMQSESGIVPDFEVGEEFHEEPKTYYELKSQPLKSRFLSPFNPPIVSEVSRVFMKS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for MITF-ARL6IP5 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for MITF-ARL6IP5 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for MITF-ARL6IP5 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
