
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:MLH1-GOLGA4 (FusionGDB2 ID:54168) |
Fusion Gene Summary for MLH1-GOLGA4 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: MLH1-GOLGA4 | Fusion gene ID: 54168 | Hgene | Tgene | Gene symbol | MLH1 | GOLGA4 | Gene ID | 4292 | 2803 |
| Gene name | mutL homolog 1 | golgin A4 | |
| Synonyms | COCA2|FCC2|HNPCC|HNPCC2|hMLH1 | CRPF46|GCP2|GOLG|MU-RMS-40.18|p230 | |
| Cytomap | 3p22.2 | 3p22.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | DNA mismatch repair protein Mlh1mutL homolog 1, colon cancer, nonpolyposis type 2 | golgin subfamily A member 4256 kDa golgin72.1 proteincentrosome-related protein F46golgi autoantigen, golgin subfamily a, 4golgin-240golgin-245protein 72.1trans-Golgi p230 | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | P40692 | Q13439 | |
| Ensembl transtripts involved in fusion gene | ENST00000492474, ENST00000231790, ENST00000435176, ENST00000455445, ENST00000458205, ENST00000536378, ENST00000539477, | ENST00000361924, ENST00000356847, ENST00000435830, ENST00000444882, | |
| Fusion gene scores | * DoF score | 2 X 3 X 2=12 | 14 X 16 X 6=1344 |
| # samples | 2 | 15 | |
| ** MAII score | log2(2/12*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(15/1344*10)=-3.16349873228288 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: MLH1 [Title/Abstract] AND GOLGA4 [Title/Abstract] AND fusion [Title/Abstract] An interstitial deletion at 3p21.3 results in the genetic fusion of MLH1 and ITGA9 in a Lynch syndrome family (pmid: 19188145) | ||
| Most frequent breakpoint | GOLGA4(37370584)-MLH1(37089010), # samples:2 GOLGA4(37343823)-MLH1(37058997), # samples:2 GOLGA4(37360685)-MLH1(37061801), # samples:2 MLH1(37050396)-GOLGA4(37376544), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | GOLGA4-MLH1 seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. GOLGA4-MLH1 seems lost the major protein functional domain in Tgene partner, which is a tumor suppressor due to the frame-shifted ORF. MLH1-GOLGA4 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. MLH1-GOLGA4 seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | GOLGA4 | GO:0043001 | Golgi to plasma membrane protein transport | 15265687 |
| Tgene | GOLGA4 | GO:0045773 | positive regulation of axon extension | 22705394 |
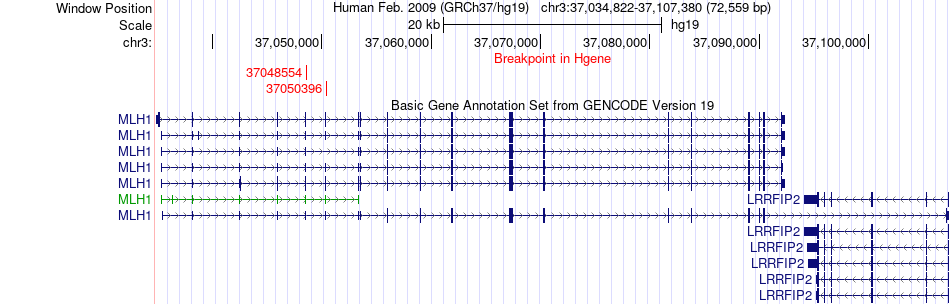
 Fusion gene breakpoints across MLH1 (5'-gene) Fusion gene breakpoints across MLH1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
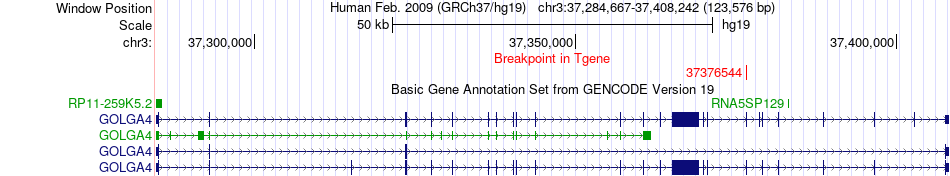
 Fusion gene breakpoints across GOLGA4 (3'-gene) Fusion gene breakpoints across GOLGA4 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-EW-A1IZ-01A | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| ChimerDB4 | BRCA | TCGA-EW-A1IZ-01A | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
Top |
Fusion Gene ORF analysis for MLH1-GOLGA4 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000492474 | ENST00000361924 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| 3UTR-3CDS | ENST00000492474 | ENST00000361924 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| 3UTR-intron | ENST00000492474 | ENST00000356847 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| 3UTR-intron | ENST00000492474 | ENST00000356847 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| 3UTR-intron | ENST00000492474 | ENST00000435830 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| 3UTR-intron | ENST00000492474 | ENST00000435830 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| 3UTR-intron | ENST00000492474 | ENST00000444882 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| 3UTR-intron | ENST00000492474 | ENST00000444882 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| 5CDS-intron | ENST00000231790 | ENST00000356847 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| 5CDS-intron | ENST00000231790 | ENST00000356847 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| 5CDS-intron | ENST00000231790 | ENST00000435830 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| 5CDS-intron | ENST00000231790 | ENST00000435830 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| 5CDS-intron | ENST00000231790 | ENST00000444882 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| 5CDS-intron | ENST00000231790 | ENST00000444882 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| 5CDS-intron | ENST00000435176 | ENST00000356847 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| 5CDS-intron | ENST00000435176 | ENST00000356847 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| 5CDS-intron | ENST00000435176 | ENST00000435830 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| 5CDS-intron | ENST00000435176 | ENST00000435830 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| 5CDS-intron | ENST00000435176 | ENST00000444882 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| 5CDS-intron | ENST00000435176 | ENST00000444882 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-3CDS | ENST00000455445 | ENST00000361924 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-3CDS | ENST00000455445 | ENST00000361924 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-3CDS | ENST00000458205 | ENST00000361924 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-3CDS | ENST00000458205 | ENST00000361924 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-3CDS | ENST00000536378 | ENST00000361924 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-3CDS | ENST00000536378 | ENST00000361924 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-3CDS | ENST00000539477 | ENST00000361924 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-intron | ENST00000455445 | ENST00000356847 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-intron | ENST00000455445 | ENST00000356847 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-intron | ENST00000455445 | ENST00000435830 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-intron | ENST00000455445 | ENST00000435830 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-intron | ENST00000455445 | ENST00000444882 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-intron | ENST00000455445 | ENST00000444882 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-intron | ENST00000458205 | ENST00000356847 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-intron | ENST00000458205 | ENST00000356847 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-intron | ENST00000458205 | ENST00000435830 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-intron | ENST00000458205 | ENST00000435830 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-intron | ENST00000458205 | ENST00000444882 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-intron | ENST00000458205 | ENST00000444882 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-intron | ENST00000536378 | ENST00000356847 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-intron | ENST00000536378 | ENST00000356847 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-intron | ENST00000536378 | ENST00000435830 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-intron | ENST00000536378 | ENST00000435830 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-intron | ENST00000536378 | ENST00000444882 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-intron | ENST00000536378 | ENST00000444882 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-intron | ENST00000539477 | ENST00000356847 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-intron | ENST00000539477 | ENST00000435830 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| 5UTR-intron | ENST00000539477 | ENST00000444882 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| Frame-shift | ENST00000231790 | ENST00000361924 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| Frame-shift | ENST00000435176 | ENST00000361924 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| In-frame | ENST00000231790 | ENST00000361924 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| In-frame | ENST00000435176 | ENST00000361924 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + |
| intron-3CDS | ENST00000539477 | ENST00000361924 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| intron-intron | ENST00000539477 | ENST00000356847 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| intron-intron | ENST00000539477 | ENST00000435830 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
| intron-intron | ENST00000539477 | ENST00000444882 | MLH1 | chr3 | 37050396 | + | GOLGA4 | chr3 | 37376544 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000231790 | MLH1 | chr3 | 37048554 | + | ENST00000361924 | GOLGA4 | chr3 | 37376544 | + | 1875 | 669 | 216 | 1169 | 317 |
| ENST00000435176 | MLH1 | chr3 | 37048554 | + | ENST00000361924 | GOLGA4 | chr3 | 37376544 | + | 1625 | 419 | 260 | 919 | 219 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000231790 | ENST00000361924 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + | 0.001236018 | 0.99876404 |
| ENST00000435176 | ENST00000361924 | MLH1 | chr3 | 37048554 | + | GOLGA4 | chr3 | 37376544 | + | 0.000439114 | 0.9995609 |
Top |
Fusion Genomic Features for MLH1-GOLGA4 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
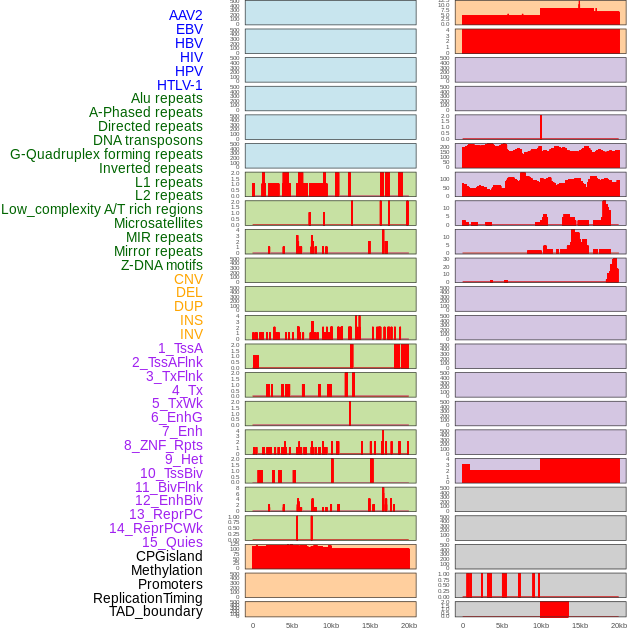
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
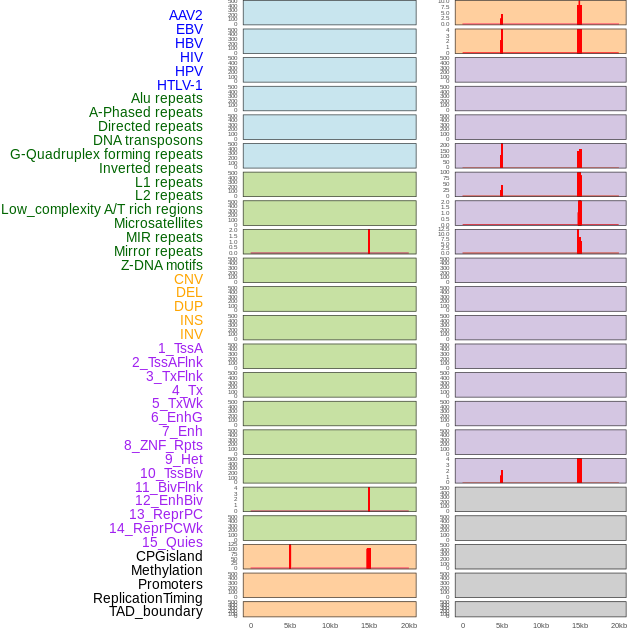
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for MLH1-GOLGA4 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:37370584/chr3:37089010) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MLH1 | GOLGA4 |
| FUNCTION: Heterodimerizes with PMS2 to form MutL alpha, a component of the post-replicative DNA mismatch repair system (MMR). DNA repair is initiated by MutS alpha (MSH2-MSH6) or MutS beta (MSH2-MSH3) binding to a dsDNA mismatch, then MutL alpha is recruited to the heteroduplex. Assembly of the MutL-MutS-heteroduplex ternary complex in presence of RFC and PCNA is sufficient to activate endonuclease activity of PMS2. It introduces single-strand breaks near the mismatch and thus generates new entry points for the exonuclease EXO1 to degrade the strand containing the mismatch. DNA methylation would prevent cleavage and therefore assure that only the newly mutated DNA strand is going to be corrected. MutL alpha (MLH1-PMS2) interacts physically with the clamp loader subunits of DNA polymerase III, suggesting that it may play a role to recruit the DNA polymerase III to the site of the MMR. Also implicated in DNA damage signaling, a process which induces cell cycle arrest and can lead to apoptosis in case of major DNA damages. Heterodimerizes with MLH3 to form MutL gamma which plays a role in meiosis. {ECO:0000269|PubMed:16873062, ECO:0000269|PubMed:18206974, ECO:0000269|PubMed:20020535, ECO:0000269|PubMed:21120944, ECO:0000269|PubMed:9311737}. | FUNCTION: Involved in vesicular trafficking at the Golgi apparatus level. May play a role in delivery of transport vesicles containing GPI-linked proteins from the trans-Golgi network through its interaction with MACF1. Involved in endosome-to-Golgi trafficking (PubMed:29084197). {ECO:0000269|PubMed:15265687, ECO:0000269|PubMed:29084197}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MLH1 | chr3:37048554 | chr3:37376544 | ENST00000231790 | + | 5 | 19 | 100_104 | 151 | 757.0 | Nucleotide binding | ATP |
| Hgene | MLH1 | chr3:37048554 | chr3:37376544 | ENST00000231790 | + | 5 | 19 | 82_84 | 151 | 757.0 | Nucleotide binding | ATP |
| Tgene | GOLGA4 | chr3:37048554 | chr3:37376544 | ENST00000356847 | 16 | 23 | 2168_2215 | 2086 | 2244.0 | Domain | GRIP | |
| Tgene | GOLGA4 | chr3:37048554 | chr3:37376544 | ENST00000361924 | 15 | 24 | 2168_2215 | 2064 | 2306.3333333333335 | Domain | GRIP |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MLH1 | chr3:37048554 | chr3:37376544 | ENST00000435176 | + | 5 | 19 | 100_104 | 53 | 659.0 | Nucleotide binding | ATP |
| Hgene | MLH1 | chr3:37048554 | chr3:37376544 | ENST00000435176 | + | 5 | 19 | 82_84 | 53 | 659.0 | Nucleotide binding | ATP |
| Hgene | MLH1 | chr3:37048554 | chr3:37376544 | ENST00000455445 | + | 5 | 19 | 100_104 | 0 | 516.0 | Nucleotide binding | ATP |
| Hgene | MLH1 | chr3:37048554 | chr3:37376544 | ENST00000455445 | + | 5 | 19 | 82_84 | 0 | 516.0 | Nucleotide binding | ATP |
| Hgene | MLH1 | chr3:37048554 | chr3:37376544 | ENST00000458205 | + | 6 | 20 | 100_104 | 0 | 516.0 | Nucleotide binding | ATP |
| Hgene | MLH1 | chr3:37048554 | chr3:37376544 | ENST00000458205 | + | 6 | 20 | 82_84 | 0 | 516.0 | Nucleotide binding | ATP |
| Hgene | MLH1 | chr3:37048554 | chr3:37376544 | ENST00000539477 | + | 5 | 18 | 100_104 | 0 | 516.0 | Nucleotide binding | ATP |
| Hgene | MLH1 | chr3:37048554 | chr3:37376544 | ENST00000539477 | + | 5 | 18 | 82_84 | 0 | 516.0 | Nucleotide binding | ATP |
| Tgene | GOLGA4 | chr3:37048554 | chr3:37376544 | ENST00000356847 | 16 | 23 | 133_2185 | 2086 | 2244.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | GOLGA4 | chr3:37048554 | chr3:37376544 | ENST00000361924 | 15 | 24 | 133_2185 | 2064 | 2306.3333333333335 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | GOLGA4 | chr3:37048554 | chr3:37376544 | ENST00000356847 | 16 | 23 | 252_2096 | 2086 | 2244.0 | Compositional bias | Note=Glu-rich | |
| Tgene | GOLGA4 | chr3:37048554 | chr3:37376544 | ENST00000361924 | 15 | 24 | 252_2096 | 2064 | 2306.3333333333335 | Compositional bias | Note=Glu-rich |
Top |
Fusion Gene Sequence for MLH1-GOLGA4 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >54168_54168_1_MLH1-GOLGA4_MLH1_chr3_37048554_ENST00000231790_GOLGA4_chr3_37376544_ENST00000361924_length(transcript)=1875nt_BP=669nt GCCGCTTCAGGGAGGGACGAAGAGACCCAGCAACCCACAGAGTTGAGAAATTTGACTGGCATTCAAGCTGTCCAATCAATAGCTGCCGCT GAAGGGTGGGGCTGGATGGCGTAAGCTACAGCTGAAGGAAGAACGTGAGCACGAGGCACTGAGGTGATTGGCTGAAGGCACTTCCGTTGA GCATCTAGACGTTTCCTTGGCTCTTCTGGCGCCAAAATGTCGTTCGTGGCAGGGGTTATTCGGCGGCTGGACGAGACAGTGGTGAACCGC ATCGCGGCGGGGGAAGTTATCCAGCGGCCAGCTAATGCTATCAAAGAGATGATTGAGAACTGTTTAGATGCAAAATCCACAAGTATTCAA GTGATTGTTAAAGAGGGAGGCCTGAAGTTGATTCAGATCCAAGACAATGGCACCGGGATCAGGAAAGAAGATCTGGATATTGTATGTGAA AGGTTCACTACTAGTAAACTGCAGTCCTTTGAGGATTTAGCCAGTATTTCTACCTATGGCTTTCGAGGTGAGGCTTTGGCCAGCATAAGC CATGTGGCTCATGTTACTATTACAACGAAAACAGCTGATGGAAAGTGTGCATACAGAGCAAGTTACTCAGATGGAAAACTGAAAGCCCCT CCTAAACCATGTGCTGGCAATCAAGGGACCCAGATCACGGCTCGTGAAGAAGAAATGACTGCAAAAGTAAGGGACCTGCAGACTCAACTT GAGGAGCTGCAGAAGAAATACCAGCAAAAGCTAGAGCAGGAGGAGAACCCTGGCAATGATAATGTAACAATTATGGAGCTACAGACACAG CTAGCACAGAAGACGACTTTAATCAGTGATTCGAAATTGAAAGAGCAAGAGTTCAGAGAACAGATTCACAATTTAGAAGACCGTTTGAAG AAATATGAAAAGAATGTATATGCAACAACTGTGGGGACACCTTACAAAGGTGGCAATTTGTACCATACGGATGTCTCACTCTTTGGAGAA CCTACCGAATTTGAGTATTTGCGAAAAGTGCTTTTTGAGTATATGATGGGTCGTGAGACTAAGACCATGGCAAAAGTTATAACCACCGTA CTGAAGTTCCCTGATGATCAGACTCAGAAAATTTTGGAAAGAGAAGATGCTCGGCTGATGTTTACTTCACCTCGCAGTGGTATCTTCTGA GTAAACCATCAGTCTGTGCTTAGTTAACATGTGTCATGGCTCCGATCTTCATCTTGAAGAAGAGTGACATTGGGTGACTGCTGCTTGGAA AACTGTCCACACTTGCTACTCTTTGAGAATGAAGTTGTCATTCAGGGCCCCTCATGTAGCCAAAAGACCAAGAAAAATCTGGCCCACAGA TAAGTTGCAGACTGCCTTTAAAATAGATTTTATCAGTGGAGAAATGGTGATAGTTTTTTCTTCAGTTTTCTCTTGGGAAGAGTTTTATGT TGTTTAAAAGATATTTTGATAACTTAACCTGCTTTATGGGCTTACATAATATTCCTTTCATCCATTCTTTTTAAAGAACGGCTTACCTTT CCTATTTATTTTTAGGGTGATTTTTTAAAAAGACTTGTGCAATACATTTTGAGGTGAAACTTAGTGGATTTTTTCTGATAAATTAGAGCA TTTAATTGACTATTTTATTCAGGTTGATCTGTTGAATATTTGCTAAAGACCAGTTCTTTAAGCTAAGACATGTAAAAAATCCCAAATGGC AGTACCTCATTGTTTACTTAGCTTTTGTACTTATATTTTTCAGAGGAAAAAACACTACTGTAAATTGTGAATAGCCAATACATAACTGTA >54168_54168_1_MLH1-GOLGA4_MLH1_chr3_37048554_ENST00000231790_GOLGA4_chr3_37376544_ENST00000361924_length(amino acids)=317AA_BP=0 MSFVAGVIRRLDETVVNRIAAGEVIQRPANAIKEMIENCLDAKSTSIQVIVKEGGLKLIQIQDNGTGIRKEDLDIVCERFTTSKLQSFED LASISTYGFRGEALASISHVAHVTITTKTADGKCAYRASYSDGKLKAPPKPCAGNQGTQITAREEEMTAKVRDLQTQLEELQKKYQQKLE QEENPGNDNVTIMELQTQLAQKTTLISDSKLKEQEFREQIHNLEDRLKKYEKNVYATTVGTPYKGGNLYHTDVSLFGEPTEFEYLRKVLF -------------------------------------------------------------- >54168_54168_2_MLH1-GOLGA4_MLH1_chr3_37048554_ENST00000435176_GOLGA4_chr3_37376544_ENST00000361924_length(transcript)=1625nt_BP=419nt GACGTGCCAGTCAGGCCTTCTCCTTTTCCGCAGACCGTGTGTTTCTTTACCGCTCTCCCCCGAGACCTTTTAAGGGTTGTTTGGAGTTTT AGATGCAAAATCCACAAGTATTCAAGTGATTGTTAAAGAGGGAGGCCTGAAGTTGATTCAGATCCAAGACAATGGCACCGGGATCAGGAA AGAAGATCTGGATATTGTATGTGAAAGGTTCACTACTAGTAAACTGCAGTCCTTTGAGGATTTAGCCAGTATTTCTACCTATGGCTTTCG AGGCTTTGGCCAGCATAAGCCATGTGGCTCATGTTACTATTACAACGAAAACAGCTGATGGAAAGTGTGCATACAGAGCAAGTTACTCAG ATGGAAAACTGAAAGCCCCTCCTAAACCATGTGCTGGCAATCAAGGGACCCAGATCACGGCTCGTGAAGAAGAAATGACTGCAAAAGTAA GGGACCTGCAGACTCAACTTGAGGAGCTGCAGAAGAAATACCAGCAAAAGCTAGAGCAGGAGGAGAACCCTGGCAATGATAATGTAACAA TTATGGAGCTACAGACACAGCTAGCACAGAAGACGACTTTAATCAGTGATTCGAAATTGAAAGAGCAAGAGTTCAGAGAACAGATTCACA ATTTAGAAGACCGTTTGAAGAAATATGAAAAGAATGTATATGCAACAACTGTGGGGACACCTTACAAAGGTGGCAATTTGTACCATACGG ATGTCTCACTCTTTGGAGAACCTACCGAATTTGAGTATTTGCGAAAAGTGCTTTTTGAGTATATGATGGGTCGTGAGACTAAGACCATGG CAAAAGTTATAACCACCGTACTGAAGTTCCCTGATGATCAGACTCAGAAAATTTTGGAAAGAGAAGATGCTCGGCTGATGTTTACTTCAC CTCGCAGTGGTATCTTCTGAGTAAACCATCAGTCTGTGCTTAGTTAACATGTGTCATGGCTCCGATCTTCATCTTGAAGAAGAGTGACAT TGGGTGACTGCTGCTTGGAAAACTGTCCACACTTGCTACTCTTTGAGAATGAAGTTGTCATTCAGGGCCCCTCATGTAGCCAAAAGACCA AGAAAAATCTGGCCCACAGATAAGTTGCAGACTGCCTTTAAAATAGATTTTATCAGTGGAGAAATGGTGATAGTTTTTTCTTCAGTTTTC TCTTGGGAAGAGTTTTATGTTGTTTAAAAGATATTTTGATAACTTAACCTGCTTTATGGGCTTACATAATATTCCTTTCATCCATTCTTT TTAAAGAACGGCTTACCTTTCCTATTTATTTTTAGGGTGATTTTTTAAAAAGACTTGTGCAATACATTTTGAGGTGAAACTTAGTGGATT TTTTCTGATAAATTAGAGCATTTAATTGACTATTTTATTCAGGTTGATCTGTTGAATATTTGCTAAAGACCAGTTCTTTAAGCTAAGACA TGTAAAAAATCCCAAATGGCAGTACCTCATTGTTTACTTAGCTTTTGTACTTATATTTTTCAGAGGAAAAAACACTACTGTAAATTGTGA ATAGCCAATACATAACTGTATTGTATGCAAATCTGTGATTGTTGGCAGTGTCATCTCTGAGAAACAGATAAATAAAGTTTATTTACTATA >54168_54168_2_MLH1-GOLGA4_MLH1_chr3_37048554_ENST00000435176_GOLGA4_chr3_37376544_ENST00000361924_length(amino acids)=219AA_BP=4 MAFEALASISHVAHVTITTKTADGKCAYRASYSDGKLKAPPKPCAGNQGTQITAREEEMTAKVRDLQTQLEELQKKYQQKLEQEENPGND NVTIMELQTQLAQKTTLISDSKLKEQEFREQIHNLEDRLKKYEKNVYATTVGTPYKGGNLYHTDVSLFGEPTEFEYLRKVLFEYMMGRET -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for MLH1-GOLGA4 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | MLH1 | chr3:37048554 | chr3:37376544 | ENST00000231790 | + | 5 | 19 | 410_650 | 151.0 | 757.0 | EXO1 |
| Hgene | MLH1 | chr3:37048554 | chr3:37376544 | ENST00000435176 | + | 5 | 19 | 410_650 | 53.0 | 659.0 | EXO1 |
| Hgene | MLH1 | chr3:37048554 | chr3:37376544 | ENST00000455445 | + | 5 | 19 | 410_650 | 0 | 516.0 | EXO1 |
| Hgene | MLH1 | chr3:37048554 | chr3:37376544 | ENST00000458205 | + | 6 | 20 | 410_650 | 0 | 516.0 | EXO1 |
| Hgene | MLH1 | chr3:37048554 | chr3:37376544 | ENST00000539477 | + | 5 | 18 | 410_650 | 0 | 516.0 | EXO1 |
| Tgene | GOLGA4 | chr3:37048554 | chr3:37376544 | ENST00000356847 | 16 | 23 | 133_203 | 2086.0 | 2244.0 | MACF1 | |
| Tgene | GOLGA4 | chr3:37048554 | chr3:37376544 | ENST00000361924 | 15 | 24 | 133_203 | 2064.0 | 2306.3333333333335 | MACF1 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for MLH1-GOLGA4 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for MLH1-GOLGA4 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
